RSVinfected Host cells
scottzijiezhang
2018-08-08
Last updated: 2019-01-23
workflowr checks: (Click a bullet for more information)-
✔ R Markdown file: up-to-date
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
-
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(20190123)The command
set.seed(20190123)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: 2342971
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Untracked files: Untracked: analysis/RSVmutant.Rmd Untracked: docs/figure/ Unstaged changes: Modified: analysis/_site.yml
Expand here to see past versions:
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 2342971 | scottzijiezhang | 2019-01-23 | wflow_publish(c(“analysis/index.Rmd”, “analysis/RSVinA549.Rmd”, “analysis/RSVinfected.Rmd”, “analysis/RSVinHela.Rmd”)) |
A549 host cell line
Count reads map to hg38 genome
library(m6Amonster)
samplenames <-c("Ctl","RSVinfect1","RSVinfect2")
gtf <- "~/Database/genome/hg38/hg38_UCSC.gtf"
RSV.A549 <- countReads(samplenames = paste(samplenames,"align_hg38",sep = "." ),
gtf = gtf,
bamFolder = "/home/zijiezhang/RSV/201803/bam_files",
outputDir = "/home/zijiezhang/RSV/201803",
modification = "m6A",
threads = 40
)
save(RSV.A549,file = "~/RSV/201803/RSV.A549_infected.RData")Call peaks
library(m6Amonster)
load("~/RSV/201803/RSV.A549_infected.RData")
#RSV.A549 <- callPeakFisher(RSV.A549,threads = 20)Report peaks for Ctl and Infected samples
CtlPeak <- reportConsistentPeak(RSV.A549,samplenames = "Ctl.align_hg38",threads = 15)Reporting peak concsistent in all samples for
Ctl.align_hg38
Hyper-thread registered: TRUE
Using 15 thread(s) to report merged report...
Time used to report peaks: 2.7153252919515 mins... InfectedPeak <- reportConsistentPeak(RSV.A549,samplenames = c("RSVinfect1.align_hg38","RSVinfect2.align_hg38"), threads = 15 )Reporting peak concsistent in all samples for
RSVinfect1.align_hg38 RSVinfect2.align_hg38
Hyper-thread registered: TRUE
Using 15 thread(s) to report merged report...
Time used to report peaks: 1.45808432896932 mins... Plot distribution of peaks on the meta-gene
plotMetaGeneMulti(list("Ctl"=CtlPeak, "RSV infected" = InfectedPeak), "~/Database/genome/hg38/hg38_UCSC.gtf")[1] "Converting BED12 to GRangesList"
[1] "It may take a few minutes"
[1] "Converting BED12 to GRangesList"
[1] "It may take a few minutes"
[1] "total 58259 transcripts extracted ..."
[1] "total 42543 transcripts left after ambiguity filter ..."
[1] "total 21293 mRNAs left after component length filter ..."
[1] "total 7986 ncRNAs left after ncRNA length filter ..."
[1] "Building Guitar Coordinates. It may take a few minutes ..."
[1] "Guitar Coordinates Built ..."
[1] "Using provided Guitar Coordinates"
[1] "resolving ambiguious features ..."
