Stim_mouse_islets
scottzijiezhang
2020-03-09
Last updated: 2020-08-26
Checks: 6 0
Knit directory: T1D_epitranscriptome/
This reproducible R Markdown analysis was created with workflowr (version 1.3.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190516) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: analysis/.Rhistory
Untracked files:
Untracked: data/m6A.batch.out.RData
Untracked: data/m6A.batchGender.out.RData
Unstaged changes:
Modified: analysis/_site.yml
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | aa2d181 | scottzijiezhang | 2020-06-05 | Build site. |
| Rmd | c02ff29 | scottzijiezhang | 2020-06-05 | wflow_publish(“Stim_mouse_islets.Rmd”) |
| html | 514bc99 | scottzijiezhang | 2020-05-08 | Build site. |
| Rmd | 5133385 | scottzijiezhang | 2020-05-08 | wflow_publish(“Stim_mouse_islets.Rmd”) |
| html | ef437e3 | scottzijiezhang | 2020-05-07 | Build site. |
| Rmd | 4fb035a | scottzijiezhang | 2020-05-07 | wflow_publish(“Stim_mouse_islets.Rmd”) |
| html | 241dfaa | scottzijiezhang | 2020-05-07 | Build site. |
| Rmd | 546bce0 | scottzijiezhang | 2020-05-07 | wflow_publish(“Stim_mouse_islets.Rmd”) |
| html | 2739184 | scottzijiezhang | 2020-03-30 | Build site. |
| Rmd | 66578ab | scottzijiezhang | 2020-03-30 | wflow_publish(“analysis/Stim_mouse_islets.Rmd”) |
| html | e1b9592 | scottzijiezhang | 2020-03-30 | Build site. |
| Rmd | 69e49cb | scottzijiezhang | 2020-03-30 | wflow_publish(“analysis/Stim_mouse_islets.Rmd”) |
Introduction
Preprocessing and alignment
from <- list.files("~/Rohit_T1D/mouse_islet/lane1/")
tmp <- gsub("CHe-SZ-42S-","",from)
tmp <- gsub("_001","",tmp)
tmp <- gsub("_S\\d+","",tmp)
repl_ID <- match( gsub("_R[0-9].fastq.gz","",tmp), paste0("T",1:42) )
to <- paste0(
c(
paste0( c( paste0("Ctl_", 1:6 ), paste0( "BIRKO_", 1:3 ), paste0( "LIRKO_", 1:4 ), paste0( "Ctl_HFD_", 1:4 ), paste0( "LIRKO_HFD_", 1:4 ) ), ".input" ),
paste0( c( paste0("Ctl_", 1:6 ), paste0( "BIRKO_", 1:3 ), paste0( "LIRKO_", 1:4 ), paste0( "Ctl_HFD_", 1:4 ), paste0( "LIRKO_HFD_", 1:4 ) ), ".m6A" )
)[repl_ID],
gsub("T\\d+","",tmp)
)
file.rename(paste0("~/Rohit_T1D/mouse_islet/lane1/",from), paste0("~/Rohit_T1D/mouse_islet/lane1/",to) )
from <- list.files("~/Rohit_T1D/mouse_islet/lane2/")
tmp <- gsub("CHe-SZ-42S-","",from)
tmp <- gsub("_001","",tmp)
tmp <- gsub("_S\\d+","",tmp)
repl_ID <- match( gsub("_R[0-9].fastq.gz","",tmp), paste0("T",1:88) )
file.rename(paste0("~/Rohit_T1D/mouse_islet/lane2/",from), paste0("~/Rohit_T1D/mouse_islet/lane2/",to) )data_name <- c(
paste0( c( paste0("Ctl_", 1:6 ), paste0( "BIRKO_", 1:3 ), paste0( "LIRKO_", 1:4 ), paste0( "Ctl_HFD_", 1:4 ), paste0( "LIRKO_HFD_", 1:4 ) ), ".input" ),
paste0( c( paste0("Ctl_", 1:6 ), paste0( "BIRKO_", 1:3 ), paste0( "LIRKO_", 1:4 ), paste0( "Ctl_HFD_", 1:4 ), paste0( "LIRKO_HFD_", 1:4 ) ), ".m6A" )
)
filePath <- "~/Rohit_T1D/mouse_islet/lane1/"
cutadapt <- paste0(" -a AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC -A AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGTGGTCGCCGTATCATT -o ",filePath,data_name,".trimmed.1.fastq.gz -p ",filePath,data_name,".trimmed.2.fastq.gz ",filePath,data_name,"_R1.fastq.gz ",filePath,data_name,"_R2.fastq.gz" )
for(i in cutadapt){
system2(command = "cutadapt", args = i , wait = F)
}
cutFirstThree <- paste0(" -u -3 -U 3 -m 20 -o ",filePath,data_name,".allTrimmed.1.fastq.gz -p ",filePath,data_name,".allTrimmed.2.fastq.gz ",filePath,data_name,".trimmed.1.fastq.gz ",filePath,data_name,".trimmed.2.fastq.gz" )
for(i in cutFirstThree){
system2(command = "cutadapt", args = i , wait = F)
}
file.remove( c(paste0(filePath,data_name,".trimmed.1.fastq.gz"),paste0(filePath,data_name,".trimmed.2.fastq.gz")) )
filePath <- "~/Rohit_T1D/mouse_islet/lane2/"
cutadapt <- paste0(" -a AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC -A AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGTGGTCGCCGTATCATT -o ",filePath,data_name,".trimmed.1.fastq.gz -p ",filePath,data_name,".trimmed.2.fastq.gz ",filePath,data_name,"_R1.fastq.gz ",filePath,data_name,"_R2.fastq.gz" )
for(i in cutadapt){
system2(command = "cutadapt", args = i , wait = F)
}
cutFirstThree <- paste0(" -u -3 -U 3 -m 20 -o ",filePath,data_name,".allTrimmed.1.fastq.gz -p ",filePath,data_name,".allTrimmed.2.fastq.gz ",filePath,data_name,".trimmed.1.fastq.gz ",filePath,data_name,".trimmed.2.fastq.gz" )
for(i in cutFirstThree){
system2(command = "cutadapt", args = i , wait = F)
}
file.remove( c(paste0(filePath,data_name,".trimmed.1.fastq.gz"),paste0(filePath,data_name,".trimmed.2.fastq.gz")) )dir.create("~/Rohit_T1D/mouse_islet/alignment_summary")
dir.create( out_dir <- "~/Rohit_T1D/mouse_islet/bam_files/" )
dir.create( unalign <- "~/Rohit_T1D/mouse_islet/unaligned_reads/" )
filePath1 <- "~/Rohit_T1D/mouse_islet/lane1/"
filePath2 <- "~/Rohit_T1D/mouse_islet/lane2/"
hisat_align <- paste0(" -x ~/Database/genome/mm10/mm10_UCSC --known-splicesite-infile ~/Database/genome/mm10/hisat2_splice_sites.txt -k 1 --un-conc-gz ",unalign,data_name,".unalign --summary-file ~/Rohit_T1D/mouse_islet/alignment_summary/",data_name,".txt -p 20 --no-discordant -1 ",
filePath1,data_name,".allTrimmed.1.fastq.gz,",filePath2,data_name,".allTrimmed.1.fastq.gz -2 ",filePath1,data_name,".allTrimmed.2.fastq.gz,",filePath2,data_name,".allTrimmed.2.fastq.gz |samtools view -bS | samtools sort -o ", out_dir,data_name,".bam ")
for(i in hisat_align){
system2(command = "hisat2", args = i,wait = T )
}Call m6A peaks across all samples for QC
load( "~/Rohit_T1D/mouse_islet/allSample_RADAR.RData")
MeRIPall <- MeRIPtools::callPeakBinomial( allSampleRADAR )
MeRIPall <- MeRIPtools::reportJointPeak(MeRIPall, joint_threshold = 4, threads = 20)
jointPeaks <- MeRIPtools::jointPeak(MeRIPall)
## motif analysis of joint peaks
write.table(jointPeaks, file = paste0("~/Rohit_T1D/mouse_islet/jointPeak_allsample.bed"),sep = "\t",row.names = F,col.names = F,quote = F)
system2(command = "bedtools", args = paste0("getfasta -fi ~/Database/genome/mm10/mm10_UCSC.fa -bed ~/Rohit_T1D/mouse_islet/jointPeak_allsample.bed -s -split > ~/Rohit_T1D/mouse_islet/jointPeak_allsample.fa ") )
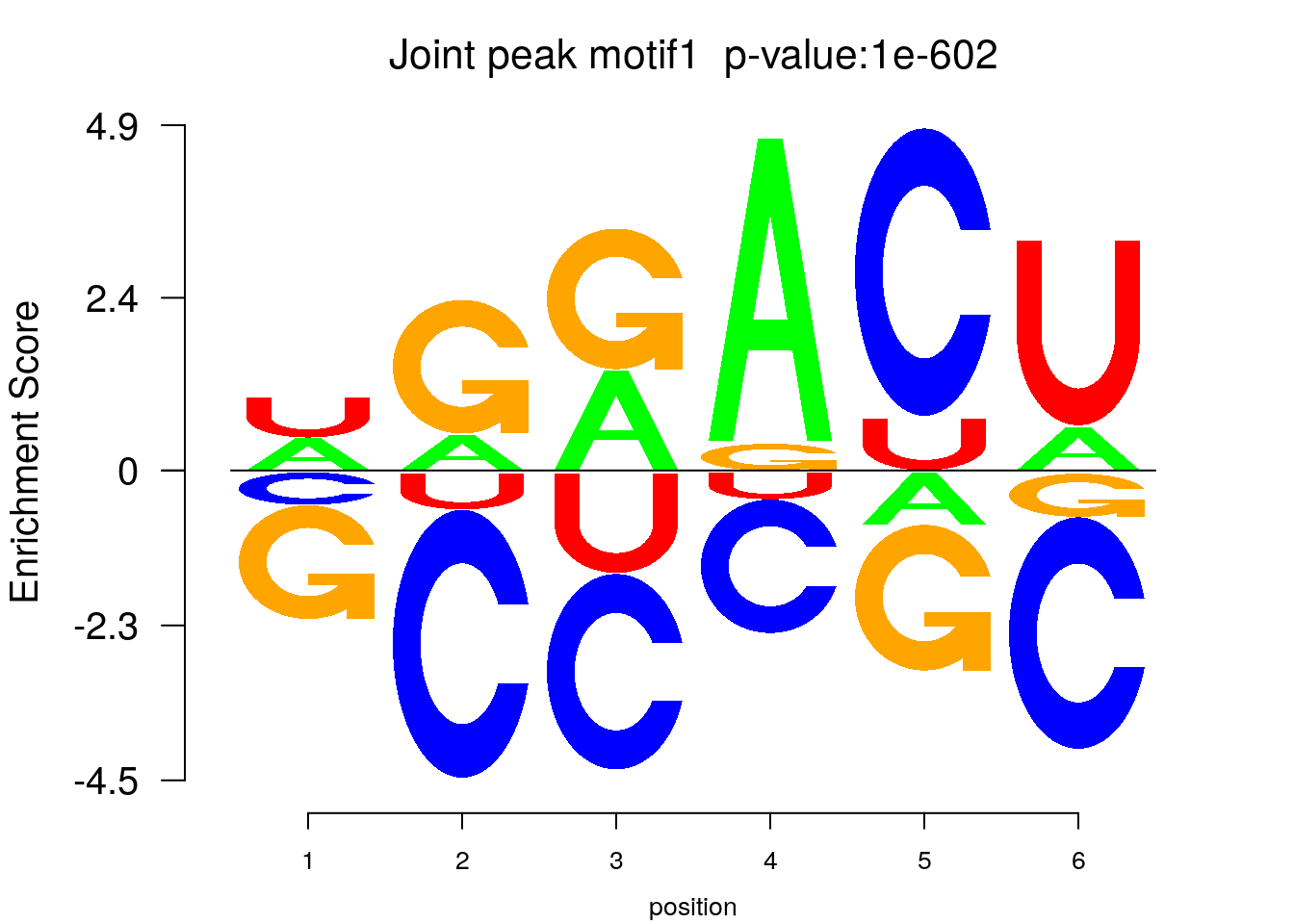
system2(command = "findMotifs.pl", args = paste0("~/Rohit_T1D/mouse_islet/jointPeak_allsample.fa fasta ~/Rohit_T1D/mouse_islet/jointPeak_allsample_Homer2 -fasta ~/Database/transcriptome/backgroud_peaks/mm10_80bpRandomPeak.fa -len 5,6 -rna -p 20 -S 5 -noknown"),wait = F )library(Logolas)
color_motif <- c( "orange", "blue", "red","green" )
for(i in 1:1){
pwm.m <- t( read.table(paste0("~/Rohit_T1D/mouse_islet/jointPeak_allsample_Homer2/homerResults/motif",i,".motif"), header = F, comment.char = ">",col.names = c("A","C","G","U")) )
motif_p <- sapply(1:1, function(x){
strsplit( as.character( read.table(paste0("~/Rohit_T1D/mouse_islet/jointPeak_allsample_Homer2/homerResults/motif",x,".motif"),comment.char = "", nrows = 1)[,12] ), split= ":")[[1]][4]
})
colnames(pwm.m) <- 1:ncol(pwm.m)
try(logomaker(pwm.m ,type = "EDLogo",colors = color_motif,color_type = "per_row" ,logo_control = list(pop_name = paste0("Joint peak motif",i," p-value:",motif_p[i])))
)
}
| Version | Author | Date |
|---|---|---|
| 514bc99 | scottzijiezhang | 2020-05-08 |
The enriched motif confirms the m6A-seq specificity.
Run RADAR pipeline for differential methylation
library(RADAR)
samplename <- c( paste0("Ctl_", 1:6 ), paste0( "BIRKO_", 1:3 ), paste0( "LIRKO_", 1:4 ), paste0( "Ctl_HFD_", 1:4 ), paste0( "LIRKO_HFD_", 1:4 ) )
allSampleRADAR <- countReads(samplenames = samplename,
gtf = "~/Database/genome/mm10/mm10_UCSC.gtf",
bamFolder = "~/Rohit_T1D/mouse_islet/bam_files/",
outputDir = "~/Rohit_T1D/mouse_islet/",
modification = "m6A",
binSize = 50,
paired = TRUE,
threads = 20
)
save(allSampleRADAR, file = "~/Rohit_T1D/mouse_islet/allSample_RADAR.RData")Compare BIRKO to Control
load( "~/Rohit_T1D/mouse_islet/allSample_RADAR.RData")
BIRKO_sample <- c( paste0("Ctl_", 1:6 ), paste0( "BIRKO_", 1:3 ) )
BIRKO_RADAR <- select( allSampleRADAR, BIRKO_sample )
BIRKO_RADAR <- normalizeLibrary( BIRKO_RADAR )
BIRKO_RADAR <- adjustExprLevel( BIRKO_RADAR )
variable( BIRKO_RADAR ) <- data.frame(genotype = c( rep( "Ctl", 6 ), rep( "BIRKO", 3 ) ),
batch = c( 0,0,0,1,1,1,0,0,1 )
)
BIRKO_RADAR <- filterBins( BIRKO_RADAR )
save(BIRKO_RADAR, file = "~/Rohit_T1D/mouse_islet/BIRKO_RADAR.RData")Check confounding factors
library(RADAR)
load( "~/Rohit_T1D/mouse_islet/BIRKO_RADAR.RData" )
plotPCAfromMatrix(BIRKO_RADAR@ip_adjExpr_filtered, variable(BIRKO_RADAR)$genotype )+scale_color_discrete(name = "Genotype")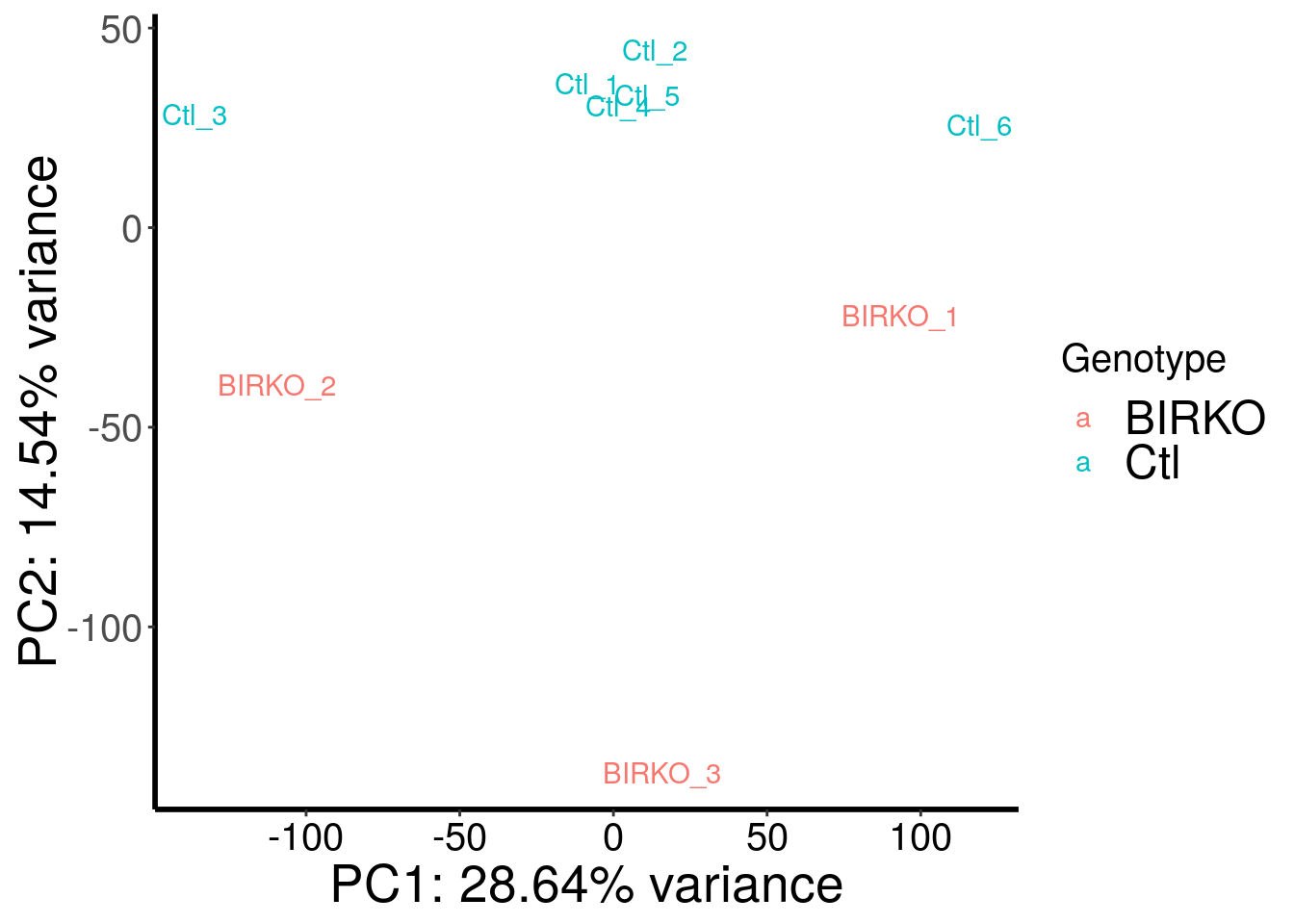
| Version | Author | Date |
|---|---|---|
| e1b9592 | scottzijiezhang | 2020-03-30 |
plotPCAfromMatrix(BIRKO_RADAR@ip_adjExpr_filtered, as.character( c( 0,0,0,1,1,1,0,0,1 ) ) )+scale_color_discrete(name = "Batch")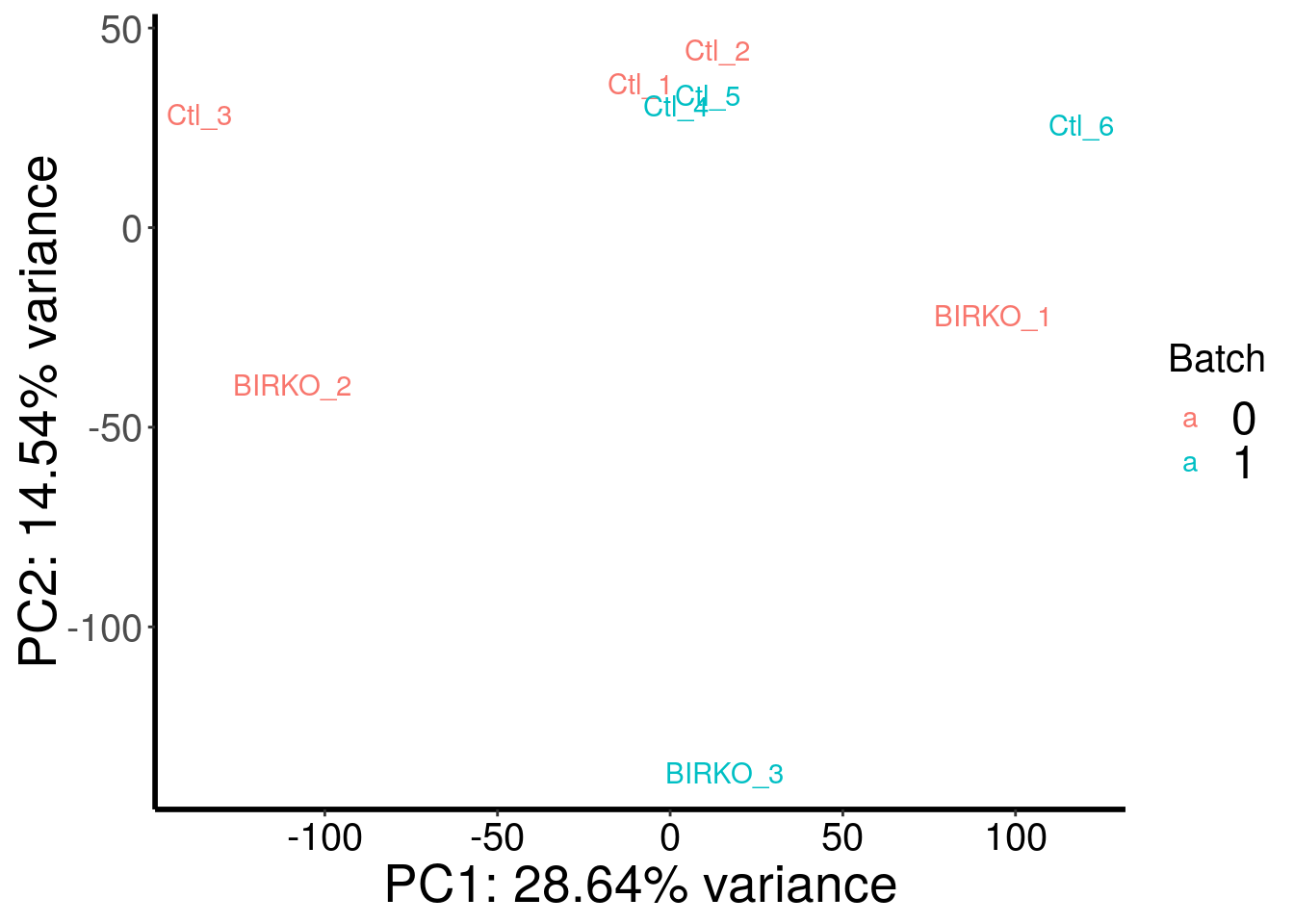
| Version | Author | Date |
|---|---|---|
| e1b9592 | scottzijiezhang | 2020-03-30 |
Samples are well separated by genotype. Batch does not contribute to the variance much.