[1] "no figure saved ..."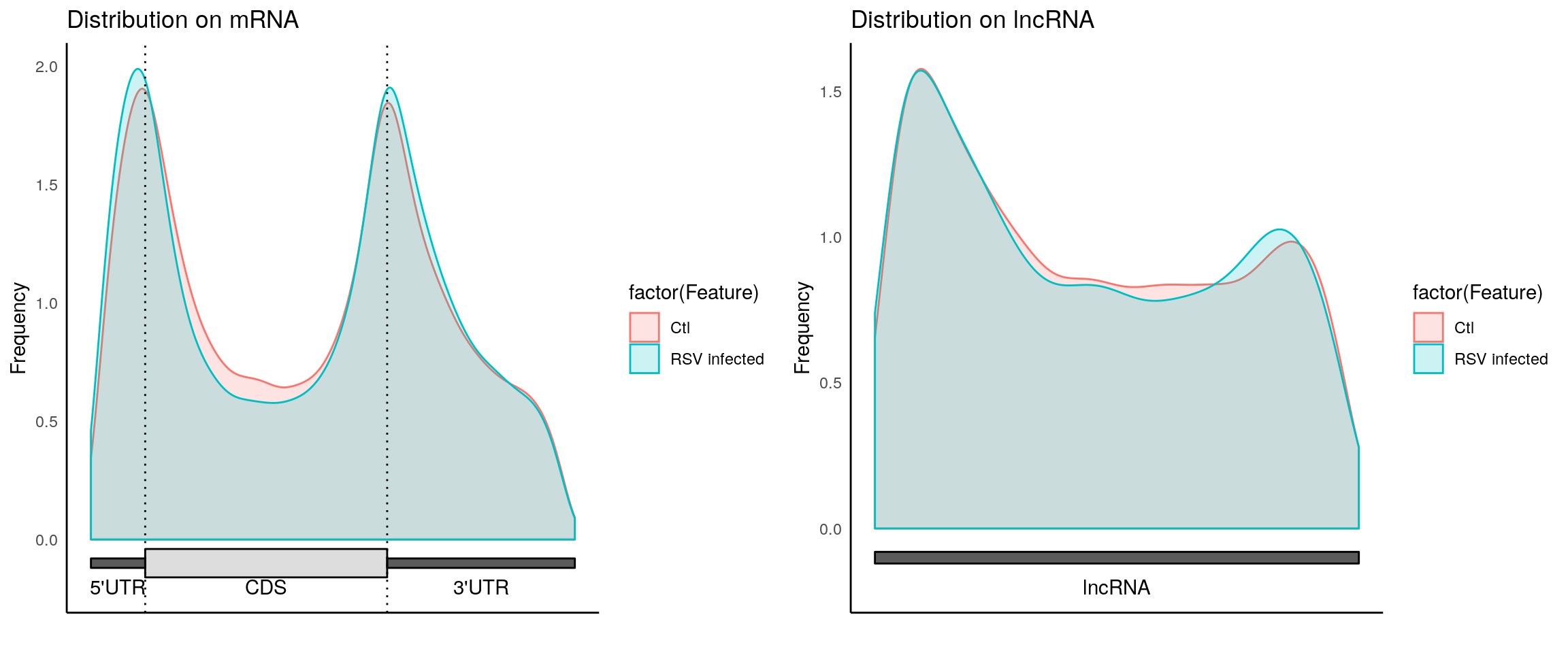
Please note: This plot only shows m6A peak distribution of both Ctl and infected samples are consistent with conanical pattern, suggesting the m6A-seq is of okay quality. It doesn’t tell any difference between the two group! The peak show up in one group but not the other group are just some boundary