DM tests with RADAR
variable( BIRKO_RADAR ) <- data.frame(genotype = c( rep( "Ctl", 6 ), rep( "BIRKO", 3 ) )
)
BIRKO_RADAR <- diffIP_parallel(BIRKO_RADAR, thread = 20)
#BIRKO_RADAR <- reportResult(BIRKO_RADAR, cutoff = 0.1, threads = 20)
save(BIRKO_RADAR, file = "~/Rohit_T1D/mouse_islet/BIRKO_RADAR.RData")
#write.table(results(BIRKO_RADAR), file = "~/Rohit_T1D/mouse_islet/BIRKO_diffPeaks_FDR0.1.xls", sep = "\t", row.names = FALSE, col.names = FALSE, quote = FALSE)No differential m6A peaks were detected at FDR cutoff of 10% (neither do 20% FDR).
Compare LIRKO to Control
load( "~/Rohit_T1D/mouse_islet/allSample_RADAR.RData")
LIRKO_sample <- c( paste0("Ctl_", 1:6 ), paste0( "LIRKO_", 1:4 ) )
LIRKO_RADAR <- select( allSampleRADAR, LIRKO_sample )
LIRKO_RADAR <- normalizeLibrary( LIRKO_RADAR )
LIRKO_RADAR <- adjustExprLevel( LIRKO_RADAR )
variable( LIRKO_RADAR ) <- data.frame(genotype = c( rep( "Ctl", 6 ), rep( "LIRKO", 4 ) ),
batch = c( 0,0,0,1,1,1,0,0,1,1 )
)
LIRKO_RADAR <- filterBins( LIRKO_RADAR )
save(LIRKO_RADAR, file = "~/Rohit_T1D/mouse_islet/LIRKO_RADAR.RData")Check confounding factors
library(RADAR)
load( "~/Rohit_T1D/mouse_islet/LIRKO_RADAR.RData" )
plotPCAfromMatrix(LIRKO_RADAR@ip_adjExpr_filtered, variable(LIRKO_RADAR)$genotype ) + scale_color_discrete(name = "Genotype")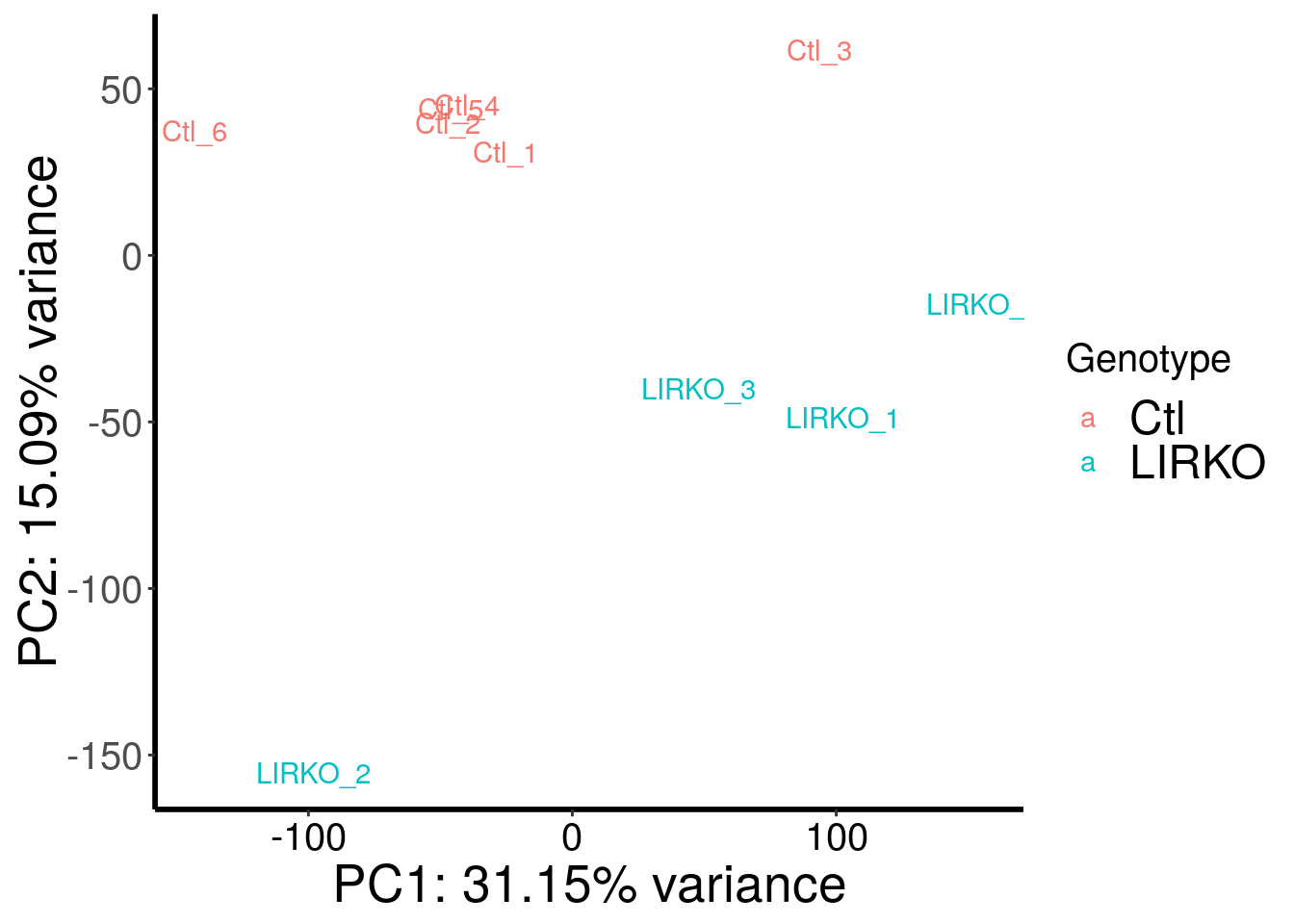
| Version | Author | Date |
|---|---|---|
| e1b9592 | scottzijiezhang | 2020-03-30 |
plotPCAfromMatrix(LIRKO_RADAR@ip_adjExpr_filtered, as.character( c( 0,0,0,1,1,1,0,0,1,1 ) ) )+scale_color_discrete(name = "Batch")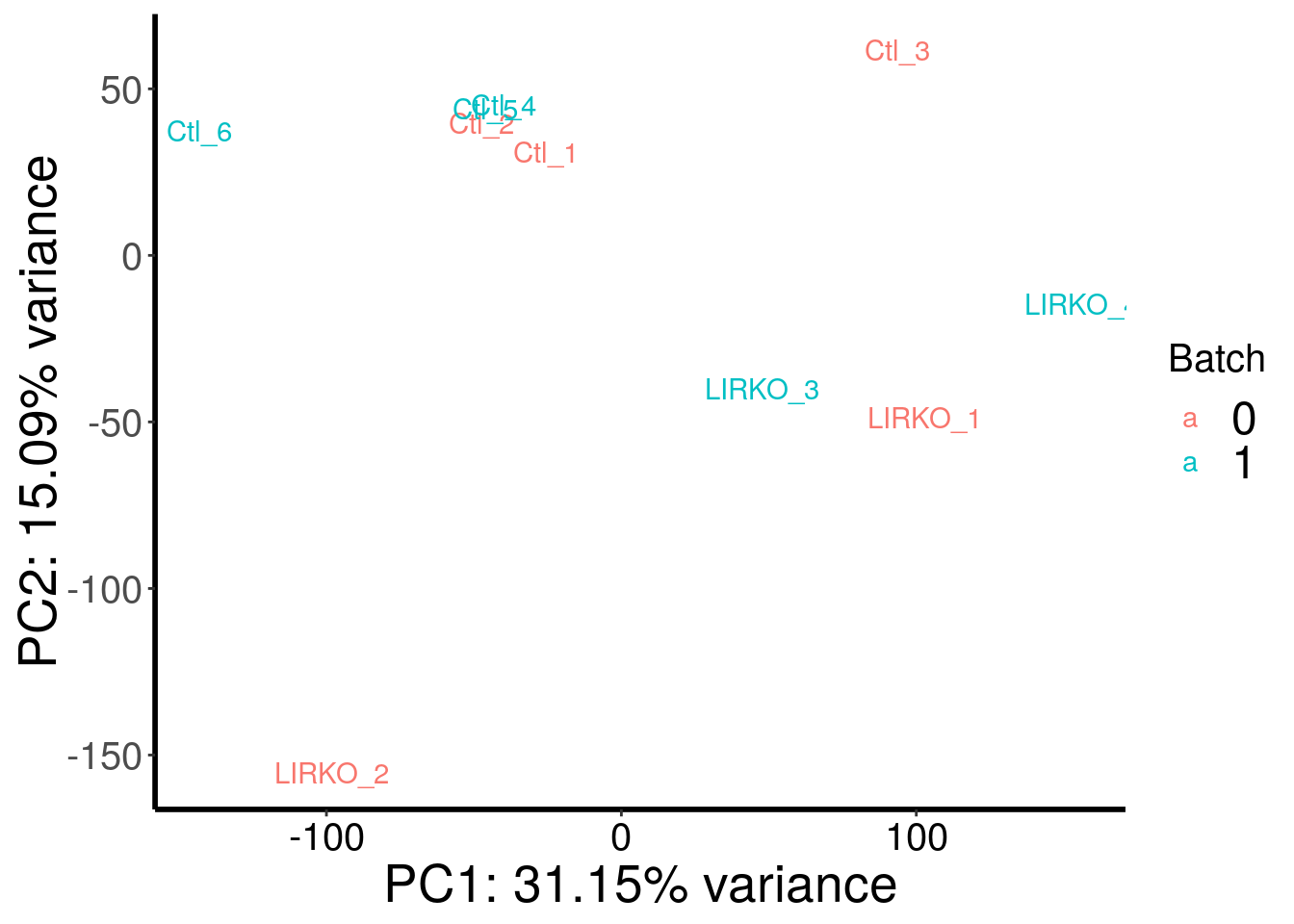
| Version | Author | Date |
|---|---|---|
| e1b9592 | scottzijiezhang | 2020-03-30 |
Samples are well separated by genotype. Batch does not contribute to the variance much.
DM tests with RADAR
variable( LIRKO_RADAR ) <- data.frame(genotype = c( rep( "Ctl", 6 ), rep( "LIRKO", 4 ) )
)
LIRKO_RADAR <- diffIP_parallel(LIRKO_RADAR, thread = 20)
LIRKO_RADAR <- reportResult(LIRKO_RADAR, cutoff = 0.1, threads = 20)
save(LIRKO_RADAR, file = "~/Rohit_T1D/mouse_islet/LIRKO_RADAR.RData")
write.table(results(LIRKO_RADAR), file = "~/Rohit_T1D/mouse_islet/LIRKO_diffPeaks_FDR0.1.xls", sep = "\t", row.names = FALSE, col.names = FALSE, quote = FALSE)Differentially methylated m6A sites at FDR 10% threshold.
load("~/Rohit_T1D/mouse_islet/LIRKO_RADAR.RData")
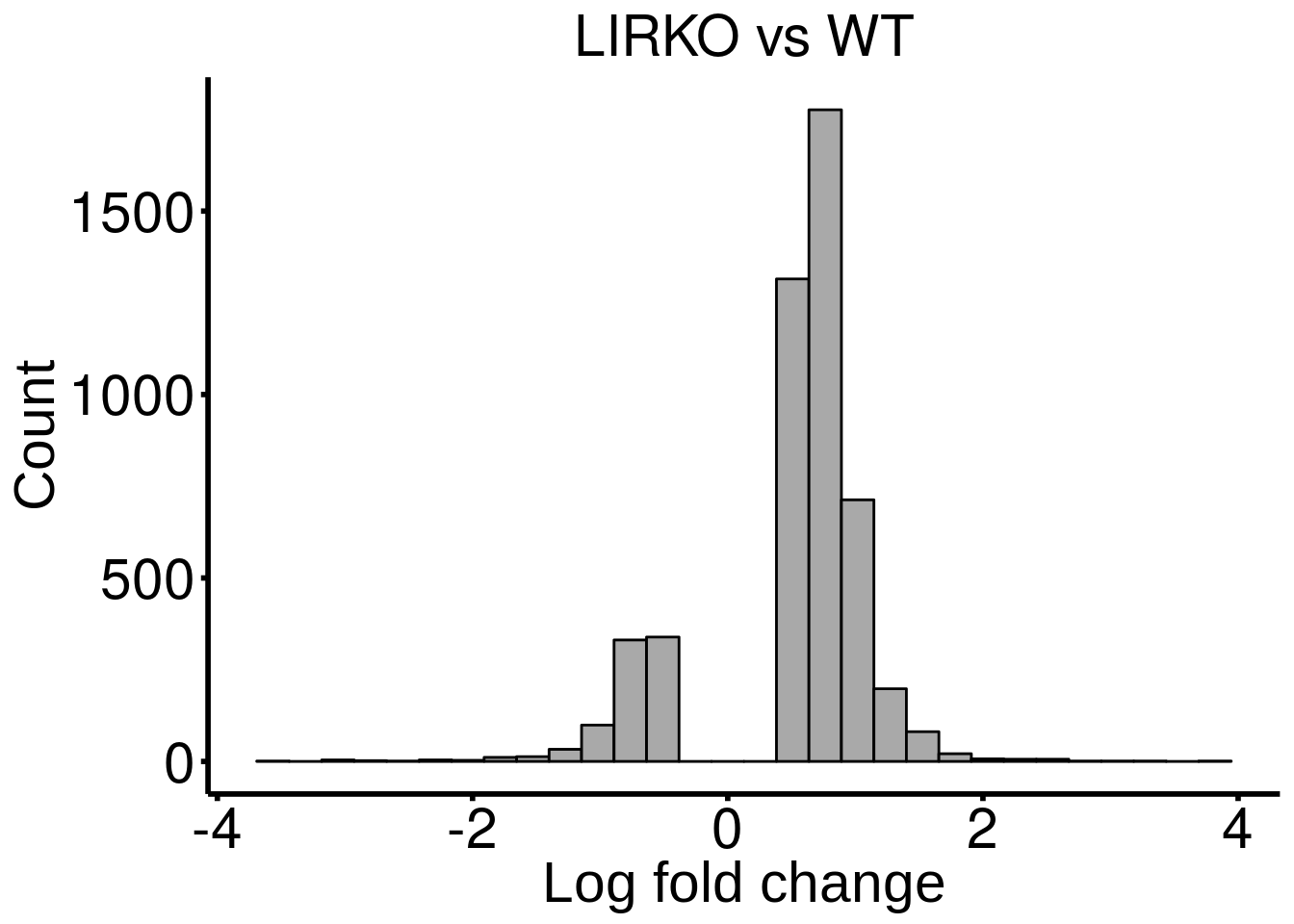
LIRKO_result <- results(LIRKO_RADAR)There are 4968 reported differential loci at FDR < 0.1 and logFoldChange > 0.5.DT::datatable( LIRKO_result , rownames = FALSE )Distribution of log fold change of significant DM sites
ggplot(LIRKO_result ,aes( x = logFC ) )+geom_histogram(color="black", fill="dark gray",bins = 30)+xlab("Log fold change")+ggtitle("LIRKO vs WT")+theme_bw() + ylab("Count")+ theme(panel.border = element_blank(), panel.grid.major = element_blank(),
panel.grid.minor = element_blank(), plot.title = element_text( size = 22,colour = "black", hjust = 0.5 ) , axis.line = element_line(colour = "black",size = 1),axis.ticks = element_line(colour = "black",size = 1), axis.text = element_text(size = 22,colour = "black"),axis.text.y = element_text(angle = 0) ,axis.title=element_text(size=22,family = "arial")
)
| Version | Author | Date |
|---|---|---|
| 514bc99 | scottzijiezhang | 2020-05-08 |
Spatial distribution of these DM sites
library(grid)
library(ggsci)
MeRIPtools::plotMetaGeneMulti( list("Hyper-methylated" = LIRKO_result[LIRKO_result$logFC>0,1:12], "Hypo-methylated" = LIRKO_result[LIRKO_result$logFC<0,1:12]), gtf = "~/Database/genome/mm10/mm10_UCSC.gtf" )[1] "Converting BED12 to GRangesList"
[1] "It may take a few minutes"
[1] "Converting BED12 to GRangesList"
[1] "It may take a few minutes"
[1] "total 35119 transcripts extracted ..."
[1] "total 31408 transcripts left after ambiguity filter ..."
[1] "total 15627 mRNAs left after component length filter ..."
[1] "total 3280 ncRNAs left after ncRNA length filter ..."
[1] "Building Guitar Coordinates. It may take a few minutes ..."
[1] "Guitar Coordinates Built ..."
[1] "Using provided Guitar Coordinates"
[1] "resolving ambiguious features ..."
[1] "no figure saved ..."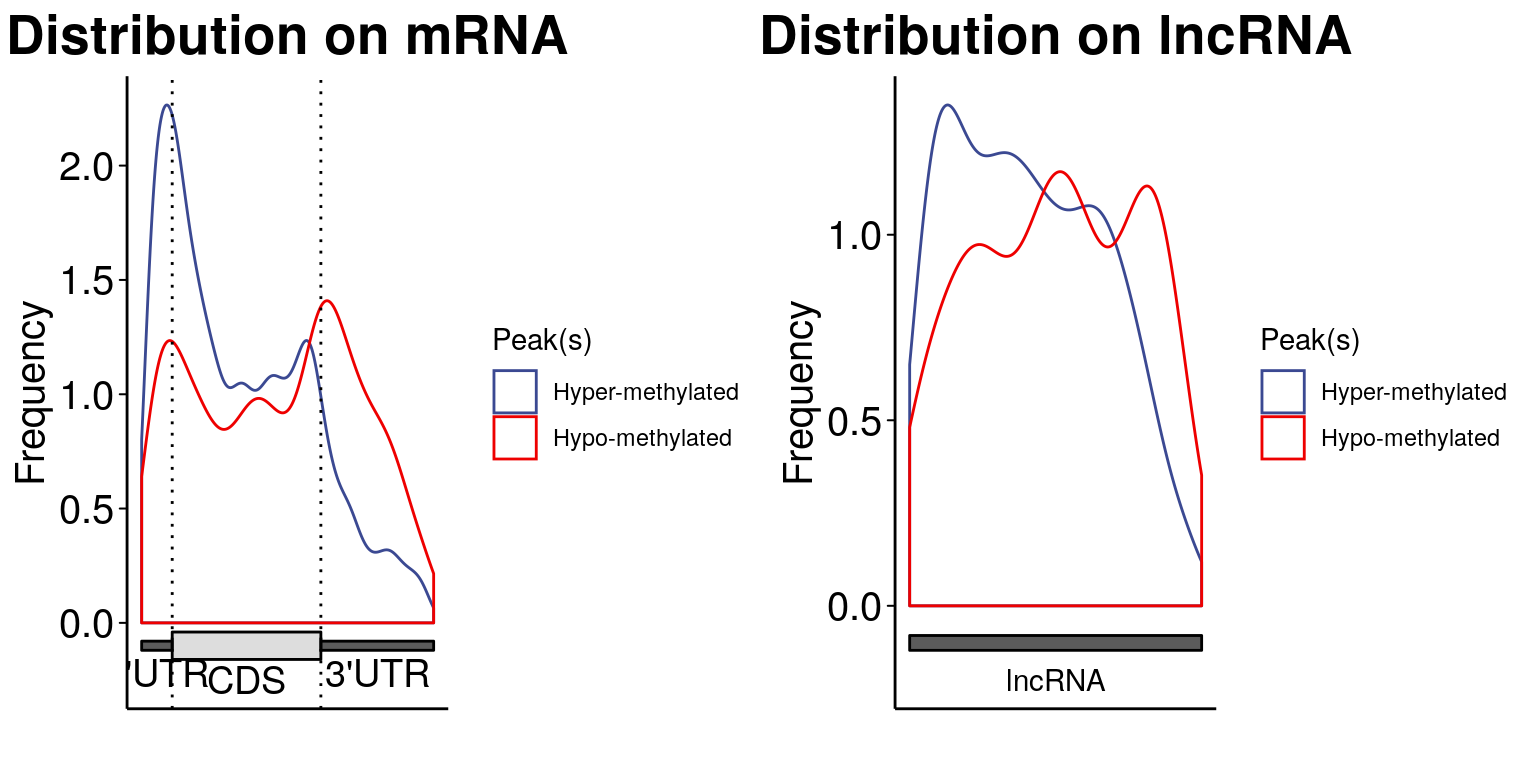
NOTE this function is a wrapper for R package "Guitar".
If you use the metaGene plot in publication, please cite the original reference:
Cui et al 2016 BioMed Research International The enriched motif learned from this DM sites set is a bit atypical.
KEGG pathway analysis of DMG
library(clusterProfiler)
eg.LIRKO <- bitr( unique(results(LIRKO_RADAR)$name), fromType="SYMBOL", toType="ENTREZID", OrgDb="org.Mm.eg.db")There are 4968 reported differential loci at FDR < 0.1 and logFoldChange > 0.5.KEGG_LIRKO <- enrichKEGG(eg.LIRKO$ENTREZID,organism = "mmu",pAdjustMethod = "fdr", pvalueCutoff = 0.1,minGSSize = 3)
emapplot(KEGG_LIRKO, vertex.label.font = 3)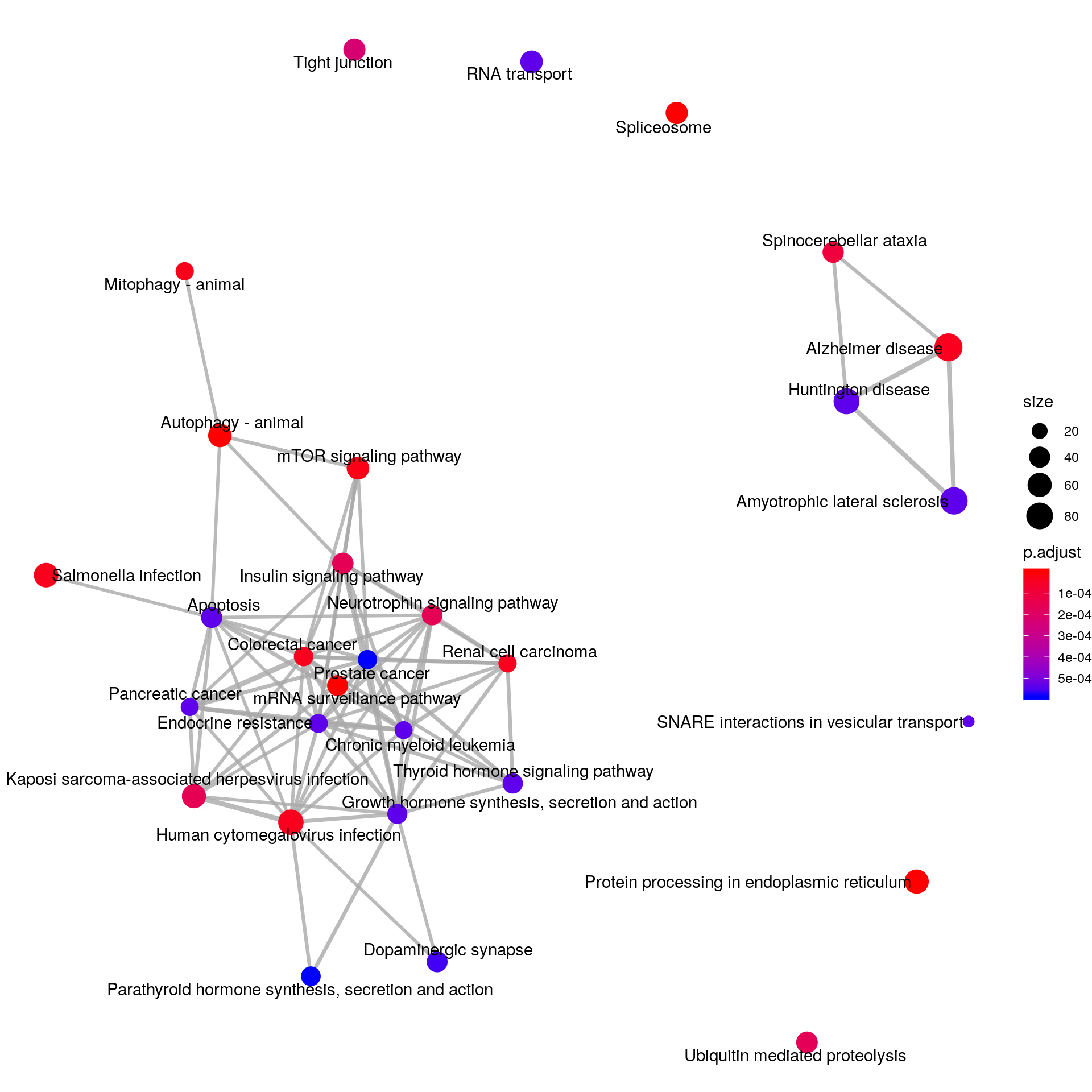
| Version | Author | Date |
|---|---|---|
| aa2d181 | scottzijiezhang | 2020-06-05 |
Intersect DMGs in LIRKO and DMGs in T2D patients.
T2D_DMpeaks <- read.table("~/Rohit_T2D/RADAR_diffPeaks_FDR0.05.xls", sep = "\t", header = TRUE)
convertMouseGeneList <- function(x){
require("biomaRt")
human = useMart("ensembl", dataset = "hsapiens_gene_ensembl")
mouse = useMart("ensembl", dataset = "mmusculus_gene_ensembl")
genesV2 = getLDS(attributes = c("mgi_symbol"), filters = "mgi_symbol", values = x , mart = mouse, attributesL = c("hgnc_symbol"), martL = human, uniqueRows=T)
return(genesV2)
}
MtoHmap <- convertMouseGeneList( unique( LIRKO_result$name ) )
LIRKO_result$HumanOrtholog <- MtoHmap$HGNC.symbol[ match(LIRKO_result$name, MtoHmap$MGI.symbol ) ]
library(Vennerable)
Vcommon_LIRKO_T2D <- Venn(list("DMGs in LIRKO" = unique(LIRKO_result$HumanOrtholog),"DMGs in T2D patient" = unique(T2D_DMpeaks$name) ) )
plot(Vcommon_LIRKO_T2D,doWeights=TRUE)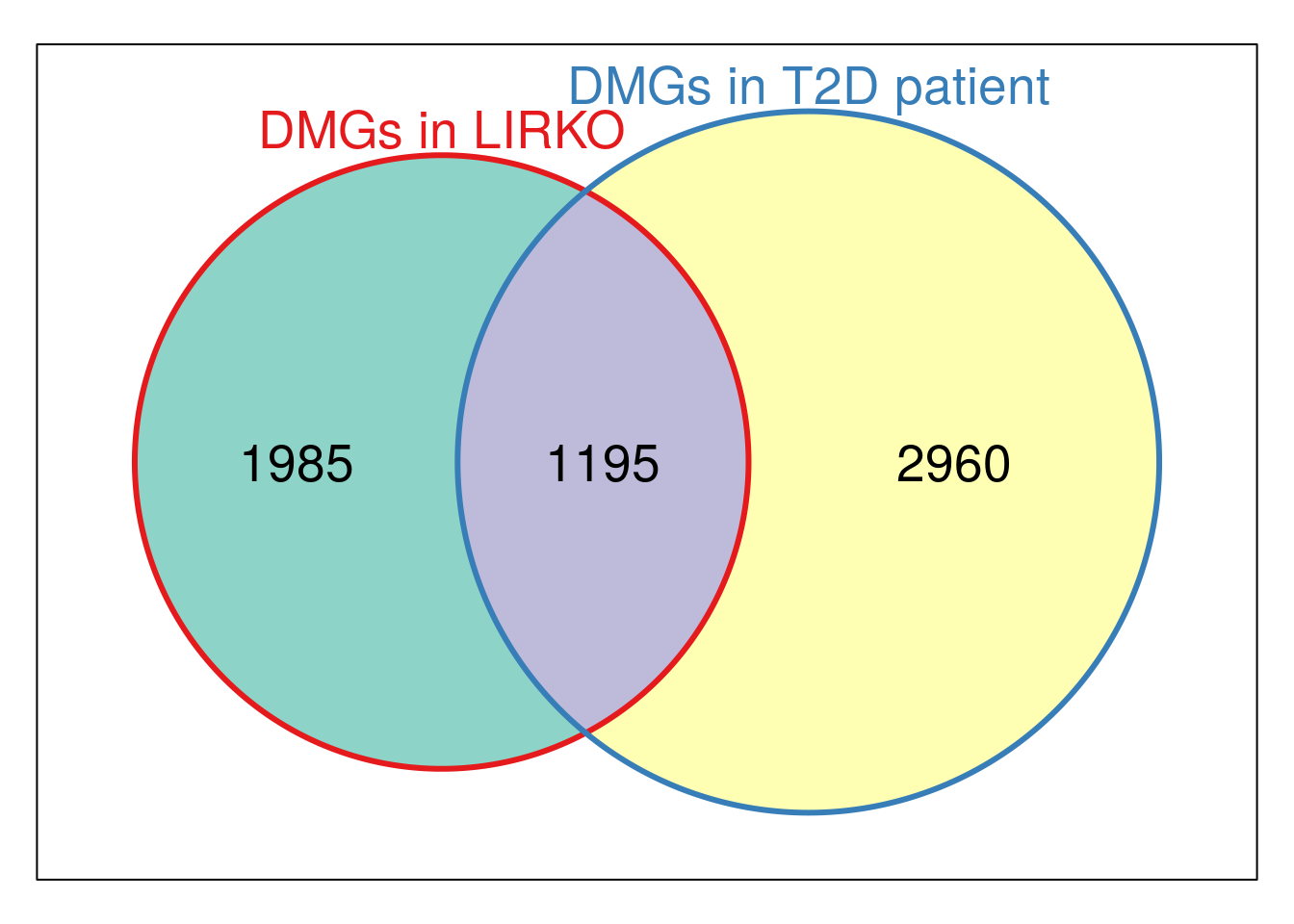
| Version | Author | Date |
|---|---|---|
| aa2d181 | scottzijiezhang | 2020-06-05 |
cat( paste0("Intersect DMGs in LIRKO and T2D patient:\n", paste(intersect( unique(LIRKO_result$HumanOrtholog), unique(T2D_DMpeaks$name) ), collapse = "\t" ) ) )Intersect DMGs in LIRKO and T2D patient:
C10orf88 KIAA0100 C11orf95 KIAA1671 KIAA0895L KIAA1109 KIAA0408 KIAA2026 C8orf48 C6orf132 C5orf51 ABCA7 ABCC5 ABCF1 ABCF3 ABHD8 ABL1 ACACA ACIN1 ACTN4 ADCY6 ADGRL1 ADORA2A ADRA2A ADRM1 AEBP2 AFF1 AFG3L2 AGAP1 AGAP3 AGRN AGTPBP1 AGTRAP AHCYL2 AHNAK AIDA AIMP1 AK1 AKAP1 AKAP11 AKAP12 AKAP13 AKAP8 AKAP9 AKIRIN2 AKT2 ALDOB ALKBH7 AMBRA1 AMFR ANKLE2 ANKRD11 ANKRD12 ANKRD17 ANKRD40 ANKRD6 ANXA4 AP2M1 AP4E1 APC APP ARAP3 ARFGAP1 ARFGEF3 ARHGAP12 ARHGAP21 ARHGAP35 ARHGEF1 ARHGEF12 ARHGEF17 ARHGEF18 ARHGEF5 ARID1A ARID1B ARIH1 ARL14 ARX ASB1 ASH1L ASNSD1 ASPH ASXL1 ASXL2 ATF7IP ATG2A ATG4D ATMIN ATP11A ATP2A2 ATP6V1E1 ATP9A ATRN ATRNL1 ATXN7L3B AUTS2 C5orf24 B3GNT3 C6orf89 C18orf32 BAHCC1 BAIAP2L1 BANP BAX BAZ2A BAZ2B BBX BCL2L13 BCL9 BDP1 BEX1 BHLHB9 BHLHE41 BIRC6 BLOC1S4 BMS1 BOD1L1 BOLA3 BPTF BRD3 BRD4 BRF2 BRPF1 BRSK1 BSN BTBD3 BTBD7 BTBD9 C1orf115 C1GALT1 C1QC CABIN1 CACNA1A CACNA1D CACNA2D1 CACUL1 CAMK1 CAND1 CAPN2 CASC3 CASR CAST CBFA2T2 CBFA2T3 CBFB CBR1 CBX6 CC2D1A CCAR1 CCDC134 CCDC50 CCDC8 CCND1 CCND3 CCNT1 CCPG1 CCSER1 CD276 CD93 CD99L2 CDC34 CDC42BPA CDC42BPB CDC42BPG CDC5L CDCP1 CDK10 CDK11B CDK13 CDK9 CDKN2C CEBPZ CELSR1 CELSR3 CENPB CEP104 CEP164 CEP170 CEP170B CEP250 CFAP36 CFLAR CGNL1 CHAMP1 CHCHD6 CHD2 CHD3 CHD5 CHD7 CHKA CHM CHP1 CHST11 CHSY1 CLIC4 CLN5 CLSTN3 CLTB CMTR1 CNNM3 CNOT3 CNTROB COA5 COASY COBL COG4 COL15A1 COL4A2 CORO1C COX20 CPD CPM CPSF7 CREBBP CREBRF CRYBG3 CSNK1G2 CTBP1 CTIF CTR9 CTSZ CUX1 CXXC4 CXXC5 CYB5RL CYP2U1 C17orf80 DAB2IP DACH1 DAPK1 DAZAP1 DCAF10 DCAF11 DCAF6 DCDC2 RSC1A1 DDIT4 DDX1 DDX21 DDX42 DDX51 DEDD DEK DENR DEPTOR DGCR8 DGKZ DHX30 DHX38 DHX8 DICER1 DIDO1 DIXDC1 DLG1 DLG3 DLG5 DMWD DMXL1 DMXL2 DNAAF2 DNAJB11 DNAJC10 DNAJC11 DNAJC2 DNAJC21 DNAJC30 DNAJC8 DNM2 DNMT3A DNTTIP2 DOT1L DROSHA DSE DST DTD2 DUSP16 DUSP18 DYNC1H1 DYNLL1 DYNLL2 DYRK1B ECE1 ECHDC1 EFNB2 EFTUD2 EGFL7 EGLN1 EGLN2 EGR1 EHBP1 EHHADH EIF2B5 EIF3CL EIF4B EIF4E3 EIF4EBP2 EIF4G3 EIF4H EIF5 ELL2 ELOVL6 EMC1 EML5 ENAH ENDOD1 ENY2 EP300 EP400 EPC1 EPS15L1 ERC1 ERLIN1 ERLIN2 ERN1 ESRRA ETS1 EWSR1 EXD2 EXO5 EXOC4 EXOSC6 EXTL3 F8A1 FADD FAM110A FAM114A2 FAM120B FAM120C FAM135B FAM193A FAM193B FAM199X FAM219B FAM222A FAM83G FAM83H FAN1 FARP1 FARP2 FBL FBXL14 FBXL16 FBXL17 FBXL19 FBXO21 FBXO38 FBXO42 FBXO45 FCHSD2 FDPS FEM1A FEM1B FGD4 FGFR1 FIS1 FKBP15 FKBP5 FKBP9 FLII FLNB FMN2 FMNL2 FN3K FNBP1L FNDC3A FNDC3B FNIP1 FOS FOXJ3 FOXO6 FOXP4 FRMD4A GAB2 GALNT2 GANAB GBF1 GGA2 GGNBP2 GINM1 GIT1 GLG1 GLIS2 GLS GMEB2 GNA11 GNA13 GNAI2 GNAZ GNG10 GNL3L GNPTG GOLGA3 GOLGA4 GOLGA7 GOLGB1 GOLM1 GOSR2 GPC1 GPR142 GPRIN1 GRIA2 GRPEL2 GSK3A GSPT1 GSTO1 GTF2A1 GTF3C1 GTPBP1 HM13 H6PD HAP1 HDAC6 HDGF HDLBP HELZ HES6 HIBADH HIP1R HIRA HIVEP2 HIVEP3 HMCES HMG20A HMGB2 HNF4A HNRNPCL1 HNRNPM HNRNPUL2-BSCL2 HOOK1 HOOK3 HPS3 HPS6 HRAS HS1BP3 HS2ST1 HSPD1 HUWE1 ICE1 ID2 IDH2 IER5 IFFO1 IGF1R IGF2 IGFBP5 IK IKBKB IL6ST IMMT INPP4A INPPL1 INSM1 INTS3 IP6K2 IPO5 IQCC IQGAP1 IRF1 IRF2BP2 IRS2 ISCA1 ITSN1 ITSN2 IWS1 JMJD1C JMJD6 JMY KANK1 KAT5 KCNK3 KCTD2 KDM1A KDM2A KDM4A KDM5A KDM5C KDR KHDRBS1 KHNYN KIF13B KIF16B KIF1A KIF1C KIF3A KIF3B KIF5B KIFC3 KL KLC1 KLF10 KLF11 KLF3 KLF5 KLF9 KLHDC1 KLHDC8B KLHL24 KLHL36 KMT2A KMT2B KMT2C KMT2D KMT2E KPNB1 KRBA1 KRT7 KSR2 LACTB LAGE3 LAMC1 LARP1 LAS1L LASP1 LBR LEMD3 LETM1 LIFR LIG4 LIMA1 LMO7 LMTK3 LPIN1 LRIG2 LRP3 LRP5 LRP6 LRRC24 LRRC58 LRRC59 LRRFIP1 LRRFIP2 LSM10 LSM14A LSR LTN1 LUC7L2 LUZP1 LYRM2 LYST LZTR1 MACF1 MAFK MAGED2 MAGI1 MAGT1 MAML3 MAN1B1 MAN1C1 MAP1A MAP1B MAP3K1 MAP3K10 MAP3K7 MAP7D1 MAPK3 MAPK8IP1 MAPT MARK3 MAZ MBD5 MCC MCF2L MCU MDC1 MDM2 MEAF6 MED13L MED22 MEF2A MEGF9 MEIS3 MEPCE METRNL METTL6 MEX3D MGA MIB2 MICAL3 MICALL1 MID2 MINK1 MIOS MLEC MLLT6 MLXIP MLXIPL MNT MNX1 MOAP1 HSPE1-MOB4 MON1B MORC3 MORF4L1 MPP5 MPRIP MRPL19 MRPL40 MRPS23 MSH6 MSL2 MSMO1 MTCL1 MTCP1 MTMR3 MTUS1 MYBBP1A MYH10 MYH14 MYL6 MYO10 MYO18A MYO5A MYO5B MYO6 MYO9A N4BP1 N4BP2L2 NAA15 NAP1L1 NAP1L3 NAV1 NAV2 NBEA NBL1 NCAPH2 NCOA3 NCOA6 NDOR1 NDUFB10 NDUFB3 NDUFS3 NEDD4L NEFH NES NEURL1 NF2 NFAT5 NFATC3 NFE2L1 NFKBIA NGDN NIN NIPBL NKAP NKIRAS2 NKTR NLRX1 