cases where p-value are close to the threshold for peak-calling. Besides, Ctl has no replicate while Infected peaks are consistent peaks of two replicates(more stringent); resulting in much fewer peaks in Infected group.
Show Ctl peaks distribution on Pie chart
peakDistribution(peak = CtlPeak, gtf = "~/Database/genome/hg38/hg38_UCSC.gtf" )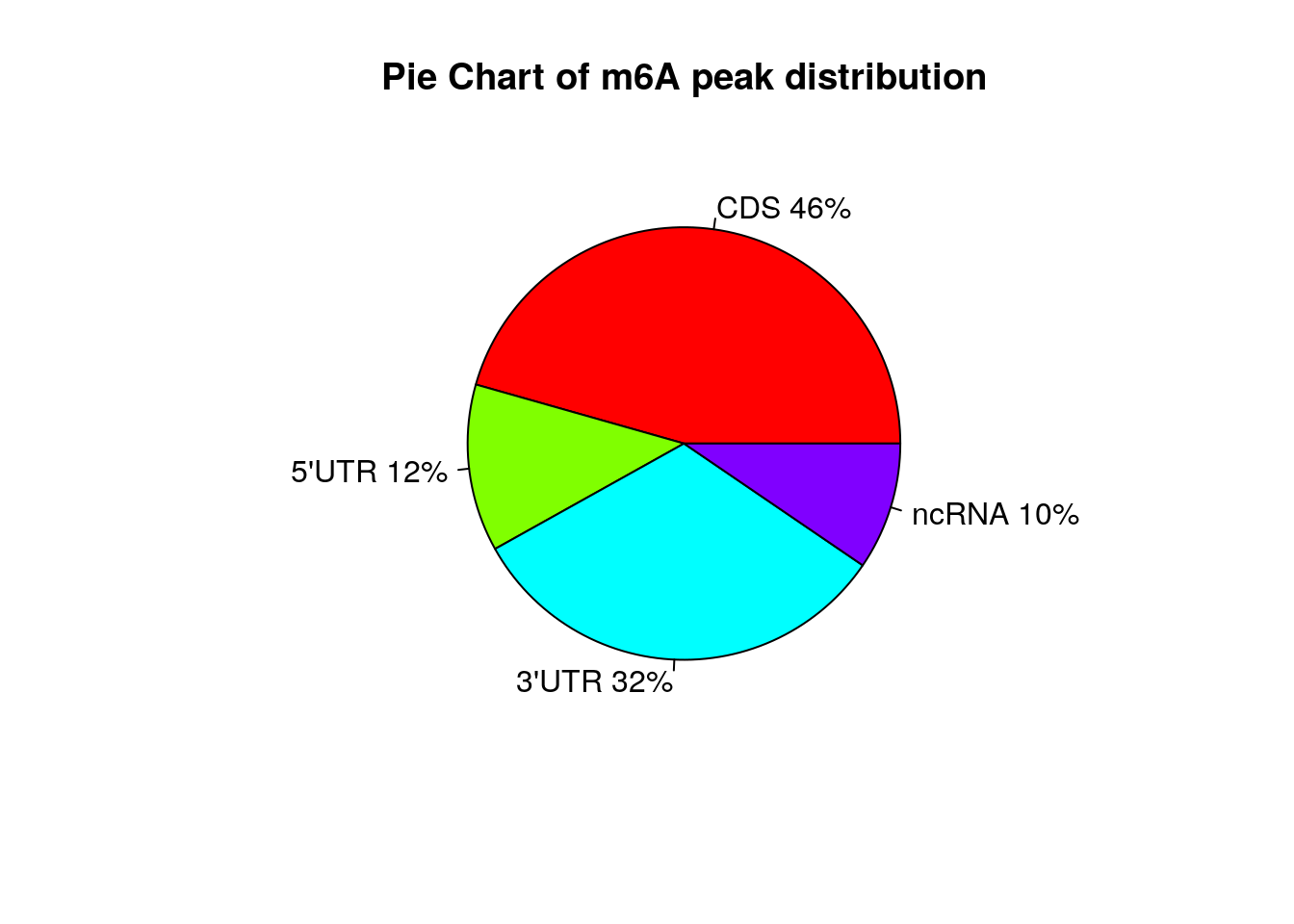
Show RSV infected peaks distribution on Pie chart
peakDistribution(peak = InfectedPeak, gtf = "~/Database/genome/hg38/hg38_UCSC.gtf" )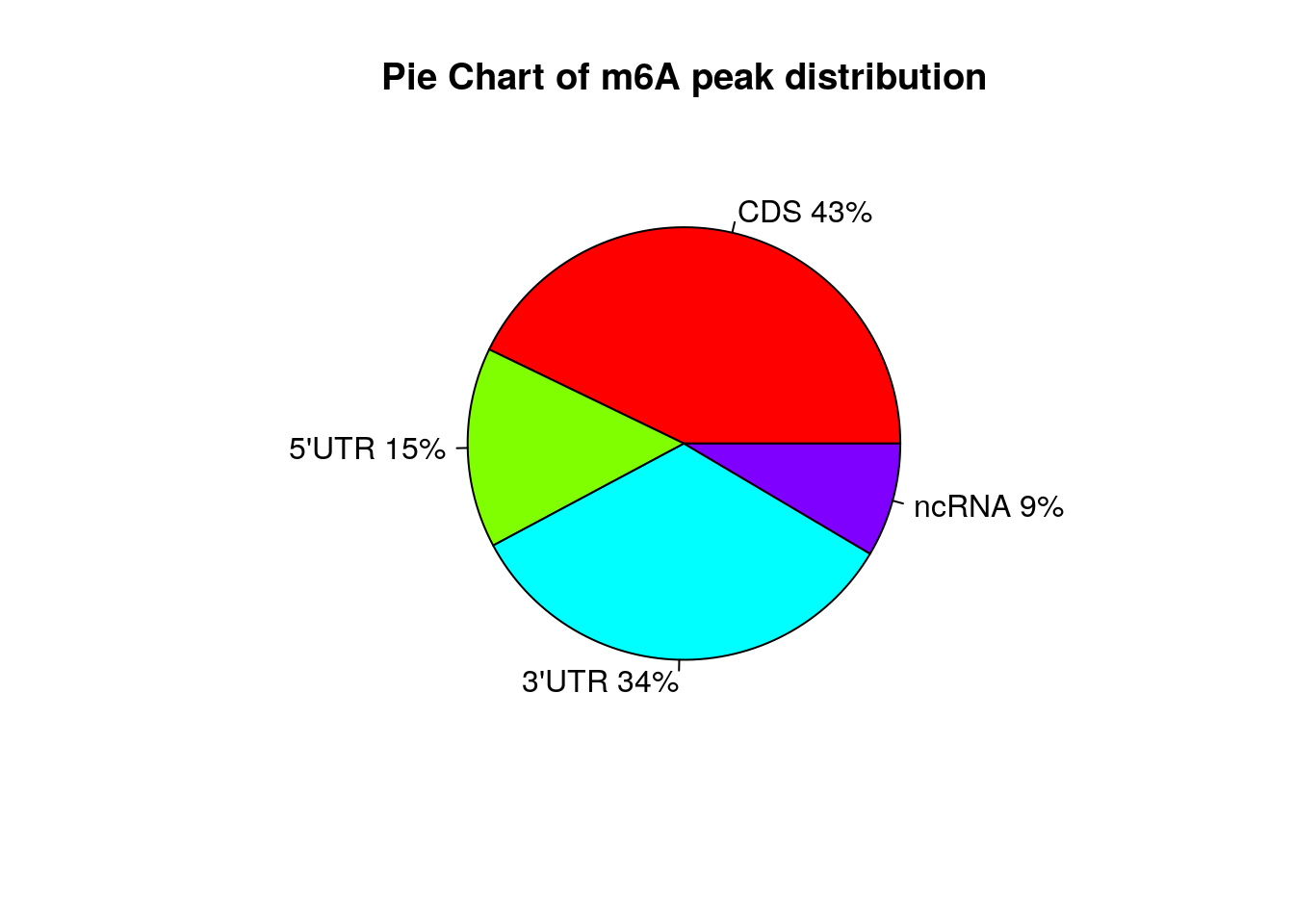
Differential methylation test with QNB
#RSV.A549 <- JointPeakCount(RSV.A549,joint_threshold = 2)
#RSV.A549 <- normalizePeak(RSV.A549,c("Ctl","Infected","Infected"))