NOC2L NOL6 NOL8 NOLC1 NOP14 NPEPPS NR2F2 NRF1 NRIP1 NT5DC3 NUB1 NUCB2 NUDT3 NUMA1 NUTF2 NVL NYAP1 NYNRIN OBSL1 OCLN OGFR OLFM1 ONECUT2 OPTN ORMDL3 OS9 OSGIN1 OXR1 P4HA1 P4HB PABPC4 PABPC5 PACSIN2 PAICS PAK3 PAPOLA PAPSS2 PARD3 PBRM1 PCDHB15 PCDHGA12 PCDHGA2 PCDHGA4 PCDHGB1 PCDHGB6 PCDHGC3 PCED1A PCF11 PCLO PCNT PCSK7 PDCD11 PDCD4 PDE4D PDE4DIP PDE5A PDE8A PDS5A PDXDC1 PDZD4 PDZD8 PDZRN3 PEG10 PEG3 PET117 PEX14 PFDN6 PGBD5 PHACTR2 PHF14 PHF8 PHKG2 PHLPP1 PI4K2A PICK1 PIGR PIN1 PITHD1 PITPNM1 PKD1 PKD2 PKHD1 PKN1 PKP4 PLCB4 PLEC PLEKHA6 PLEKHG3 PLEKHM2 PLS1 PLVAP PLXNA2 PLXNA3 PMM2 PNPLA8 POLR3E POM121 POR PPAN-P2RY11 PPARD PPFIA1 PPIL2 PPIL4 PPP1CB PPP1R10 PPP1R1A PPP1R26 PPP1R37 PPP1R9B PPP2R5B PPP2R5C PPP2R5D PPP2R5E PPP3CA PRADC1 PRELID1 PRICKLE2 PRKAR2A PROX1 PRPF31 PRPF4B PRPF6 PRPS1 PRR14 PRR14L PRRC2B PRRC2C PRSS23 PRUNE2 PSMC5 PSMD2 PTP4A3 PTPN11 PTPN12 PTPN23 PTPRA PTRH2 PURA PURB PWWP2A QSOX1 RAB10 RAB11FIP1 RAB11FIP2 RAB43 RABGAP1L RAI1 RAI14 RALBP1 RALGAPB RANGAP1 RAP1GAP2 RAP2B RAPH1 RASSF3 RBM12 RBM12B RBM14 RBM15B RBM17 RBM26 RBM5 RBMXL1 RCAN2 RCAN3 RCN1 RDX REEP3 REPIN1 REST REV3L REXO1 RGL1 RGS9 RHOBTB2 RHOG RIC8B RING1 RIPK4 RLF RLIM RMND5A RNASET2 RNF145 RNF20 RNF217 RNF26 RNF31 RNF40 RNF7 ROCK2 ROR2 RPL36 RPL36A RPRD1B RPTOR RRAGD RRAS RRBP1 RREB1 RRP12 RRP1B RSPH3 RSRC1 RTN2 RUFY1 RUNDC3A RXRB SAFB SAMD4B SAP30BP SAP30L SART1 SASH1 SBNO1 SCAF1 SCAF11 SCAPER SCLY SCRT1 SDCCAG8 SDHA SEC62 SEMA4B SEMA6A SEMA6D SENP6 SERPING1 SETX SF3A3 SF3B1 SFPQ SFRP5 SH3GLB1 SH3KBP1 SH3PXD2A SHROOM2 SHROOM3 SIPA1L3 SIRT7 SKIV2L SLC10A7 SLC12A4 SLC12A7 SLC19A2 SLC25A1 SLC25A23 SLC25A28 SLC25A29 SLC25A38 SLC25A42 SLC25A46 SLC25A52 SLC30A4 SLC38A10 SLC39A3 SLC3A2 SLC44A2 SLC6A6 SLC7A5 SLK SLMAP SLX4 SMAD7 SMARCA2 SMC1A SMC3 SMC5 SMCR8 SMG7 SMIM12 SNAP29 SNAPC2 SNAPC3 SNAPC4 SNPH SNRK SNRNP70 SOBP SOCS4 SOGA1 SOGA3 SON SOWAHC SOX13 SP1 SPAG9 SPECC1 SPECC1L SPEG SPEN SPIN1 SPIRE1 SPTAN1 SPTBN1 SPTBN2 SPTY2D1 SQLE SRCAP SRF SRGAP1 SRRM1 SRRM2 SSBP3 SSTR3 STAG2 STIM1 STOX2 STRN3 STX6 STXBP5L SUB1 SUCO SUPT5H SUPT6H SURF2 SYMPK SYNE2 SYNGAP1 SYNPO SYT7 SYTL1 TAB2 TACC1 TACC2 TADA2B TANC1 TANC2 TAOK1 TAOK3 TBC1D10B TBC1D15 TBC1D16 TBC1D8 TBCB TBRG1 TBX3 TCEA1 TCEAL6 TCF20 TCF3 TEAD1 TELO2 TENM3 TERF2 TGS1 THEMIS2 TIMM10B TIMM44 TJAP1 TJP1 TJP2 TLE1 TLK1 TLN1 TM9SF4 TMCC2 TMED9 TMEM126A TMEM184A TMEM185B TMEM222 TMEM30A TMEM63C TMEM80 TMEM8B TMEM97 TMF1 TMOD2 TMPO TMTC2 TNIP2 TNK2 TNKS1BP1 TNPO1 TNRC6A TNRC6B TNRC6C TNS2 TOMM34 TOP1 TOP3A TOR1A TOR1AIP2 TPCN1 TPM1 TPR TRABD TRAPPC10 TRAPPC5 TRIM2 TRIM25 TRIM41 TRIM56 TRIO TRIOBP TRIP11 TRIP12 TRIP4 TRMT12 TRMT6 TRPM3 TSC1 TSC22D1 TSC22D4 TSEN34 TSHZ1 TTBK2 TTC17 TTC22 TTC28 TTC3 TTC30B TUFT1 TWSG1 TXLNA TXLNG TXNDC15 TXNRD1 U2SURP UACA UAP1L1 UBA5 UBAP2 UBASH3B UBE2D2 UBE2G2 UBE2O UBE2Q1 UBE2Z UBL3 UBN1 UBN2 UBR3 UBTF UBXN10 UGGT1 UIMC1 UNC13B UPF3A UPF3B URGCP URI1 USH1C USP1 USP16 USP51 USP8 USP9X UTP14C VAMP2 VEGFA VEZF1 VPS18 VPS26B VPS35 VPS36 VTI1A VWA5B2 WAC WDR20 WDR43 WDR59 WDR81 WDR82 WIZ WNK1 WNK2 WSCD1 XBP1 XPC XPO6 XRN2 XYLT2 YKT6 YLPM1 YTHDF3 YY1 ZBTB1 ZBTB11 ZBTB18 ZBTB38 ZBTB41 ZBTB44 ZC3H10 ZC3H11A ZC3H4 ZCCHC18 ZCCHC2 ZCCHC3 ZDHHC14 ZFHX3 ZNF106 ZNF12 ZNF205 ZNF142 ZNF148 ZNF174 ZNF217 ZNF248 ZNF281 ZNF282 ZFP30 ZNF316 ZNF322 ZNF346 ZFP36L2 ZNF570 ZNF446 ZNF507 ZNF516 ZNF518A ZNF524 ZNF598 ZNF780A ZNF609 ZNF628 ZNF638 ZNF646 ZNF653 ZNF654 ZNF664 ZNF667 ZNF687 ZNF688 ZNF697 ZNF703 ZNF704 ZNF775 ZNF845 ZNF770 ZNF771 ZNF420 ZNF787 ZFP91 ZFR ZFX ZGPAT ZHX1 ZKSCAN4 ZKSCAN7 ZMAT1 ZMAT5 ZMYM1 ZMYM4 ZMYND11 ZNFX1 ZSCAN12 ZSCAN21KEGG pathway analysis of overlapping DMGs
eg.LIRKO_T2D <- bitr( intersect( unique( LIRKO_result$HumanOrtholog), unique(T2D_DMpeaks$name) ) , fromType="SYMBOL", toType="ENTREZID", OrgDb="org.Hs.eg.db")
KEGGLIRKO_T2D <- enrichKEGG(eg.LIRKO_T2D$ENTREZID,organism = "hsa",pAdjustMethod = "fdr", pvalueCutoff = 0.1,minGSSize = 3)
dotplot(KEGGLIRKO_T2D, showCategory=20 )+theme(
panel.grid.minor = element_blank(), axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=22, color = "black", hjust=0.5,family = "arial"),
axis.title.y=element_text(size=22, color = "black", vjust=0.4, angle=90,family = "arial"),
legend.title=element_text(size=15, color = "black"),legend.text = element_text(size = 20, color = "black",family = "arial"),
axis.text.x = element_text(size = 15,color = "black",family = "arial") ,axis.text.y = element_text(size = 15,color = "black",family = "arial"))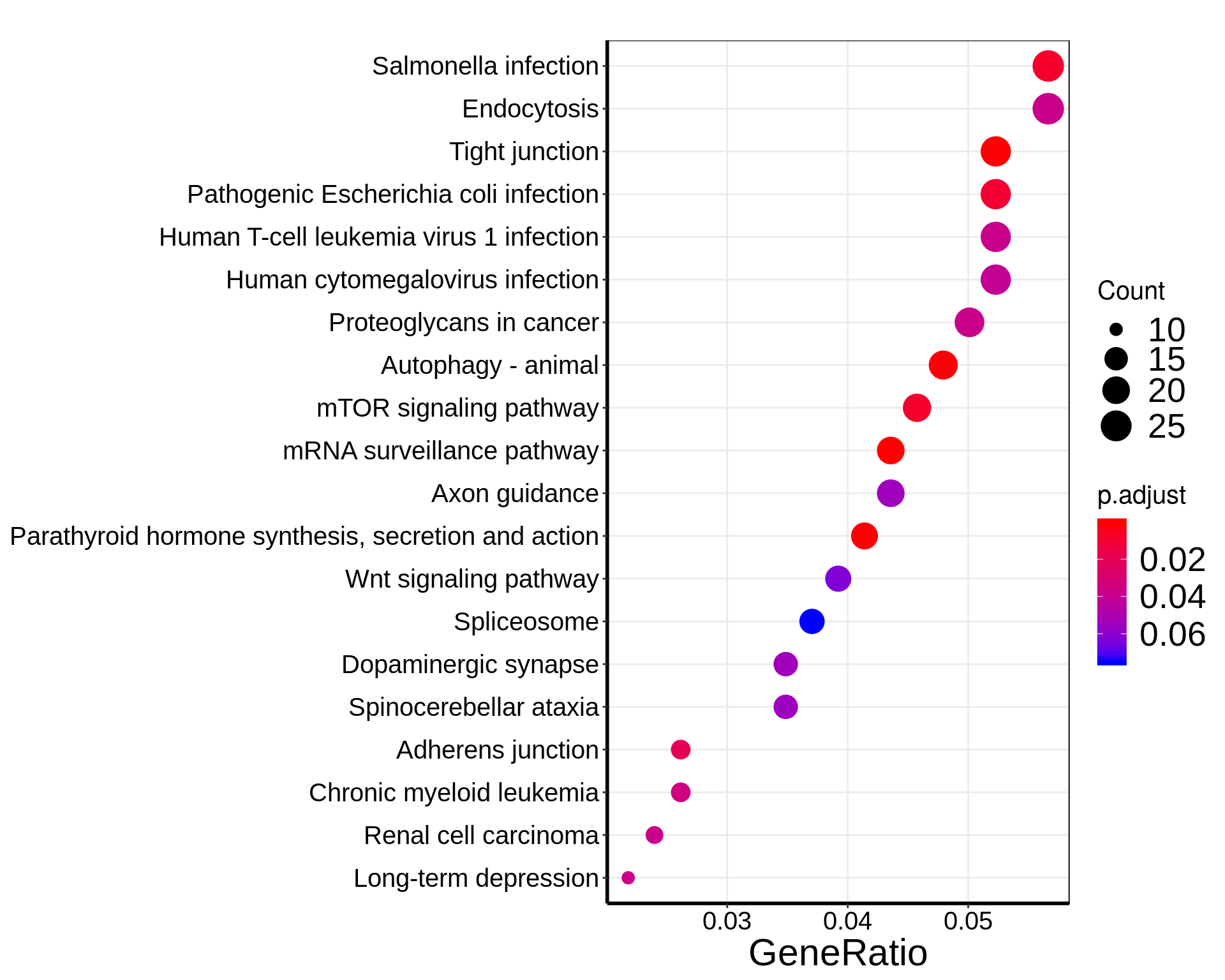
| Version | Author | Date |
|---|---|---|
| aa2d181 | scottzijiezhang | 2020-06-05 |
emapplot(KEGGLIRKO_T2D, vertex.label.font = 3)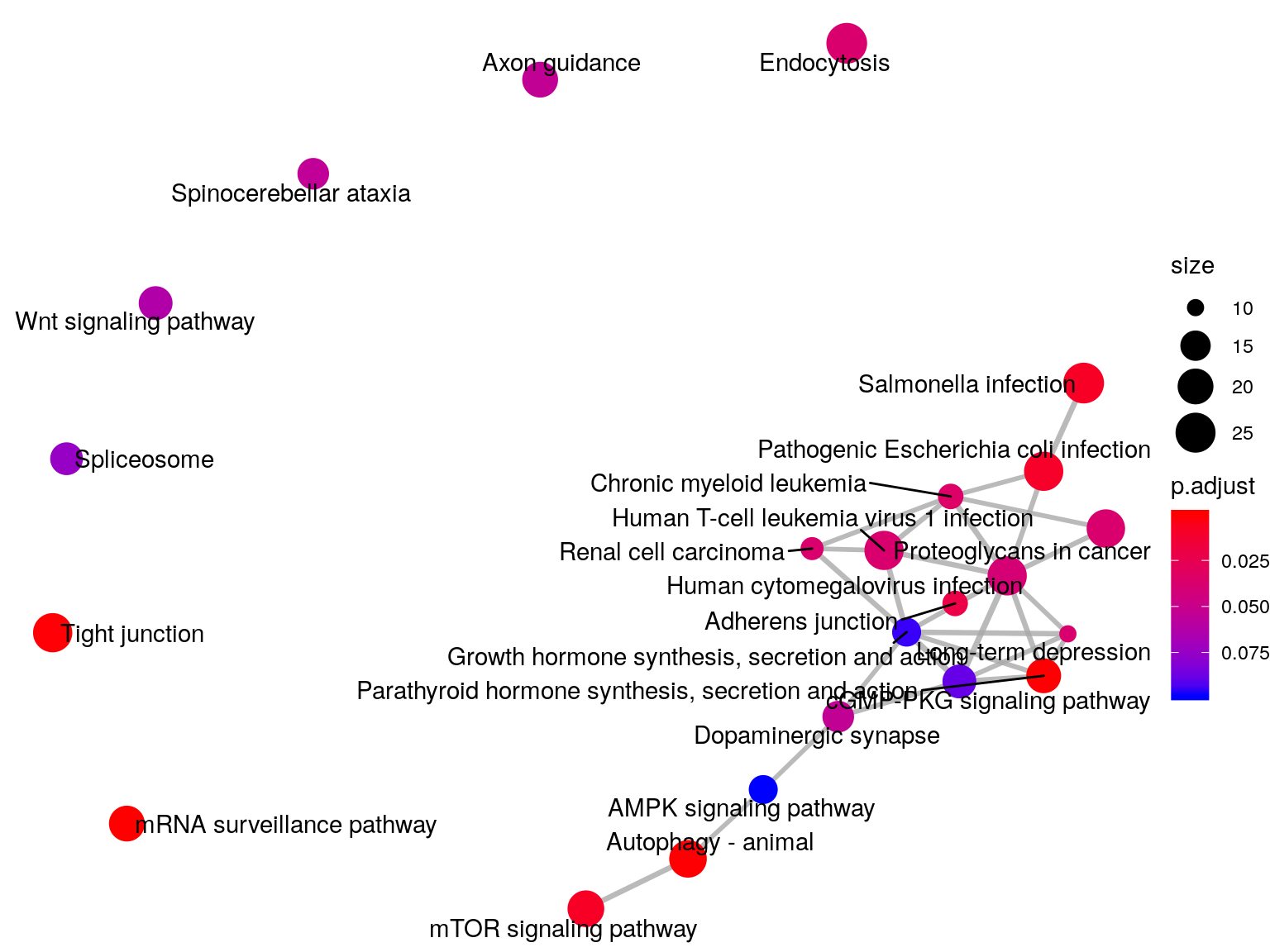
| Version | Author | Date |
|---|---|---|
| aa2d181 | scottzijiezhang | 2020-06-05 |
Compare High fat diet to Control
load( "~/Rohit_T1D/mouse_islet/allSample_RADAR.RData")
HFD_sample <- c( paste0("Ctl_", 1:6 ), paste0( "Ctl_HFD_", 1:4 ) )
HFD_RADAR <- select( allSampleRADAR, HFD_sample )
HFD_RADAR <- normalizeLibrary( HFD_RADAR )
HFD_RADAR <- adjustExprLevel( HFD_RADAR )
variable( HFD_RADAR ) <- data.frame(genotype = c( rep( "Ctl", 6 ), rep( "HFD", 4 ) ),
batch = c( 0,0,0,1,1,1,0,0,1,1 )
)
HFD_RADAR <- filterBins( HFD_RADAR )
save(HFD_RADAR, file = "~/Rohit_T1D/mouse_islet/HFD_RADAR.RData")Check confounding factors
library(RADAR)
load( "~/Rohit_T1D/mouse_islet/HFD_RADAR.RData" )
plotPCAfromMatrix(HFD_RADAR@ip_adjExpr_filtered, variable(HFD_RADAR)$genotype )+scale_color_discrete(name = "Genotype")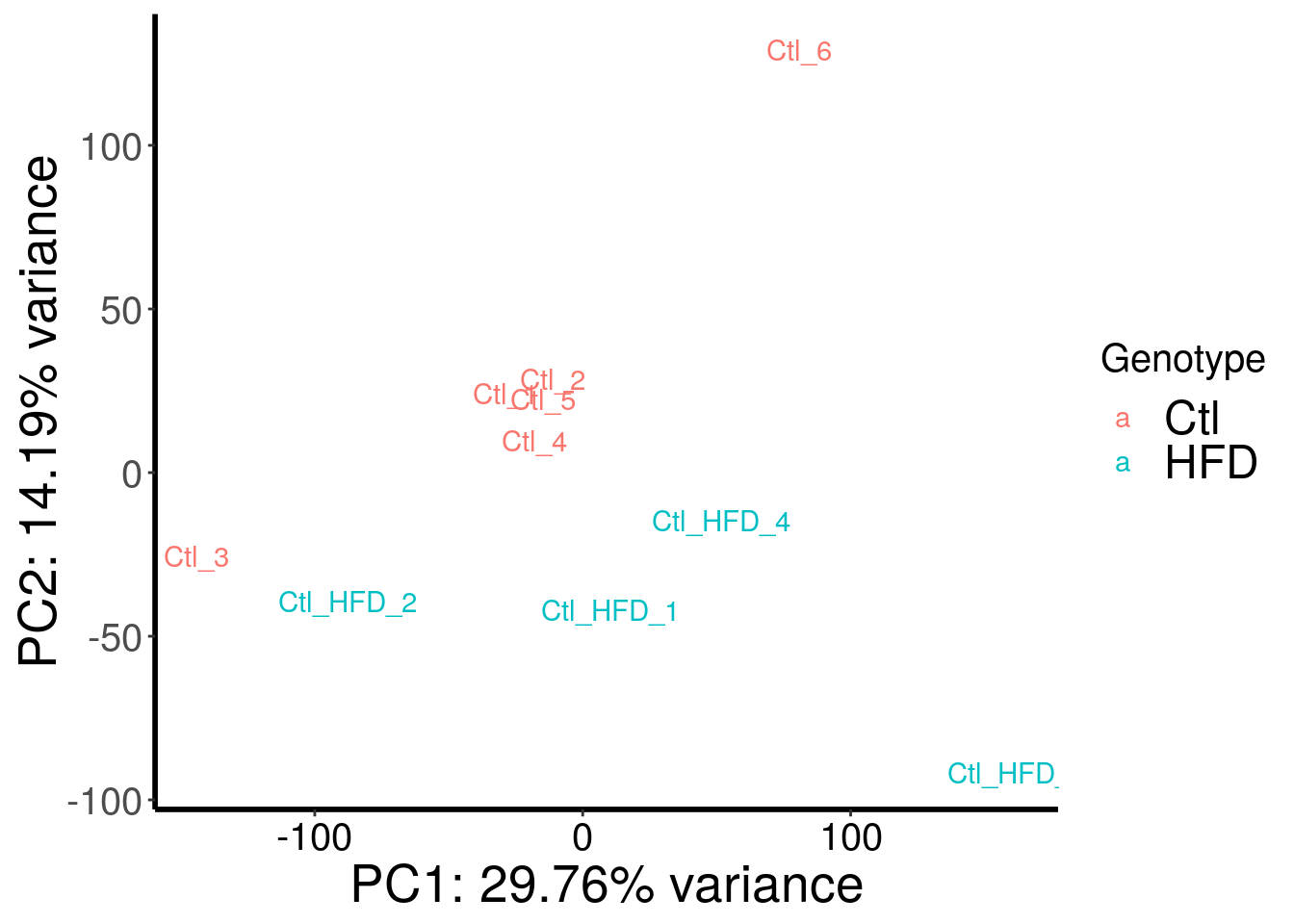
| Version | Author | Date |
|---|---|---|
| e1b9592 | scottzijiezhang | 2020-03-30 |
plotPCAfromMatrix(HFD_RADAR@ip_adjExpr_filtered, as.character( c( 0,0,0,1,1,1,0,0,1,1 ) ) )+scale_color_discrete(name = "Batch")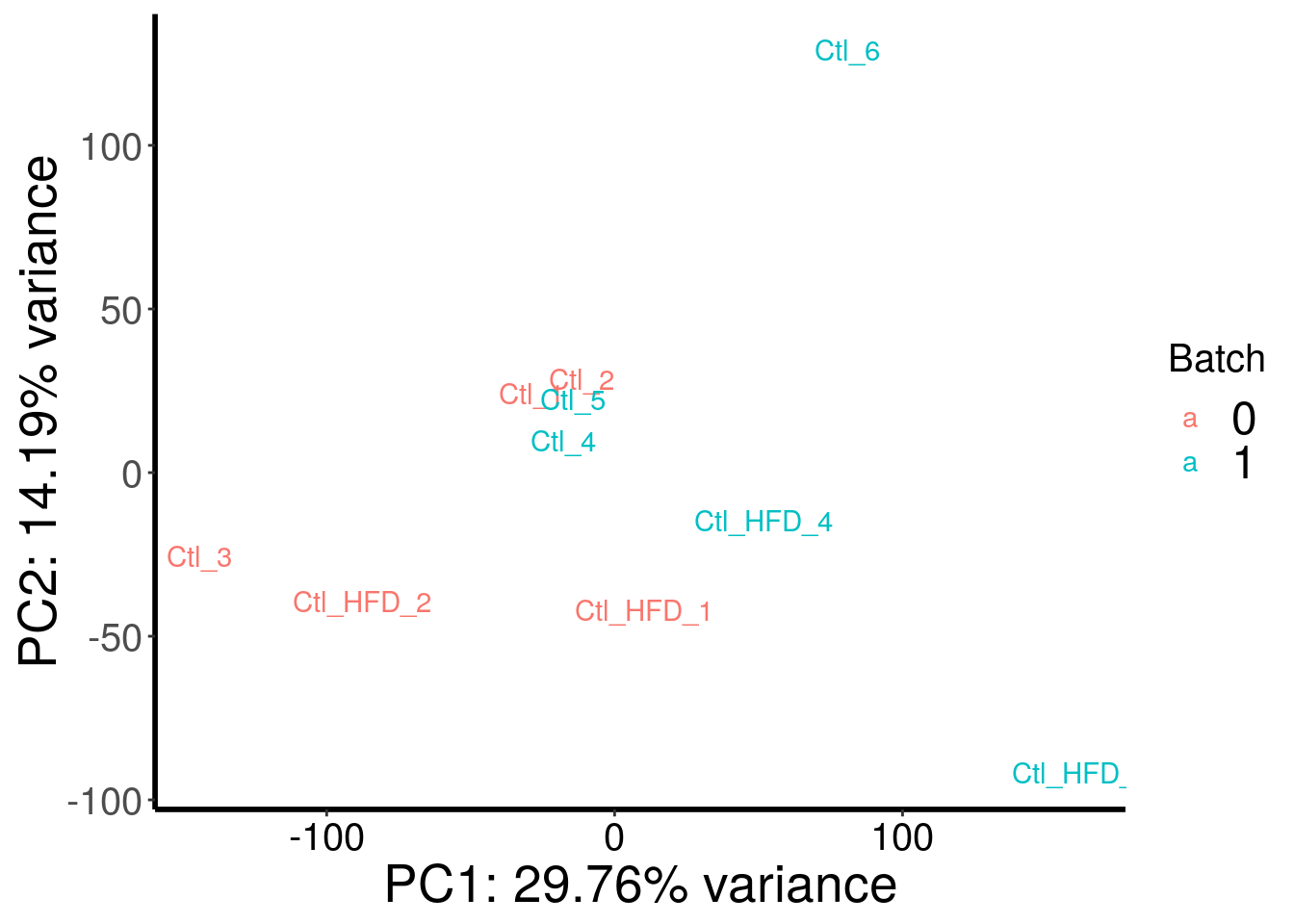
| Version | Author | Date |
|---|---|---|
| e1b9592 | scottzijiezhang | 2020-03-30 |
Samples are separated by genotype. Batch also contributed a bit to the variance. Therefore, we included batch as a covariate in this test.