#RSV.A549 <- QNBtest(RSV.A549)
hist(qvalue::qvalue(RSV.A549$all.est.peak$p_value))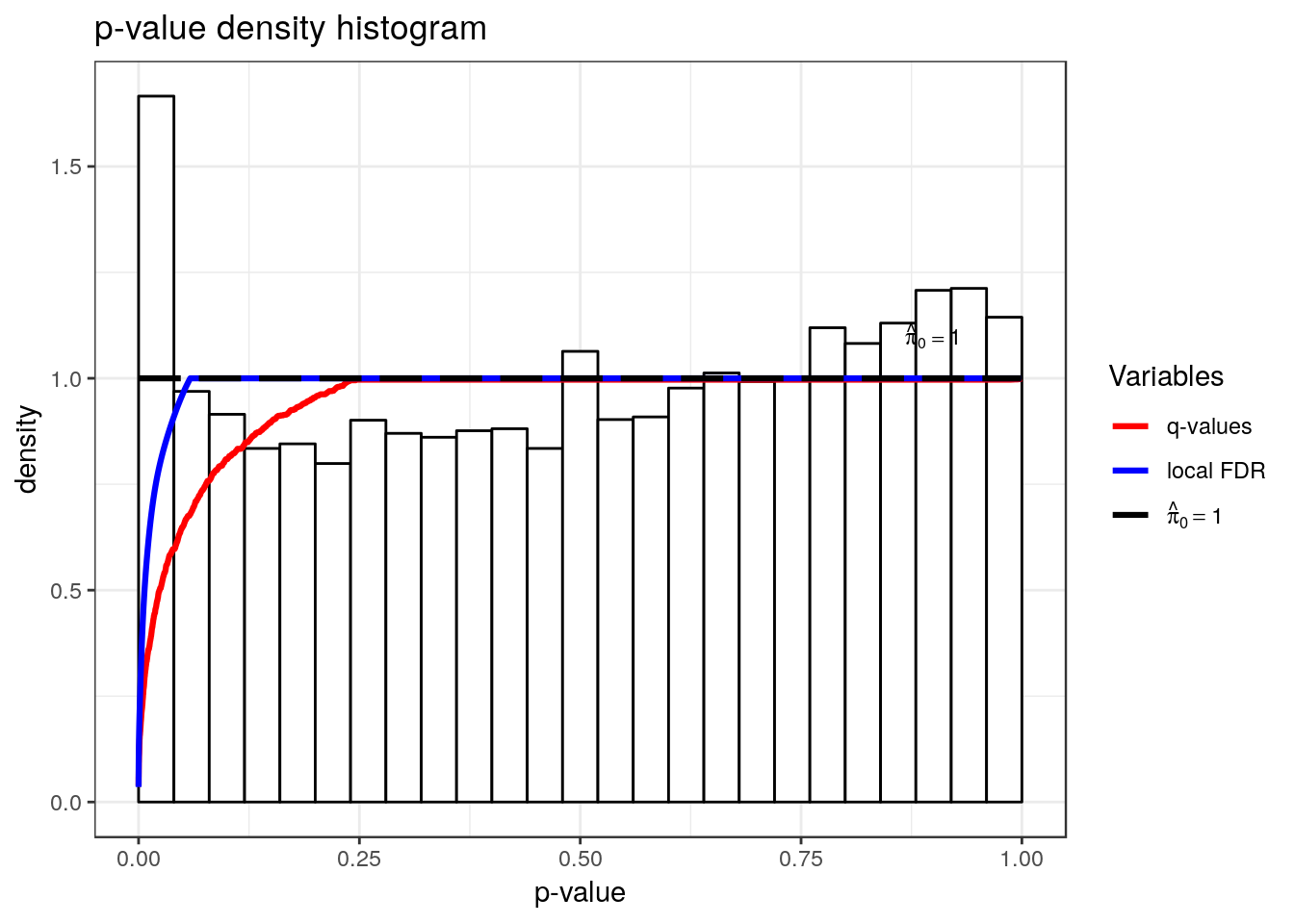
Display differential methylated peaks
jointPeak <- reportJointPeak(RSV.A549,threads = 10)Reporting joint peak with PoissonGamma test statistics...
Reporting 16156 peaks...
Hyper-thread registered: TRUE
Using 10 thread(s) to report merged report...
Time used to report peaks: 4.10002266565959 mins... #write.table(jointPeak[which(jointPeak$fdr<0.1 & abs(jointPeak$logFC)>0.5 ),], file = "~/RSV/201803/Host_diff_meth.xls",sep = "\t", quote = F,row.names = F)
knitr::kable(jointPeak[which(jointPeak$fdr<0.1 & abs(jointPeak$logFC)>0.5 ),], format = "pandoc", row.names = F)| chr | start | end | name | score | strand | thickStart | thickEnd | itemRgb | blockCount | blockSizes | blockStarts | logFC | p_value | fdr |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chr22 | 35658721 | 35659569 | APOL6 | 0 | + | 35658721 | 35659569 | 0 | 1 | 848 | 0 | -0.7543814 | 0.0000580 | 0.0590205 |
| chr11 | 64989641 | 64990150 | BATF2 | 0 | - | 64989641 | 64990150 | 0 | 2 | 172,77 | 0,433 | -1.2247847 | 0.0002172 | 0.0779511 |
| chr2 | 110674354 | 110678007 | BUB1 | 0 | - | 110674354 | 110678007 | 0 | 2 | 12,38 | 0,3616 | 1.4177365 | 0.0000677 | 0.0590205 |
| chr17 | 40555670 | 40560214 | CCR7 | 0 | - | 40555670 | 40560214 | 0 | 3 | 149,50,1 | 0,3223,4544 | -Inf | 0.0001685 | 0.0716152 |
| chr19 | 45409420 | 45409719 | CD3EAP | 0 | + | 45409420 | 45409719 | 0 | 1 | 299 | 0 | 1.0097749 | 0.0000340 | 0.0456997 |
| chr1 | 37707021 | 37708377 | CDCA8 | 0 | + | 37707021 | 37708377 | 0 | 2 | 44,56 | 0,1301 | 0.5939262 | 0.0001340 | 0.0663984 |
| chr3 | 45082572 | 45083119 | CDCP1 | 0 | - | 45082572 | 45083119 | 0 | 1 | 547 | 0 | -1.3914161 | 0.0000236 | 0.0422699 |
| chr3 | 143121872 | 143122319 | CHST2 | 0 | + | 143121872 | 143122319 | 0 | 1 | 447 | 0 | -1.2996244 | 0.0001768 | 0.0732160 |
| chr3 | 39146698 | 39153511 | CSRNP1 | 0 | - | 39146698 | 39153511 | 0 | 2 | 25,74 | 0,6740 | -Inf | 0.0000133 | 0.0422699 |
| chr6 | 131949074 | 131949422 | CTGF | 0 | - | 131949074 | 131949422 | 0 | 1 | 348 | 0 | -0.9295820 | 0.0003029 | 0.0938555 |
| chr12 | 89348873 | 89349172 | DUSP6 | 0 | - | 89348873 | 89349172 | 0 | 1 | 299 | 0 | -0.7198940 | 0.0004031 | 0.0999217 |
| chr12 | 89352211 | 89352510 | DUSP6 | 0 | - | 89352211 | 89352510 | 0 | 1 | 299 | 0 | -1.0344784 | 0.0003970 | 0.0999217 |
| chr3 | 194685893 | 194686291 | FAM43A | 0 | + | 194685893 | 194686291 | 0 | 1 | 398 | 0 | -1.4067112 | 0.0003800 | 0.0974025 |
| chr20 | 38951921 | 38952168 | FAM83D | 0 | + | 38951921 | 38952168 | 0 | 1 | 247 | 0 | -0.5599304 | 0.0004084 | 0.0999217 |
| chr11 | 61796894 | 61797043 | FEN1 | 0 | + | 61796894 | 61797043 | 0 | 1 | 149 | 0 | -0.5691996 | 0.0001398 | 0.0663984 |
| chr15 | 26547839 | 26547987 | GABRB3 | 0 | - | 26547839 | 26547987 | 0 | 1 | 148 | 0 | 0.5370579 | 0.0002764 | 0.0910725 |
| chr20 | 63562606 | 63562904 | HELZ2 | 0 | - | 63562606 | 63562904 | 0 | 1 | 298 | 0 | -0.9334816 | 0.0000997 | 0.0596632 |
| chr20 | 63565101 | 63565699 | HELZ2 | 0 | - | 63565101 | 63565699 | 0 | 1 | 598 | 0 | -0.9156383 | 0.0000177 | 0.0422699 |
| chr20 | 63565749 | 63565998 | HELZ2 | 0 | - | 63565749 | 63565998 | 0 | 1 | 249 | 0 | -1.1795725 | 0.0000298 | 0.0445201 |