DM tests with RADAR
HFD_RADAR <- diffIP_parallel(HFD_RADAR, thread = 20)
HFD_RADAR <- reportResult(HFD_RADAR, cutoff = 0.1, threads = 20)
save(HFD_RADAR, file = "~/Rohit_T1D/mouse_islet/HFD_RADAR.RData")
write.table(results(HFD_RADAR), file = "~/Rohit_T1D/mouse_islet/HFD_diffPeaks_FDR0.1.xls", sep = "\t", row.names = FALSE, col.names = FALSE, quote = FALSE)load("~/Rohit_T1D/mouse_islet/HFD_RADAR.RData")
HFD_result <- results(HFD_RADAR)There are 1257 reported differential loci at FDR < 0.1 and logFoldChange > 0.5.DT::datatable( HFD_result , rownames = FALSE )Distribution of log fold change of significant DM sites
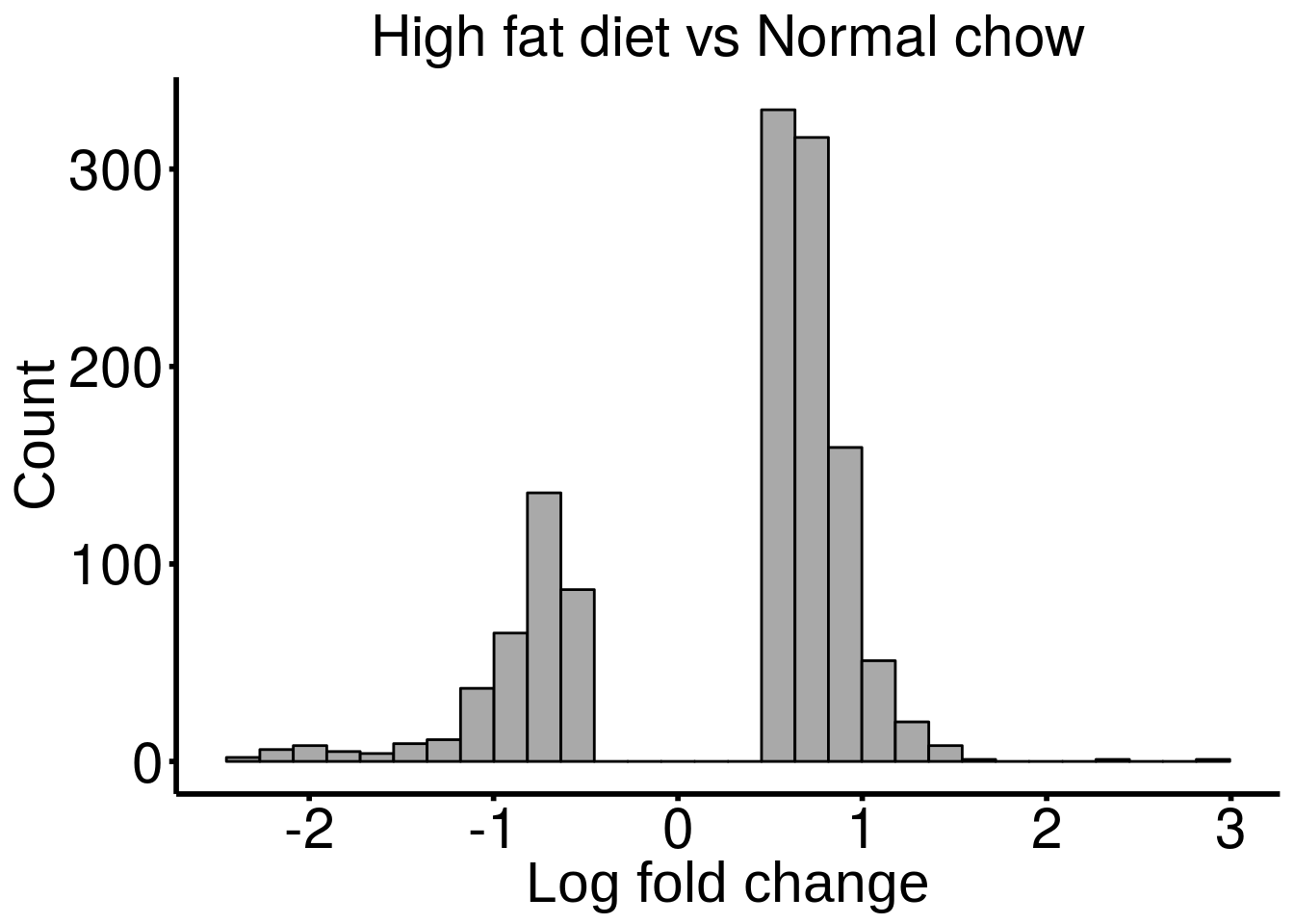
ggplot( HFD_result ,aes( x = logFC ) )+geom_histogram(color="black", fill="dark gray",bins = 30)+xlab("Log fold change")+ggtitle("High fat diet vs Normal chow")+theme_bw() + ylab("Count")+ theme(panel.border = element_blank(), panel.grid.major = element_blank(),
panel.grid.minor = element_blank(), plot.title = element_text( size = 22,colour = "black", hjust = 0.5 ) , axis.line = element_line(colour = "black",size = 1),axis.ticks = element_line(colour = "black",size = 1), axis.text = element_text(size = 22,colour = "black"),axis.text.y = element_text(angle = 0) ,axis.title=element_text(size=22,family = "arial")
)
| Version | Author | Date |
|---|---|---|
| 514bc99 | scottzijiezhang | 2020-05-08 |
Spatial distribution of these DM sites
MeRIPtools::plotMetaGeneMulti( list("Hyper-methylated" = HFD_result[HFD_result$logFC>0,1:12], "Hypo-methylated" = HFD_result[HFD_result$logFC<0,1:12]), gtf = "~/Database/genome/mm10/mm10_UCSC.gtf" )[1] "Converting BED12 to GRangesList"
[1] "It may take a few minutes"
[1] "Converting BED12 to GRangesList"
[1] "It may take a few minutes"
[1] "total 35119 transcripts extracted ..."
[1] "total 31408 transcripts left after ambiguity filter ..."
[1] "total 15627 mRNAs left after component length filter ..."
[1] "total 3280 ncRNAs left after ncRNA length filter ..."
[1] "Building Guitar Coordinates. It may take a few minutes ..."
[1] "Guitar Coordinates Built ..."
[1] "Using provided Guitar Coordinates"
[1] "resolving ambiguious features ..."
[1] "no figure saved ..."
NOTE this function is a wrapper for R package "Guitar".
If you use the metaGene plot in publication, please cite the original reference:
Cui et al 2016 BioMed Research International ####KEGG pathway analysis of DMG
eg.HFD <- bitr( unique(HFD_result$name), fromType="SYMBOL", toType="ENTREZID", OrgDb="org.Mm.eg.db")
KEGG_HFD <- enrichKEGG(eg.HFD$ENTREZID,organism = "mmu",pAdjustMethod = "fdr", pvalueCutoff = 0.1,minGSSize = 3)
dotplot(KEGG_HFD)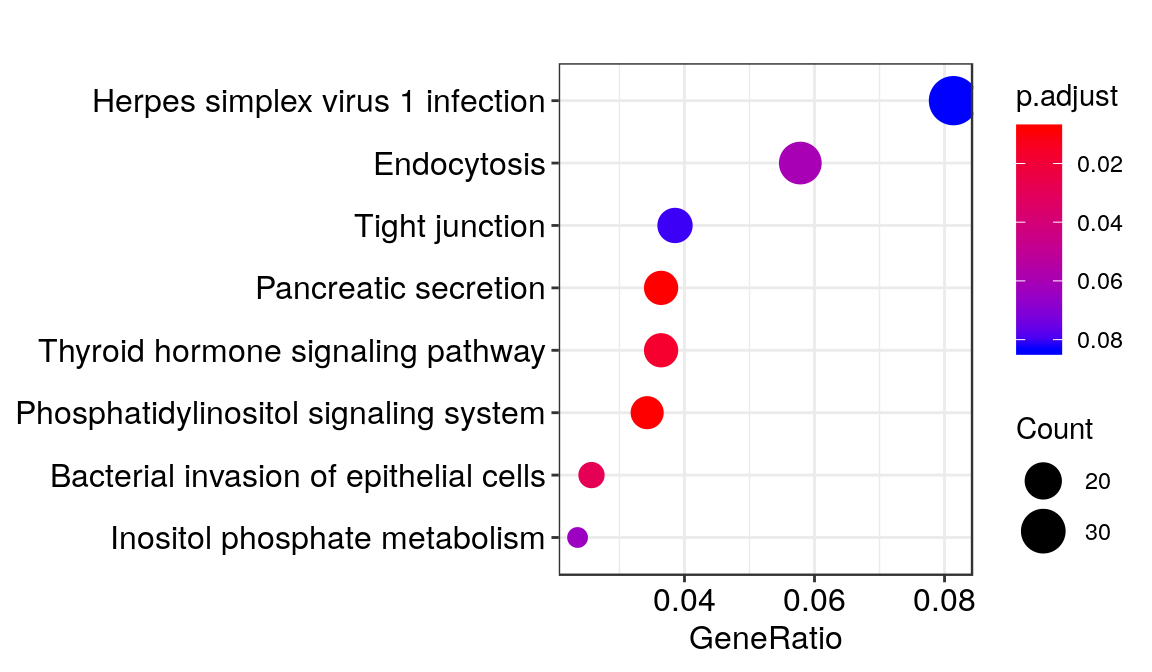
Genes included in some of these pathways
KEGG_HFD.df <- as.data.frame(KEGG_HFD)
KEGG_HFD.df$geneID <- unlist( lapply(strsplit(KEGG_HFD.df$geneID,"/"),function(x){ paste( bitr(x,fromType="ENTREZID", toType="SYMBOL",OrgDb="org.Mm.eg.db")$SYMBOL, collapse = "/") }) )
DT::datatable( KEGG_HFD.df[,c("Description", "geneID")] , rownames = FALSE )Compare High-fat-diet LIRKO vs. Normal chow WT
load( "~/Rohit_T1D/mouse_islet/allSample_RADAR.RData")
HFD_LIRKO_norWT_sample <- c( paste0("Ctl_", 1:6 ), paste0( "LIRKO_HFD_", 1:4 ) )
HFD_LIRKO_norWT_RADAR <- select( allSampleRADAR, HFD_LIRKO_norWT_sample )
HFD_LIRKO_norWT_RADAR <- normalizeLibrary( HFD_LIRKO_norWT_RADAR )
HFD_LIRKO_norWT_RADAR <- adjustExprLevel( HFD_LIRKO_norWT_RADAR )
variable( HFD_LIRKO_norWT_RADAR ) <- data.frame(genotype = c( rep( "Ctl", 6 ), rep( "HFD_LIRKO", 4 ) ),
batch = c( 0,0,0,1,1,1,0,0,1,1 )
)
HFD_LIRKO_norWT_RADAR <- filterBins( HFD_LIRKO_norWT_RADAR )
save(HFD_LIRKO_norWT_RADAR, file = "~/Rohit_T1D/mouse_islet/HFD_LIRKO_norWT_RADAR.RData")Check confounding factors
library(RADAR)
load( "~/Rohit_T1D/mouse_islet/HFD_LIRKO_norWT_RADAR.RData" )
plotPCAfromMatrix(HFD_LIRKO_norWT_RADAR@ip_adjExpr_filtered, variable(HFD_LIRKO_norWT_RADAR)$genotype )+scale_color_discrete(name = "Genotype")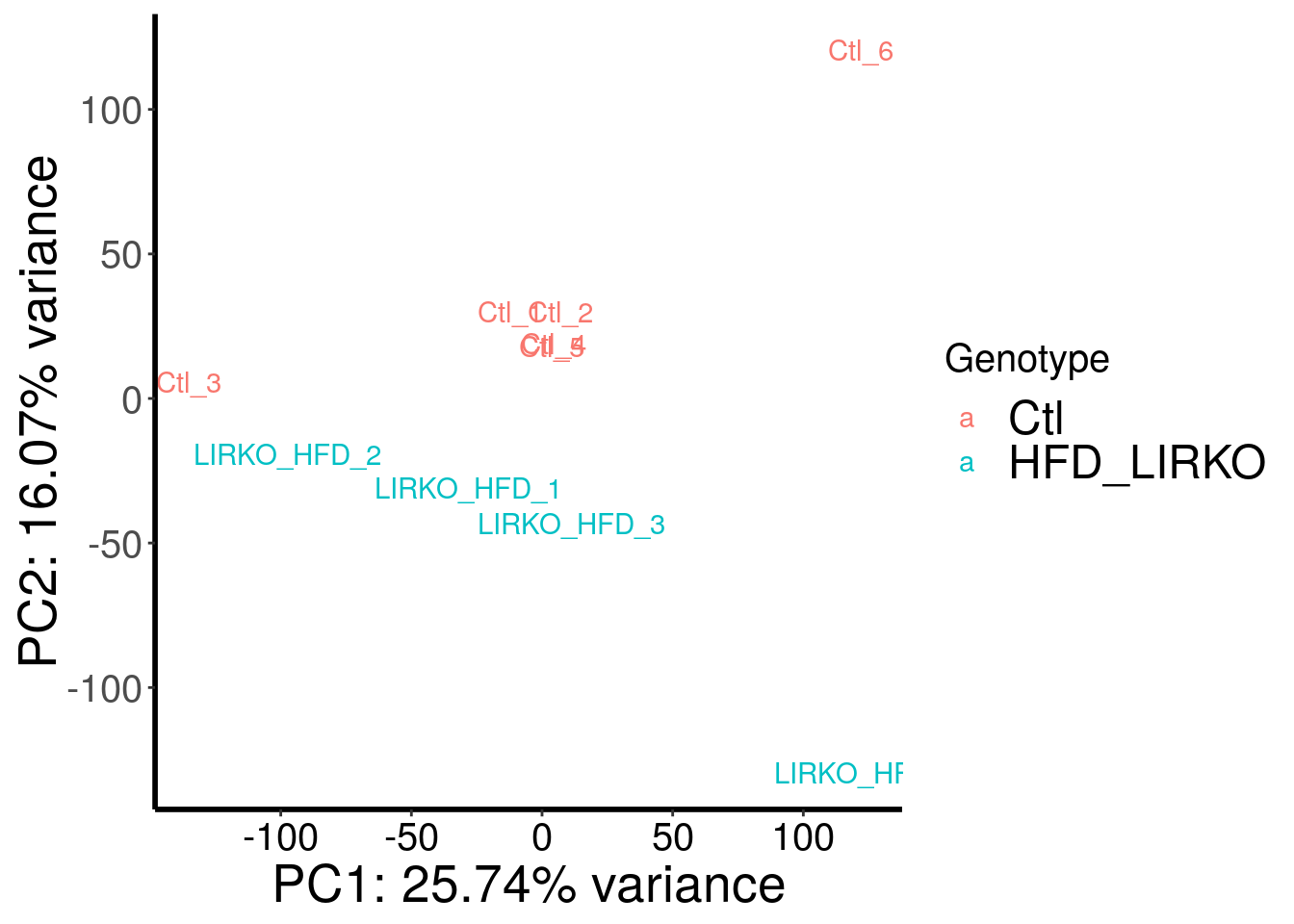
| Version | Author | Date |
|---|---|---|
| 241dfaa | scottzijiezhang | 2020-05-07 |
plotPCAfromMatrix(HFD_LIRKO_norWT_RADAR@ip_adjExpr_filtered, as.character( c( 0,0,0,1,1,1,0,0,1,1 ) ) )+scale_color_discrete(name = "Batch")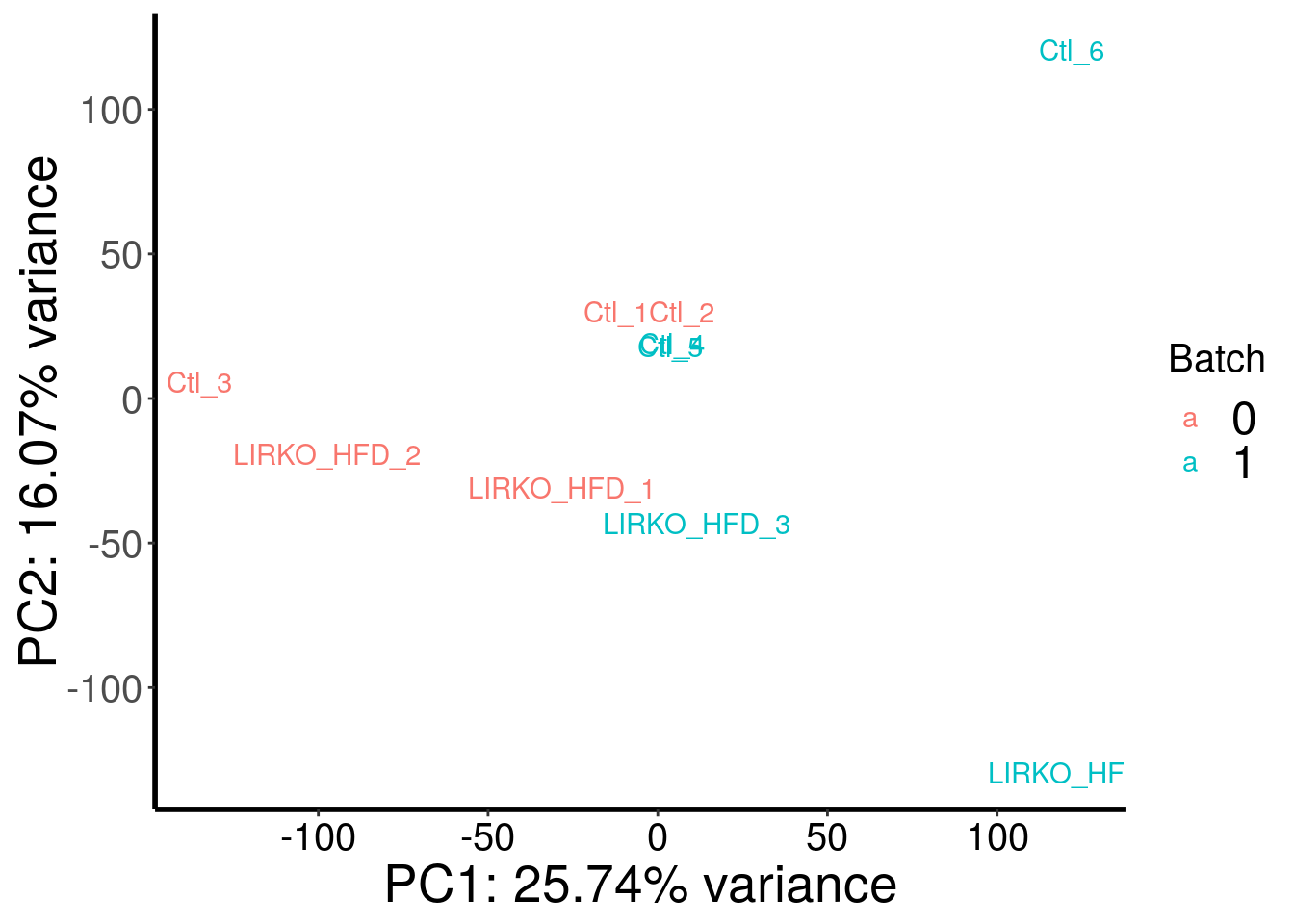
| Version | Author | Date |
|---|---|---|
| 241dfaa | scottzijiezhang | 2020-05-07 |
Samples are separated by genotype. Batch also contributed substantial portion to the variance. Therefore, we included batch as a covariate in this test.
DM tests with RADAR
HFD_LIRKO_norWT_RADAR <- diffIP_parallel(HFD_LIRKO_norWT_RADAR, thread = 20)
HFD_LIRKO_norWT_RADAR <- reportResult(HFD_LIRKO_norWT_RADAR, cutoff = 0.1, threads = 20)
save(HFD_LIRKO_norWT_RADAR, file = "~/Rohit_T1D/mouse_islet/HFD_LIRKO_norWT_RADAR.RData")
write.table(results(HFD_LIRKO_norWT_RADAR), file = "~/Rohit_T1D/mouse_islet/HFD_LIRKO_vs_normWT_diffPeaks_FDR0.1.xls", sep = "\t", row.names = FALSE, col.names = FALSE, quote = FALSE)load("~/Rohit_T1D/mouse_islet/HFD_LIRKO_norWT_RADAR.RData")
HFD_LIRKO_norWT_result <- results(HFD_LIRKO_norWT_RADAR)There are 1141 reported differential loci at FDR < 0.1 and logFoldChange > 0.5.DT::datatable( HFD_LIRKO_norWT_result , rownames = FALSE )Distribution of log fold change of significant DM sites
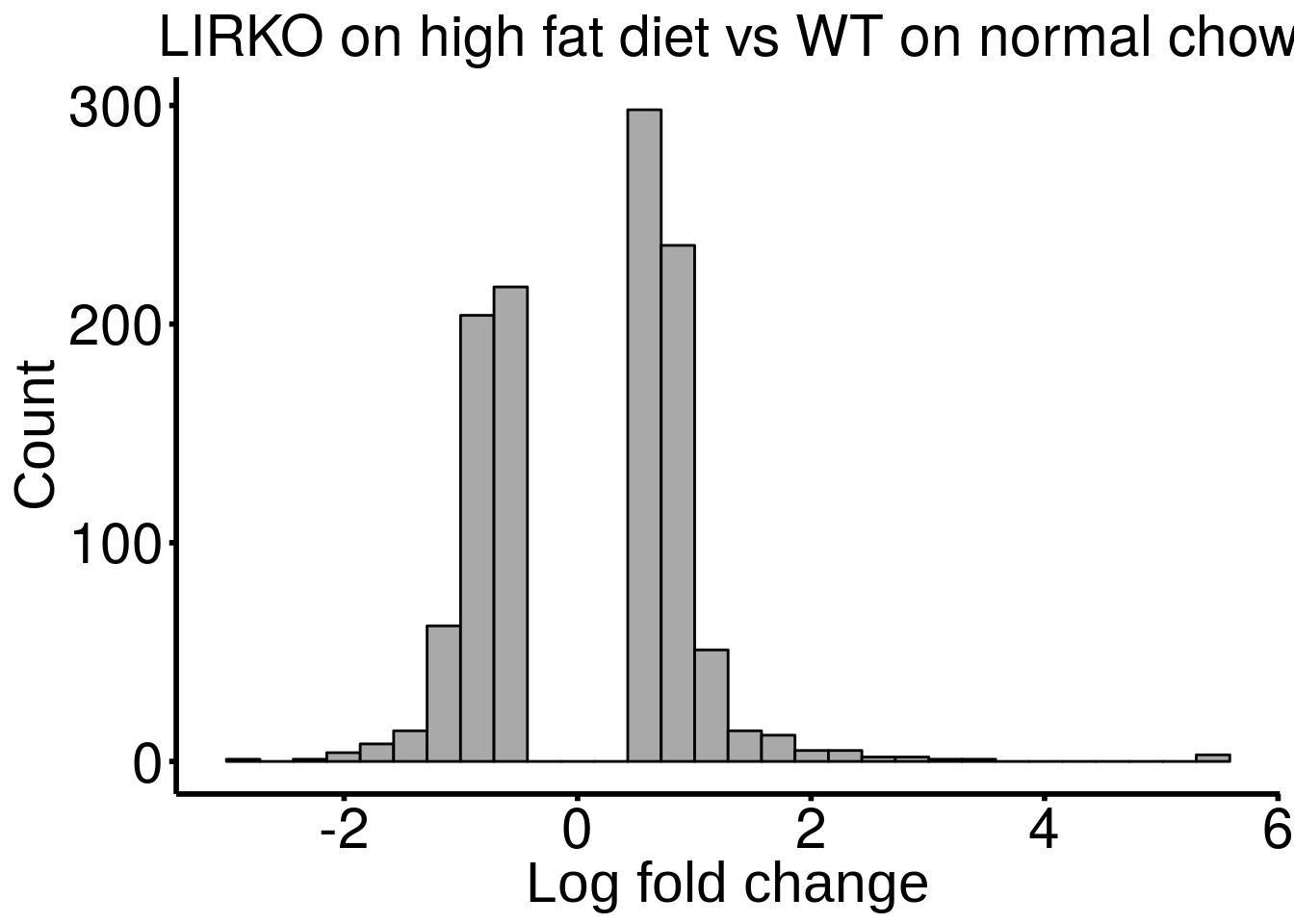
ggplot( HFD_LIRKO_norWT_result ,aes( x = logFC ) )+geom_histogram(color="black", fill="dark gray",bins = 30)+xlab("Log fold change")+ggtitle("LIRKO on high fat diet vs WT on normal chow")+theme_bw() + ylab("Count")+ theme(panel.border = element_blank(), panel.grid.major = element_blank(),
panel.grid.minor = element_blank(), plot.title = element_text( size = 22,colour = "black", hjust = 0.5 ) , axis.line = element_line(colour = "black",size = 1),axis.ticks = element_line(colour = "black",size = 1), axis.text = element_text(size = 22,colour = "black"),axis.text.y = element_text(angle = 0) ,axis.title=element_text(size=22,family = "arial")
)
| Version | Author | Date |
|---|---|---|
| 514bc99 | scottzijiezhang | 2020-05-08 |
Spatial distribution of these DM sites
MeRIPtools::plotMetaGeneMulti( list("Hyper-methylated" = HFD_LIRKO_norWT_result[HFD_LIRKO_norWT_result$logFC>0,1:12], "Hypo-methylated" = HFD_LIRKO_norWT_result[HFD_LIRKO_norWT_result$logFC<0,1:12]), gtf = "~/Database/genome/mm10/mm10_UCSC.gtf" )[1] "Converting BED12 to GRangesList"
[1] "It may take a few minutes"
[1] "Converting BED12 to GRangesList"
[1] "It may take a few minutes"
[1] "total 35119 transcripts extracted ..."
[1] "total 31408 transcripts left after ambiguity filter ..."
[1] "total 15627 mRNAs left after component length filter ..."
[1] "total 3280 ncRNAs left after ncRNA length filter ..."
[1] "Building Guitar Coordinates. It may take a few minutes ..."
[1] "Guitar Coordinates Built ..."
[1] "Using provided Guitar Coordinates"
[1] "resolving ambiguious features ..."
[1] "no figure saved ..."
NOTE this function is a wrapper for R package "Guitar".
If you use the metaGene plot in publication, please cite the original reference:
Cui et al 2016 BioMed Research International KEGG pathway analysis of DMG
eg.HFD_LIRKO_norWT <- bitr( unique(results(HFD_LIRKO_norWT_RADAR)$name), fromType="SYMBOL", toType="ENTREZID", OrgDb="org.Mm.eg.db")There are 1141 reported differential loci at FDR < 0.1 and logFoldChange > 0.5.KEGG_HFD_LIRKO_norWT <- enrichKEGG(eg.HFD_LIRKO_norWT$ENTREZID,organism = "mmu",pAdjustMethod = "fdr", pvalueCutoff = 0.1,minGSSize = 3)
dotplot(KEGG_HFD_LIRKO_norWT)+theme(
panel.grid.minor = element_blank(), axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=22, color = "black", hjust=0.5,family = "arial"),
axis.title.y=element_text(size=22, color = "black", vjust=0.4, angle=90,family = "arial"),
legend.title=element_text(size=15, color = "black"),legend.text = element_text(size = 20, color = "black",family = "arial"),
axis.text.x = element_text(size = 15,color = "black",family = "arial") ,axis.text.y = element_text(size = 15,color = "black",family = "arial"))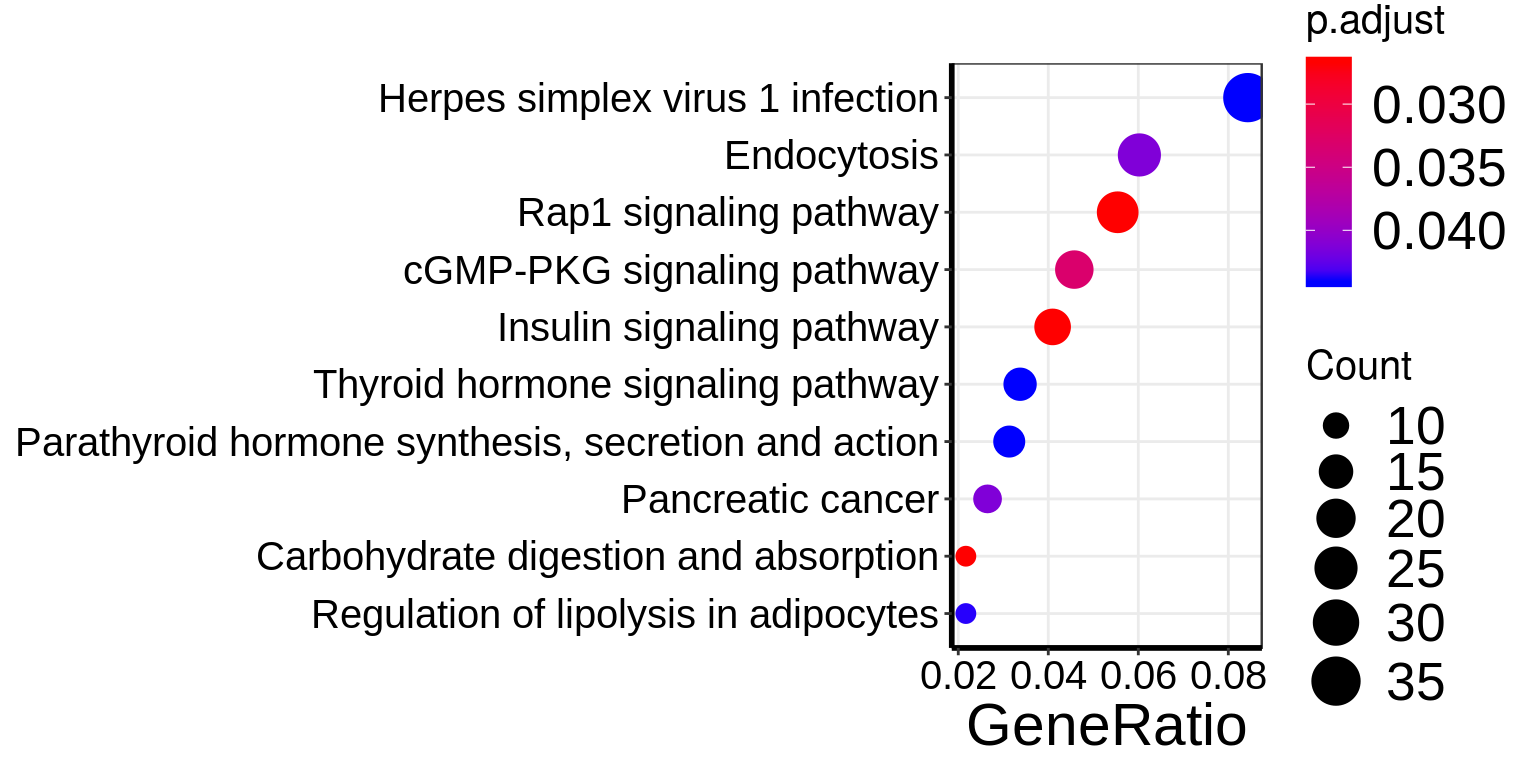
emapplot(KEGG_HFD_LIRKO_norWT, vertex.label.font = 3)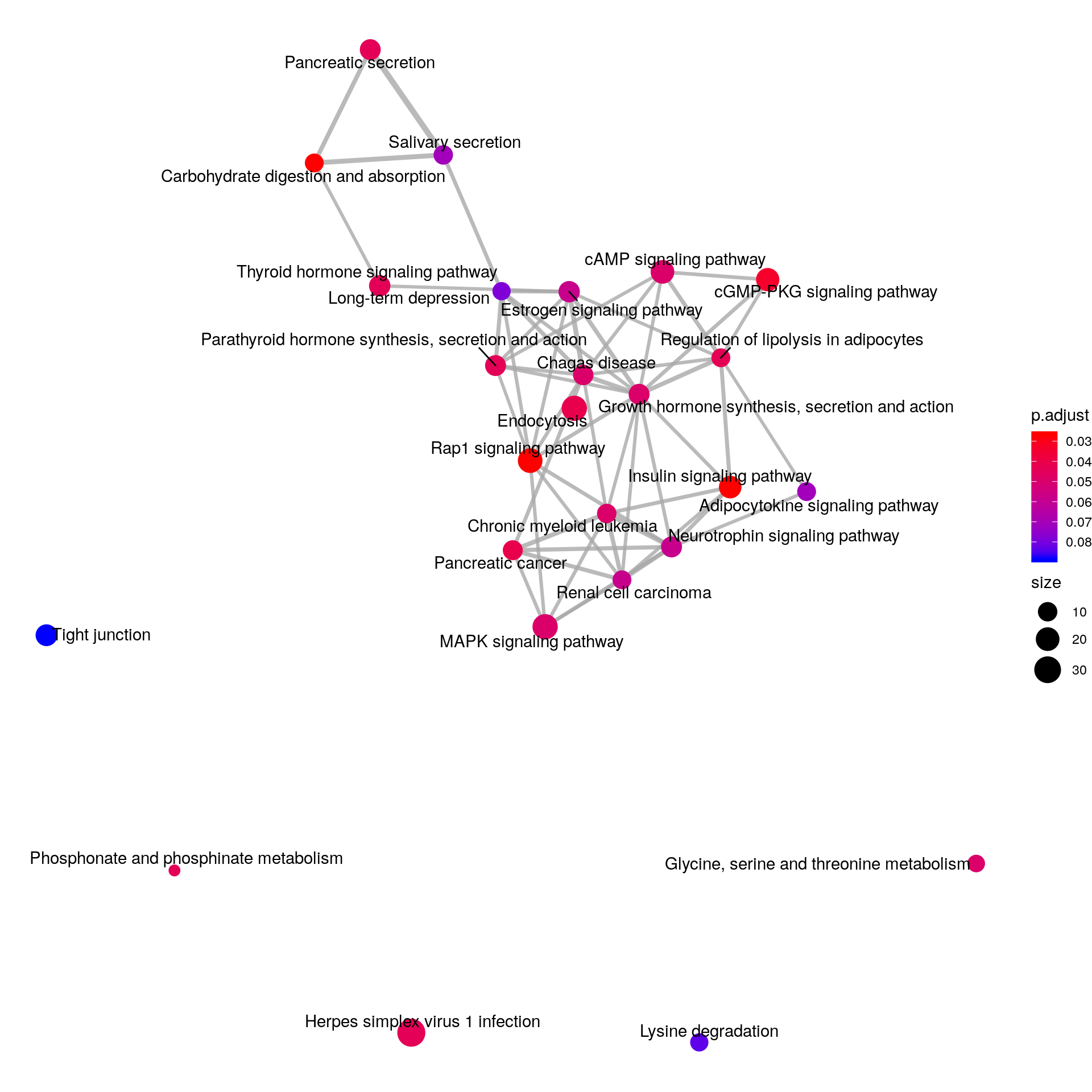
Genes included in some of these pathways
KEGG_HFD_LIRKO_norWT.df <- as.data.frame(KEGG_HFD_LIRKO_norWT)
KEGG_HFD_LIRKO_norWT.df$geneID <- unlist( lapply(strsplit(KEGG_HFD_LIRKO_norWT.df$geneID,"/"),function(x){ paste( bitr(x,fromType="ENTREZID", toType="SYMBOL",OrgDb="org.Mm.eg.db")$SYMBOL, collapse = "/") }) )
DT::datatable( KEGG_HFD_LIRKO_norWT.df[c(1:8,10,12:15,17:19),c("Description", "geneID")] , rownames = FALSE)Intersect DMGs in LIRKO_HFD and DMGs in T2D patients.
convertMouseGeneList <- function(x){
require("biomaRt")
human = useMart("ensembl", dataset = "hsapiens_gene_ensembl")
mouse = useMart("ensembl", dataset = "mmusculus_gene_ensembl")
genesV2 = getLDS(attributes = c("mgi_symbol"), filters = "mgi_symbol", values = x , mart = mouse, attributesL = c("hgnc_symbol"), martL = human, uniqueRows = T )
return(genesV2)
}
MtoHmap_LIRKO_HFD <- convertMouseGeneList( as.character( unique( HFD_LIRKO_norWT_result$name ) ) )
HFD_LIRKO_norWT_result$HumanOrtholog <- MtoHmap_LIRKO_HFD$HGNC.symbol[ match(HFD_LIRKO_norWT_result$name, MtoHmap_LIRKO_HFD$MGI.symbol ) ]
Vcommon_LIRKO_HFD_T2D <- Venn(list("DMGs in LIRKO_HFD" = unique(HFD_LIRKO_norWT_result$HumanOrtholog),"DMGs in T2D patient" = unique(T2D_DMpeaks$name) ) )
plot(Vcommon_LIRKO_HFD_T2D,doWeights=TRUE)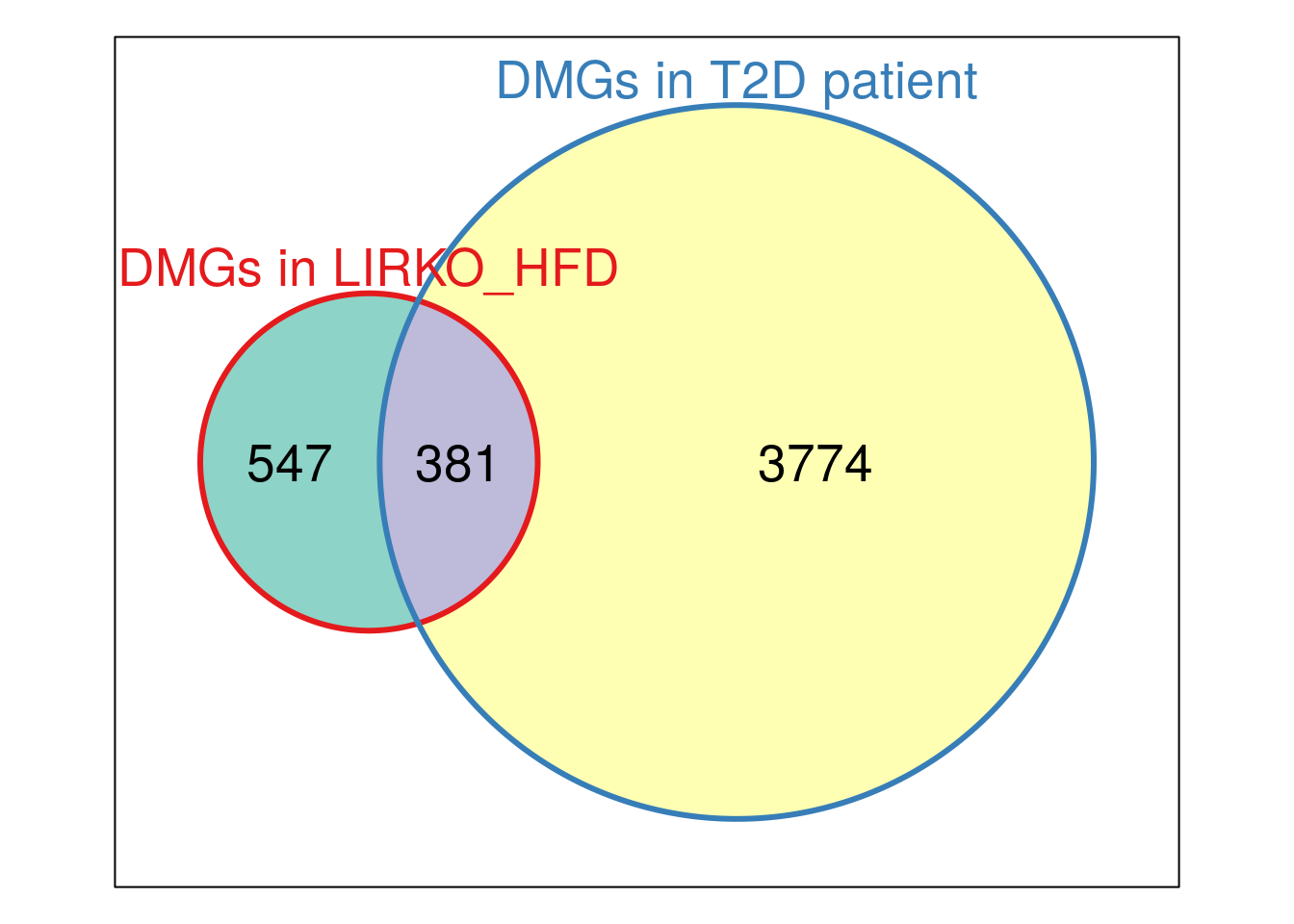
| Version | Author | Date |
|---|---|---|
| aa2d181 | scottzijiezhang | 2020-06-05 |
cat( paste0("Intersect DMGs in LIRKO w/ HFD and T2D patient:\n", paste(intersect( unique(HFD_LIRKO_norWT_result$HumanOrtholog), unique(T2D_DMpeaks$name) ), collapse = "\t" ) ) )Intersect DMGs in LIRKO w/ HFD and T2D patient:
C10orf88 C7orf50 C6orf141 KIAA2026 ACHE ACOT1 ACTN4 ADO ADRA2A AHNAK AKAP13 AKAP7 AKAP8 AKT2 ANKH ANKRD40 ANKRD42 AP3D1 ARFGEF1 ARHGEF18 ARHGEF3 ARID1B ARRDC4 ASAP1 ASH1L ASPH ASXL2 ATP2A2 ATP7A AUTS2 AZGP1 BAIAP2L1 BCR BHLHB9 BICC1 BTBD3 C1GALT1 C2CD2L CACNA1C CACUL1 CAMLG CAPN2 CAST CBX2 CCDC71L CCDC93 CD99L2 CDC42BPG CDC42EP3 CDH1 CDIPT CEBPD CHAC1 CHD5 CHD7 CHKA CHMP2B CHST12 CHST15 CHSY1 CITED4 CNNM2 COL13A1 COL15A1 CPM CPSF7 CREBRF CRIM1 CRKL CSF1 CX3CL1 CYS1 DCDC2 DDX6 DGCR2 DHCR7 DICER1 DISP2 DLC1 DNAAF2 DNAJB1 DNAJB12 DNAJC5 DNMBP DST DTX2 DYNLL2 EEA1 EEF1D EGFL7 EHBP1 EIF4E2 EP400 EPS8L2 ERC1 ESF1 EXO5 EXOSC2 FAF2 FAM83G FARP2 FBXO38 FBXO9 FEM1A FIZ1 FKBP5 FLII FNBP4 FOS FOXJ2 FOXO1 FOXQ1 FRY FYN GAS8 GFER GINM1 GJB1 GNAO1 GOLM1 GSPT1 GSTO1 H6PD HCFC1 HERC2 HIRIP3 HIVEP2 HNF1B HPS6 HSPA1A IGFBP3 IGFBP5 IKBKB INAFM2 IQGAP2 IRF2BPL IRS2 ITPRIP ITSN1 JMJD6 KCTD15 KDM2A KDM3B KDM4B KIF16B KIF1B KL KLF5 KLHL15 KMT2D LARP1 LENG8 LIMCH1 LNX2 LRP6 LRRC8E LRRFIP2 LZTS2 MAGI1 MAP1A MAP3K10 MAP7D1 MAPT MAST3 MCF2L MDC1 METTL9 MFHAS1 MFSD8 MGA MICAL3 MICALL1 MIER1 MINK1 MLXIPL MOCS3 MON1B MPHOSPH8 MRPL40 MSL2 MUC1 MYH10 MYH14 NAP1L1 NAV1 NAV2 NBEA NCOA3 NCOR2 NDUFB10 NEFH NEU3 NF2 NFAT5 NFYA NFYC NKD2 NOC3L NOL4 NOLC1 NPNT NPTXR NR2F2 NUDT3 NUFIP2 NUP50 NXF1 OPTN OXNAD1 OXSR1 P3H4 PABPC4 PALM PANK3 PAPOLA PARD3 PCDHB2 PCDHB15 PCDHGB2 PCSK7 PDE4DIP PHF14 PHF8 PHLDB1 PHRF1 PIGR PIK3R2 PITPNM1 PKD1 PLCB4 PLEC PLEKHA6 PLEKHG3 PLS1 PLXNA1 PLXNA2 PML PPFIBP1 PPP3CA PREPL PRR14L PRRC2C PRUNE2 PSD3 PSIP1 PTPRA QSOX1 RAB40C RAB9B RABEP1 RAI1 RALBP1 RALGAPB RBM14 RBM26 RELA RERE REV3L REXO1 RFX6 RIMBP2 RNF139 RNF169 RNF26 RNF7 RORC RRBP1 RRP1 RRP12 RSRC2 RWDD3 SAMD4B SAP30 SCARA3 SERF2 SETD1B SETD5 SF3B1 SH3GLB1 SH3PXD2A SIPA1L1 SLC19A2 SLC30A6 SLC35A2 SLC39A3 SLC3A1 SLC4A1AP SLC6A6 SLC9A8 SLK SLTM SMAD7 SMARCA2 SMC1A SMC3 SMCR8 SMG7 SMIM19 SNAPC2 SNRK SOX13 SPECC1 SPPL3 SPRED1 SPTB SPTBN1 SRF SRRM1 SWSAP1 SYDE2 TACC1 TAGAP TAOK3 TEF TGFA TGS1 THBD THG1L TICAM1 TIMMDC1 TIPARP TJAP1 TJP1 TLE1 TLE3 TMED7-TICAM2 TMEM97 TNRC6A TRAK1 TRAPPC5 TRIL TRIOBP TRMT10B TRPM3 TSEN54 TSHZ1 TSPAN13 TSPAN3 TTBK2 TTC28 TTC3 U2SURP UBTF UIMC1 UNC13A UNC50 UNC80 UNC93B1 URI1 USP36 UTRN VAPA VDR VPS18 VPS26B WDR6 WNK1 WWP2 ZADH2 ZC3H14 ZC3H15 ZC3HAV1 ZDHHC14 ZDHHC3 ZNF217 ZFP30 ZNF316 ZFP36L2 ZNF570 ZNF446 ZNF518A ZNF574 ZNF667 ZNF704 ZNF845 ZNF771 ZNF777 ZNF839 ZKSCAN1 ZSCAN21 ZSWIM6KEGG pathway analysis of overlapping DMGs
eg.HFD_LIRKO_norWT_T2D <- bitr( intersect( unique(HFD_LIRKO_norWT_result$HumanOrtholog), unique(T2D_DMpeaks$name) ) , fromType="SYMBOL", toType="ENTREZID", OrgDb="org.Hs.eg.db")
KEGG_HFD_LIRKO_norWT_T2D <- enrichKEGG(eg.HFD_LIRKO_norWT_T2D$ENTREZID,organism = "hsa",pAdjustMethod = "fdr", pvalueCutoff = 0.1,minGSSize = 3)
dotplot(KEGG_HFD_LIRKO_norWT_T2D, showCategory=20 )+theme(
panel.grid.minor = element_blank(), axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=22, color = "black", hjust=0.5,family = "arial"),
axis.title.y=element_text(size=22, color = "black", vjust=0.4, angle=90,family = "arial"),
legend.title=element_text(size=15, color = "black"),legend.text = element_text(size = 20, color = "black",family = "arial"),
axis.text.x = element_text(size = 15,color = "black",family = "arial") ,axis.text.y = element_text(size = 15,color = "black",family = "arial"))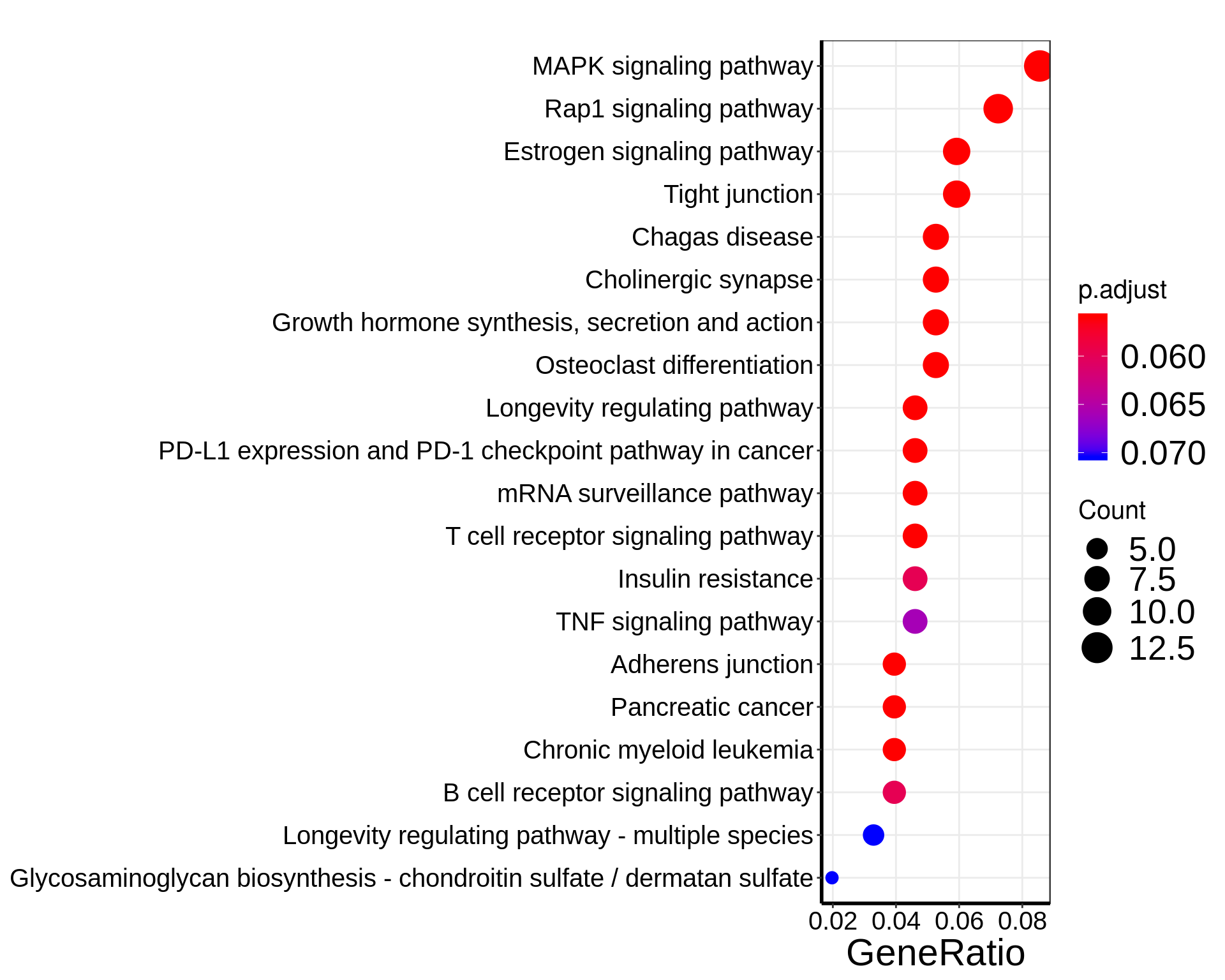
| Version | Author | Date |
|---|---|---|
| aa2d181 | scottzijiezhang | 2020-06-05 |
emapplot(KEGG_HFD_LIRKO_norWT_T2D, vertex.label.font = 3)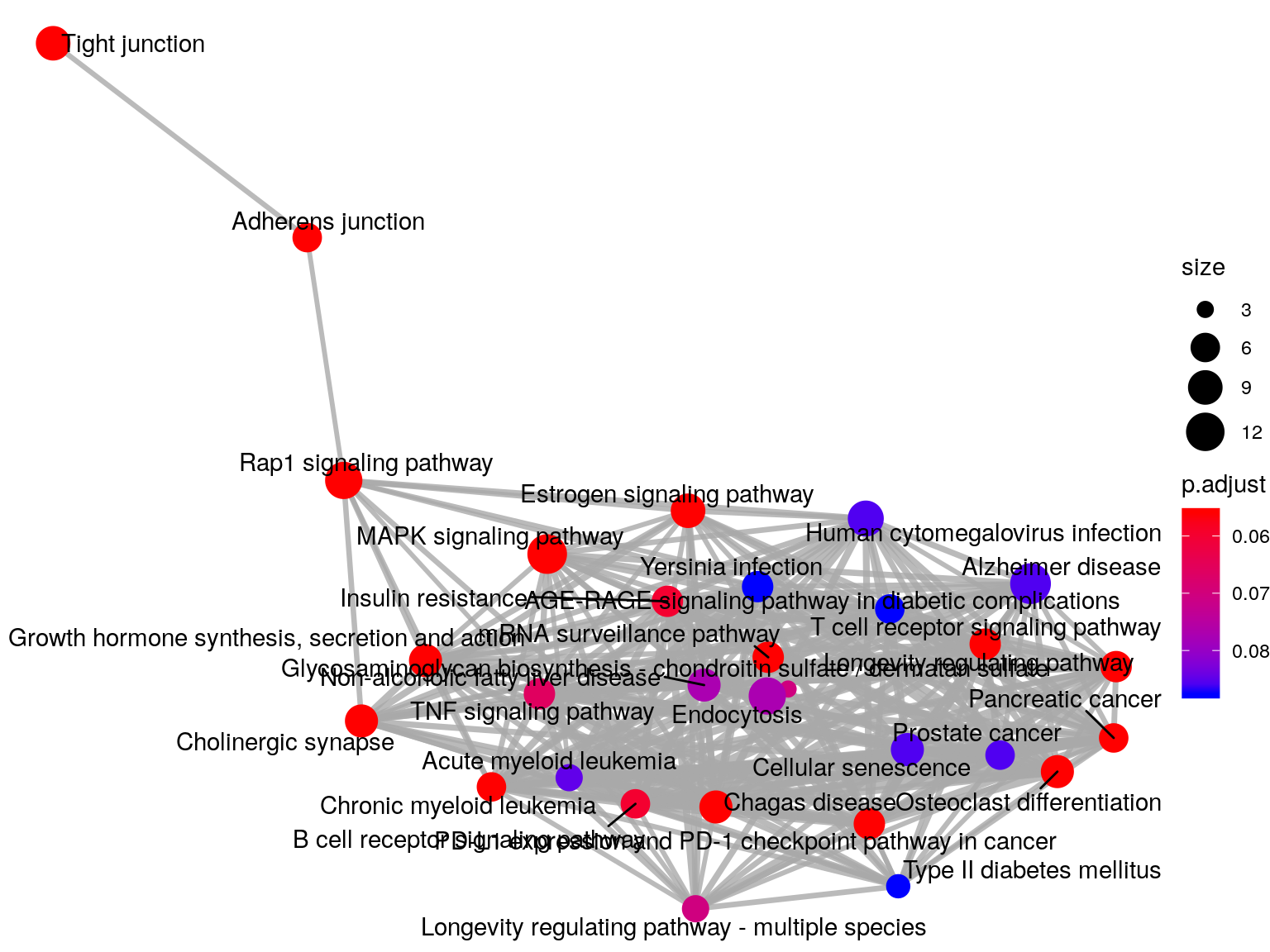
| Version | Author | Date |
|---|---|---|
| aa2d181 | scottzijiezhang | 2020-06-05 |
Intersect DMGs in LIRKO w/ HFD, DMGs in LIRKO and DMGs in T2D patients.
In above analysis, we noticed that overlap between LIRKO & T2D are not enriched for “T2D pathway” while overlap between LIRKO w/ HFD & T2D are enriched for “T2D” and “Insulin resistance” pathway.
Here I compare all three set of genes.
Vcommon_LIRKOhfd_LIRKO_T2D <- Venn(list("DMGs in LIRKO w/ HFD" = unique(HFD_LIRKO_norWT_result$HumanOrtholog),"DMGs in LIRKO" = unique(LIRKO_result$HumanOrtholog),"DMGs in T2D patient" = unique(T2D_DMpeaks$name) ) )
plot(Vcommon_LIRKOhfd_LIRKO_T2D,doWeights=TRUE)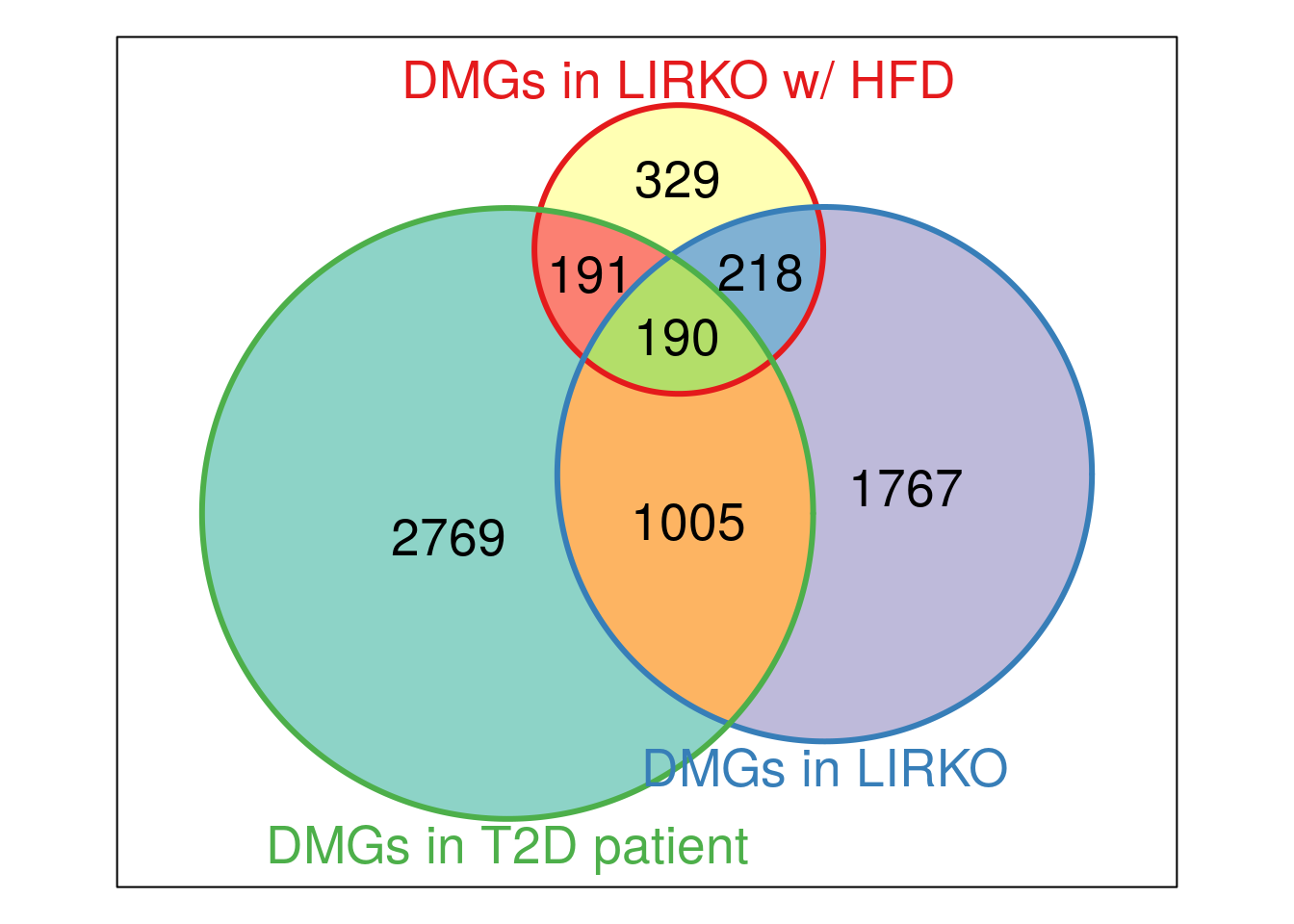
| Version | Author | Date |
|---|---|---|
| aa2d181 | scottzijiezhang | 2020-06-05 |
Compare enriched pathway (KEGG)
DMGall <- list("DMGs in LIRKO w/ HFD overlap T2D" = eg.HFD_LIRKO_norWT_T2D$ENTREZID ,"DMGs in LIRKO overlap T2D" = eg.LIRKO_T2D$ENTREZID )
ck <- compareCluster(geneCluster = DMGall, fun = "enrichKEGG", organism = "hsa", pAdjustMethod = "fdr", pvalueCutoff = 0.1, minGSSize = 3)
dotplot(ck,showCategory=30)+theme(
panel.grid.minor = element_blank(), axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=22, hjust=0.5,family = "arial"),
axis.title.y=element_text(size=22, vjust=0.4, angle=90, ),
legend.title=element_text(size=15 ),legend.text = element_text(size = 20),
axis.text.x = element_text(size = 15,angle = -20, vjust = 0.3 ) ,axis.text.y = element_text(size = 15 ) )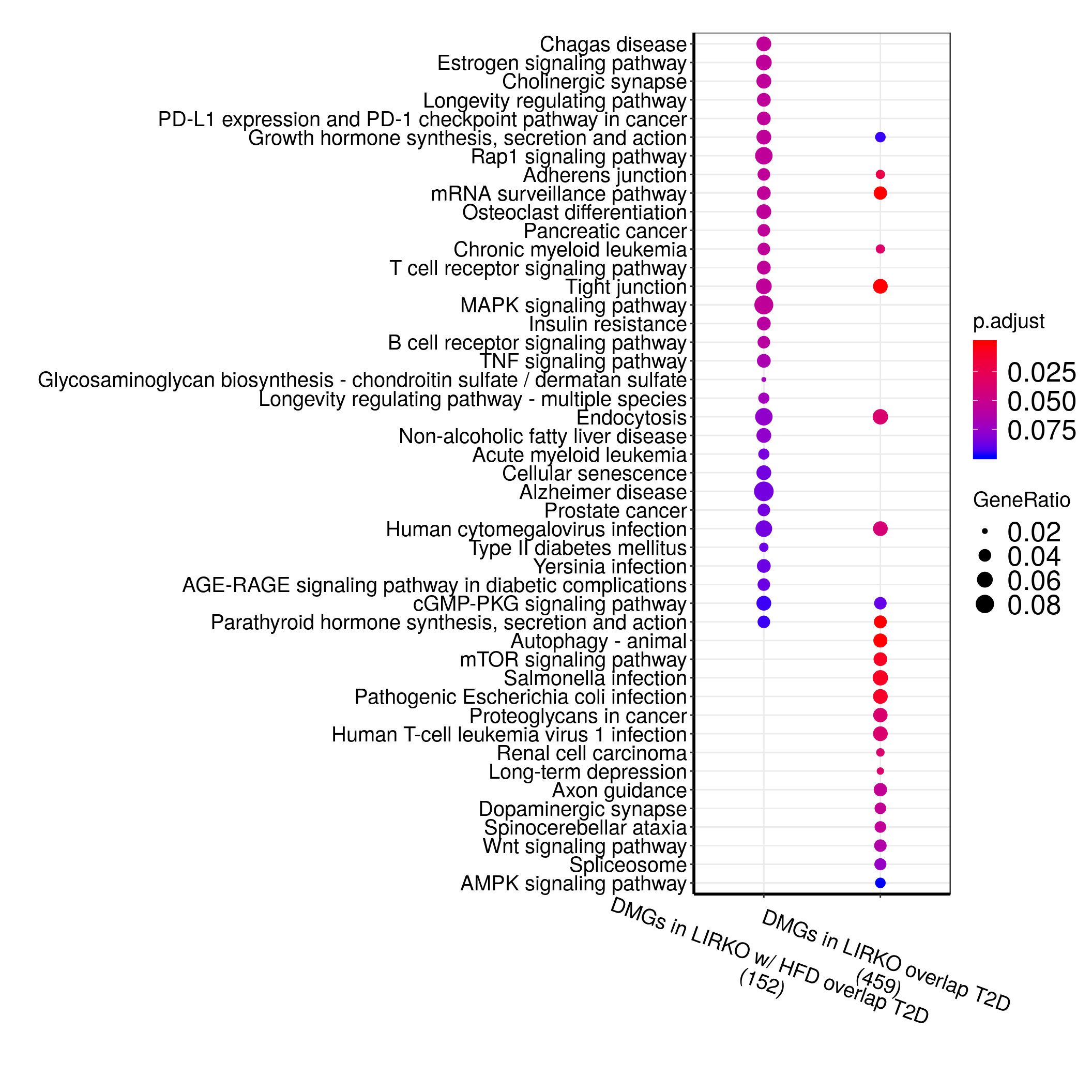
| Version | Author | Date |
|---|---|---|
| aa2d181 | scottzijiezhang | 2020-06-05 |
ckKEGG.df <- as.data.frame(ck)
ckKEGG.df$geneID <- unlist( lapply(strsplit(ckKEGG.df$geneID,"/"),function(x){ paste( bitr(x,fromType="ENTREZID", toType="SYMBOL",OrgDb="org.Hs.eg.db")$SYMBOL, collapse = "/") }) )
ckGOtoShow <- ckKEGG.df[ which( ckKEGG.df$ID %in% c("hsa04935", "hsa04931", "hsa04660", "hsa04010", "hsa04662", "hsa04930", "hsa04935") ),c("Description","Cluster", "geneID")]
DT::datatable(ckGOtoShow, rownames = FALSE)Compare High-fat-diet LIRKO vs. High-fat-diet WT
load( "~/Rohit_T1D/mouse_islet/allSample_RADAR.RData")
HFD_LIRKO_sample <- c( paste0("Ctl_HFD_", 1:4 ), paste0( "LIRKO_HFD_", 1:4 ) )
HFD_LIRKO_RADAR <- select( allSampleRADAR, HFD_LIRKO_sample )
HFD_LIRKO_RADAR <- normalizeLibrary( HFD_LIRKO_RADAR )
HFD_LIRKO_RADAR <- adjustExprLevel( HFD_LIRKO_RADAR )
variable( HFD_LIRKO_RADAR ) <- data.frame(genotype = c( rep( "Ctl_HFD_", 4 ), rep( "HFD_LIRKO", 4 ) ),
batch = c( 0,0,1,1,0,0,1,1 )
)
HFD_LIRKO_RADAR <- filterBins( HFD_LIRKO_RADAR )
save(HFD_LIRKO_RADAR, file = "~/Rohit_T1D/mouse_islet/HFD_LIRKO_RADAR.RData")Check confounding factors
library(RADAR)
load( "~/Rohit_T1D/mouse_islet/HFD_LIRKO_RADAR.RData" )
plotPCAfromMatrix(HFD_LIRKO_RADAR@ip_adjExpr_filtered, variable(HFD_LIRKO_RADAR)$genotype )+scale_color_discrete(name = "Genotype")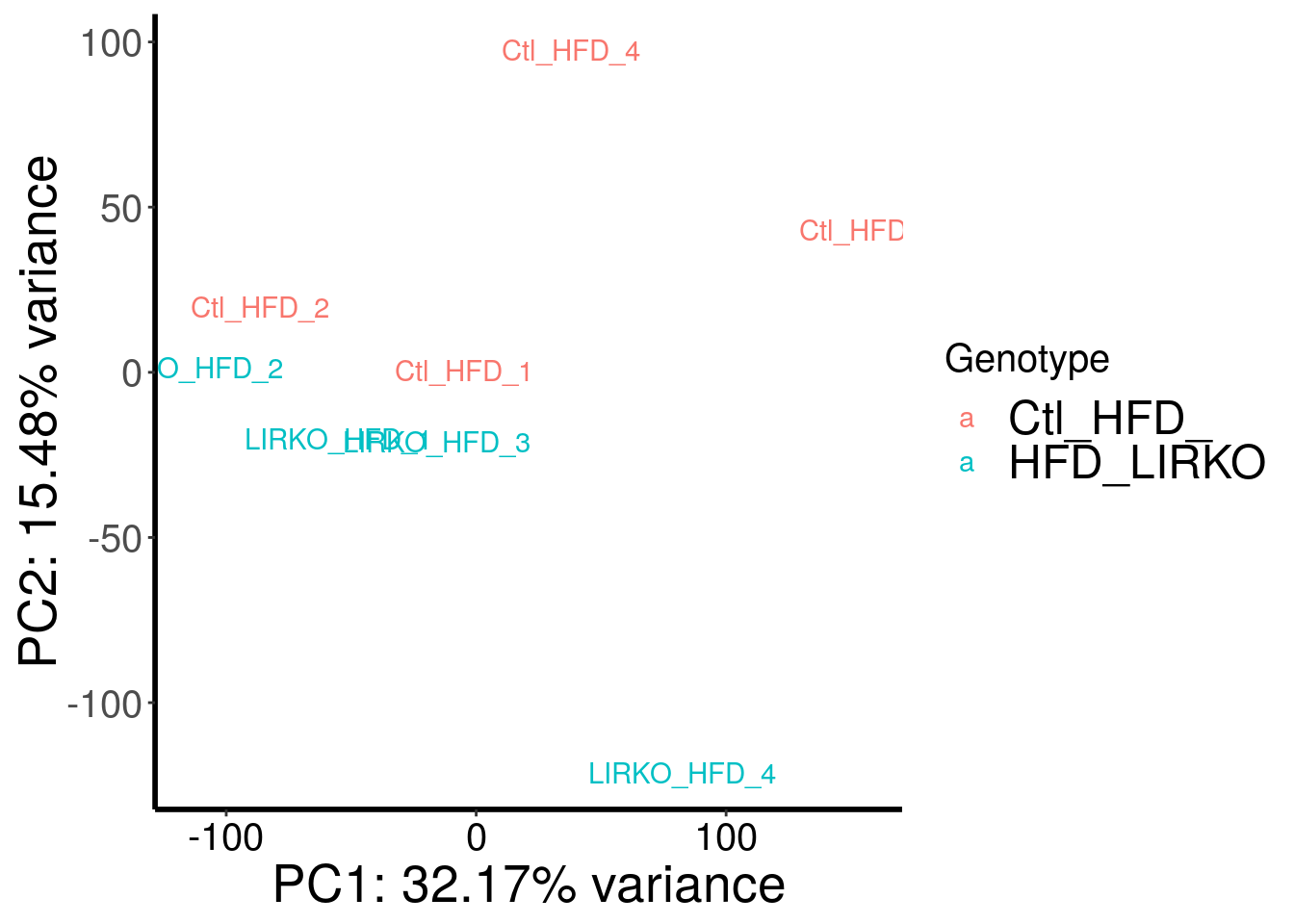
| Version | Author | Date |
|---|---|---|
| e1b9592 | scottzijiezhang | 2020-03-30 |
plotPCAfromMatrix(HFD_LIRKO_RADAR@ip_adjExpr_filtered, as.character( c( 0,0,1,1,0,0,1,1 ) ) )+scale_color_discrete(name = "Batch")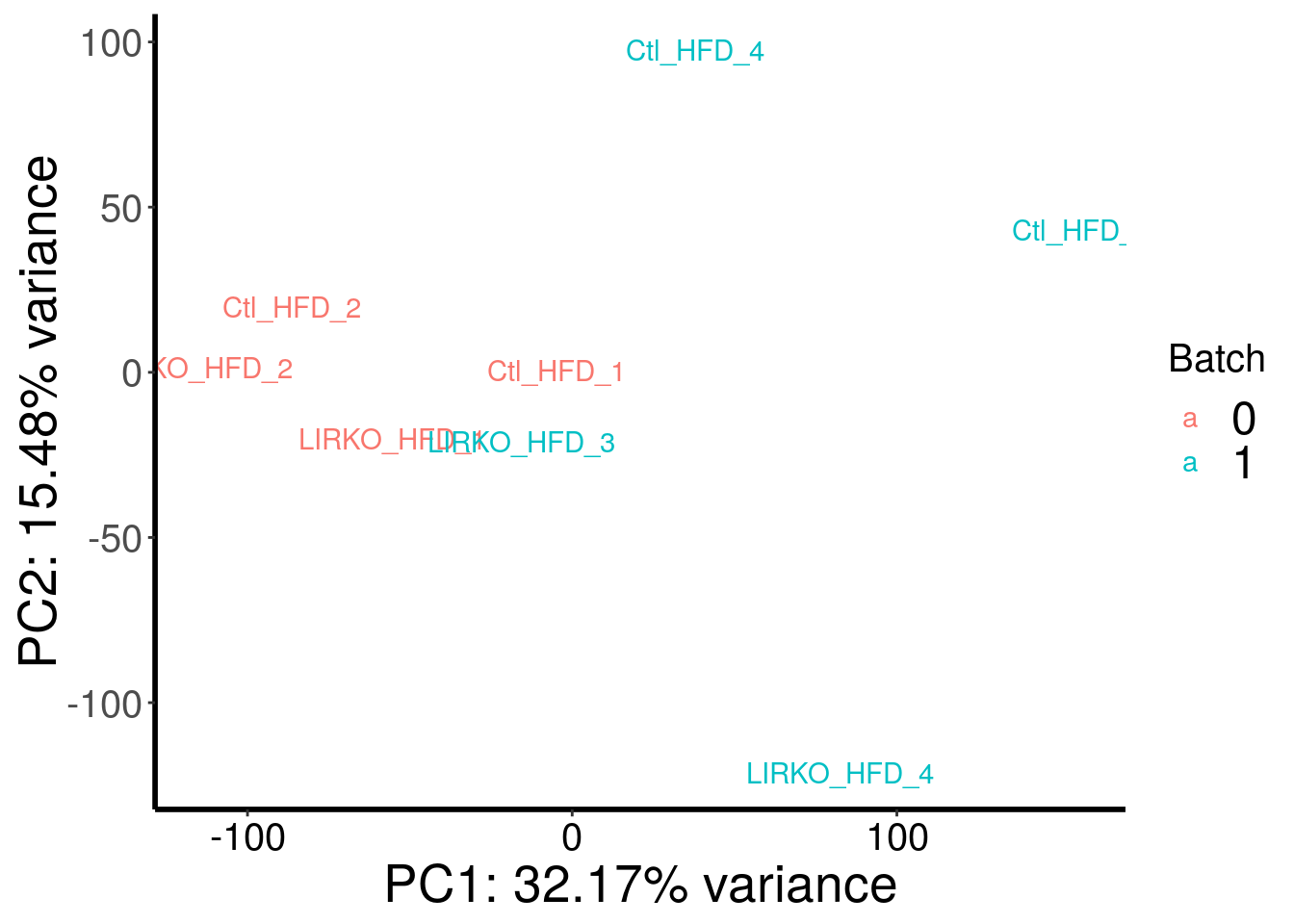
| Version | Author | Date |
|---|---|---|
| e1b9592 | scottzijiezhang | 2020-03-30 |
Samples are separated by genotype. Batch seems also contributed a bit to the variance. Therefore, we included batch as a covariate in this test.
DM tests with RADAR
HFD_LIRKO_RADAR <- diffIP_parallel(HFD_LIRKO_RADAR, thread = 20)
HFD_LIRKO_RADAR <- reportResult(HFD_LIRKO_RADAR, cutoff = 0.1, threads = 20)
save(HFD_LIRKO_RADAR, file = "~/Rohit_T1D/mouse_islet/HFD_LIRKO_RADAR.RData")
write.table(results(HFD_LIRKO_RADAR), file = "~/Rohit_T1D/mouse_islet/HFD_LIRKO_diffPeaks_FDR0.1.xls", sep = "\t", row.names = FALSE, col.names = FALSE, quote = FALSE)load("~/Rohit_T1D/mouse_islet/HFD_LIRKO_RADAR.RData")
DT::datatable( results(HFD_LIRKO_RADAR) , rownames = FALSE )There are 882 reported differential loci at FDR < 0.1 and logFoldChange > 0.5.KEGG pathway analysis of DMG
eg.HFD_LIRKO <- bitr( unique(results(HFD_LIRKO_RADAR)$name), fromType="SYMBOL", toType="ENTREZID", OrgDb="org.Mm.eg.db")There are 882 reported differential loci at FDR < 0.1 and logFoldChange > 0.5.KEGG_HFD_LIRKO <- enrichKEGG(eg.HFD_LIRKO$ENTREZID,organism = "mmu",pAdjustMethod = "fdr", pvalueCutoff = 0.1,minGSSize = 3)
dotplot(KEGG_HFD_LIRKO)+theme(
panel.grid.minor = element_blank(), axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=22, color = "black", hjust=0.5,family = "arial"),
axis.title.y=element_text(size=22, color = "black", vjust=0.4, angle=90,family = "arial"),
legend.title=element_text(size=15, color = "black"),legend.text = element_text(size = 20, color = "black",family = "arial"),
axis.text.x = element_text(size = 15,color = "black",family = "arial") ,axis.text.y = element_text(size = 15,color = "black",family = "arial"))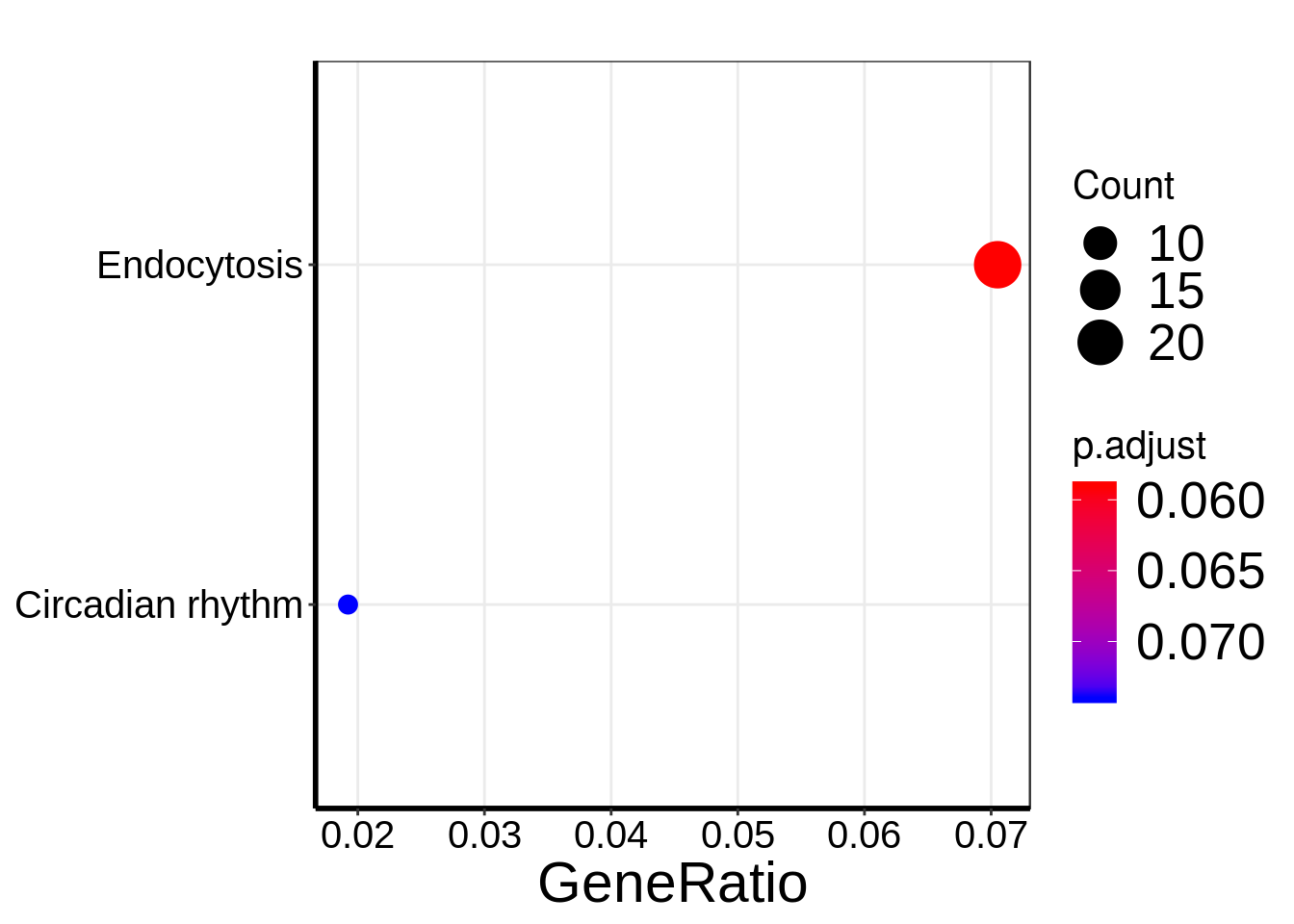
sessionInfo()R version 3.5.3 (2019-03-11)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 17.10
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/libopenblasp-r0.2.20.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] grid stats4 parallel stats graphics grDevices utils
[8] datasets methods base
other attached packages:
[1] org.Hs.eg.db_3.7.0 Vennerable_3.1.0.9000
[3] biomaRt_2.38.0 org.Mm.eg.db_3.7.0
[5] clusterProfiler_3.10.1 ggsci_2.9
[7] RADAR_0.2.3 qvalue_2.14.1
[9] RcppArmadillo_0.9.400.2.0 Rcpp_1.0.1
[11] RColorBrewer_1.1-2 gplots_3.0.1.1
[13] doParallel_1.0.14 iterators_1.0.10
[15] foreach_1.4.4 ggplot2_3.1.1
[17] Rsamtools_1.34.1 Biostrings_2.50.2
[19] XVector_0.22.0 GenomicFeatures_1.34.8
[21] AnnotationDbi_1.44.0 Biobase_2.42.0
[23] GenomicRanges_1.34.0 GenomeInfoDb_1.18.2
[25] IRanges_2.16.0 S4Vectors_0.20.1
[27] BiocGenerics_0.28.0 Logolas_1.6.0
loaded via a namespace (and not attached):
[1] tidyselect_0.2.5 RSQLite_2.1.1
[3] htmlwidgets_1.3 BiocParallel_1.16.6
[5] munsell_0.5.0 codetools_0.2-16
[7] DT_0.5.1 withr_2.1.2
[9] colorspace_1.4-1 GOSemSim_2.8.0
[11] knitr_1.22 rstudioapi_0.10
[13] DOSE_3.8.2 Guitar_1.20.1
[15] labeling_0.3 git2r_0.25.2
[17] exomePeak_2.16.0 urltools_1.7.3
[19] GenomeInfoDbData_1.2.0 polyclip_1.10-0
[21] bit64_0.9-7 farver_1.1.0
[23] rprojroot_1.3-2 generics_0.0.2
[25] xfun_0.6 R6_2.4.0
[27] locfit_1.5-9.1 bitops_1.0-6
[29] fgsea_1.8.0 gridGraphics_0.3-0
[31] DelayedArray_0.8.0 assertthat_0.2.1
[33] promises_1.0.1 scales_1.0.0
[35] pinfsc50_1.1.0 ggraph_1.0.2
[37] nnet_7.3-12 enrichplot_1.2.0
[39] gtable_0.3.0 workflowr_1.3.0
[41] rlang_0.4.0 genefilter_1.64.0
[43] splines_3.5.3 rtracklayer_1.42.2
[45] lazyeval_0.2.2 acepack_1.4.1
[47] broom_0.5.2 europepmc_0.3
[49] checkmate_1.9.1 yaml_2.2.0
[51] reshape2_1.4.3 crosstalk_1.0.0
[53] backports_1.1.4 httpuv_1.5.1
[55] Hmisc_4.2-0 RBGL_1.58.2
[57] tools_3.5.3 ggplotify_0.0.3
[59] gridBase_0.4-7 gamlss.data_5.1-3
[61] ggridges_0.5.1 gamlss_5.1-3
[63] plyr_1.8.4 base64enc_0.1-3
[65] progress_1.2.0 zlibbioc_1.28.0
[67] purrr_0.3.2 RCurl_1.95-4.12
[69] prettyunits_1.0.2 rpart_4.1-13
[71] viridis_0.5.1 cowplot_0.9.4
[73] SummarizedExperiment_1.12.0 ggrepel_0.8.0
[75] cluster_2.0.7-1 fs_1.3.0
[77] magrittr_1.5 data.table_1.12.2
[79] DO.db_2.9 triebeard_0.3.0
[81] SQUAREM_2017.10-1 whisker_0.3-2
[83] matrixStats_0.54.0 hms_0.4.2
[85] mime_0.6 evaluate_0.13
[87] xtable_1.8-4 XML_3.98-1.19
[89] gridExtra_2.3 MeRIPtools_0.1.8
[91] vcfR_1.8.0 compiler_3.5.3
[93] tibble_2.1.1 KernSmooth_2.23-15
[95] crayon_1.3.4 htmltools_0.3.6
[97] mgcv_1.8-28 later_0.8.0
[99] Formula_1.2-3 tidyr_0.8.3
[101] geneplotter_1.60.0 DBI_1.0.0
[103] tweenr_1.0.1 MASS_7.3-51.4
[105] Matrix_1.2-17 permute_0.9-5
[107] gdata_2.18.0 igraph_1.2.4.1
[109] pkgconfig_2.0.2 rvcheck_0.1.3
[111] GenomicAlignments_1.18.1 foreign_0.8-71
[113] xml2_1.2.0 annotate_1.60.1
[115] gamlss.dist_5.1-3 stringr_1.4.0
[117] digest_0.6.18 vegan_2.5-4
[119] QNB_1.1.11 graph_1.60.0
[121] rmarkdown_1.12 fastmatch_1.1-0
[123] htmlTable_1.13.1 curl_3.3
[125] shiny_1.3.2 gtools_3.8.1
[127] nlme_3.1-137 jsonlite_1.6
[129] viridisLite_0.3.0 BSgenome_1.50.0
[131] pillar_1.3.1 lattice_0.20-38
[133] httr_1.4.0 survival_2.44-1.1
[135] GO.db_3.7.0 glue_1.3.1
[137] UpSetR_1.3.3 bit_1.1-14
[139] ggforce_0.2.2 stringi_1.4.3
[141] blob_1.1.1 DESeq2_1.22.2
[143] latticeExtra_0.6-28 caTools_1.17.1.2
[145] memoise_1.1.0 dplyr_0.8.0.1
[147] ape_5.3