| chr2 | 233840654 | 233840802 | HJURP | 0 | - | 233840654 | 233840802 | 0 | 1 | 148 | 0 | -0.5548283 | 0.0000652 | 0.0590205 |
| chr2 | 233841351 | 233841550 | HJURP | 0 | - | 233841351 | 233841550 | 0 | 1 | 199 | 0 | -0.7097939 | 0.0000186 | 0.0422699 |
| chr10 | 89402706 | 89402755 | IFIT1 | 0 | + | 89402706 | 89402755 | 0 | 1 | 49 | 0 | -0.5413852 | 0.0001635 | 0.0713520 |
| chr9 | 107485448 | 107485597 | KLF4 | 0 | - | 107485448 | 107485597 | 0 | 1 | 149 | 0 | -0.8803973 | 0.0001256 | 0.0663984 |
| chr9 | 107485747 | 107485895 | KLF4 | 0 | - | 107485747 | 107485895 | 0 | 1 | 148 | 0 | -0.5215939 | 0.0002422 | 0.0832117 |
| chr9 | 107487347 | 107487545 | KLF4 | 0 | - | 107487347 | 107487545 | 0 | 1 | 198 | 0 | -0.7457239 | 0.0000857 | 0.0596632 |
| chr2 | 135840722 | 135840771 | MCM6 | 0 | - | 135840722 | 135840771 | 0 | 1 | 49 | 0 | 0.8385662 | 0.0002954 | 0.0935330 |
| chr11 | 1244507 | 1244706 | MUC5B | 0 | + | 1244507 | 1244706 | 0 | 1 | 199 | 0 | -2.9314777 | 0.0000303 | 0.0445201 |
| chr2 | 205776455 | 205776852 | NRP2 | 0 | + | 205776455 | 205776852 | 0 | 1 | 397 | 0 | -3.4848766 | 0.0000130 | 0.0422699 |
| chr10 | 23440306 | 23440948 | OTUD1 | 0 | + | 23440306 | 23440948 | 0 | 1 | 642 | 0 | -0.8041484 | 0.0002145 | 0.0779511 |
| chr8 | 143984821 | 143985267 | PARP10 | 0 | - | 143984821 | 143985267 | 0 | 1 | 446 | 0 | -0.9763381 | 0.0002943 | 0.0935330 |
| chr3 | 122555394 | 122555742 | PARP9 | 0 | - | 122555394 | 122555742 | 0 | 1 | 348 | 0 | -1.3021122 | 0.0003507 | 0.0943841 |
| chr6 | 106099522 | 106105242 | PRDM1 | 0 | + | 106099522 | 106105242 | 0 | 2 | 31,418 | 0,5303 | -1.9640802 | 0.0000633 | 0.0590205 |
| chr17 | 17494788 | 17495087 | RASD1 | 0 | - | 17494788 | 17495087 | 0 | 1 | 299 | 0 | -Inf | 0.0001288 | 0.0663984 |
| chr1 | 206586855 | 206587300 | RASSF5 | 0 | + | 206586855 | 206587300 | 0 | 1 | 445 | 0 | -0.7995462 | 0.0000710 | 0.0590205 |
| chr17 | 18328596 | 18328743 | SHMT1 | 0 | - | 18328596 | 18328743 | 0 | 1 | 147 | 0 | 1.1006248 | 0.0000211 | 0.0422699 |
| chr17 | 76386972 | 76387317 | SPHK1 | 0 | + | 76386972 | 76387317 | 0 | 1 | 345 | 0 | -1.0352626 | 0.0003403 | 0.0943841 |
| chr2 | 119263327 | 119263426 | STEAP3 | 0 | + | 119263327 | 119263426 | 0 | 1 | 99 | 0 | 1.0807028 | 0.0003649 | 0.0965847 |
| chr9 | 120903039 | 120903287 | TRAF1 | 0 | - | 120903039 | 120903287 | 0 | 1 | 248 | 0 | -1.4932589 | 0.0001011 | 0.0596632 |
| chr9 | 120903586 | 120903834 | TRAF1 | 0 | - | 120903586 | 120903834 | 0 | 1 | 248 | 0 | -1.4527720 | 0.0001612 | 0.0713520 |
| chr9 | 120903984 | 120904133 | TRAF1 | 0 | - | 120903984 | 120904133 | 0 | 1 | 149 | 0 | -1.4295287 | 0.0001035 | 0.0596632 |
| chr3 | 36833409 | 36833707 | TRANK1 | 0 | - | 36833409 | 36833707 | 0 | 1 | 298 | 0 | -0.8984218 | 0.0003795 | 0.0974025 |
| chr5 | 917112 | 917161 | TRIP13 | 0 | + | 917112 | 917161 | 0 | 1 | 49 | 0 | 3.5698784 | 0.0000994 | 0.0596632 |
| chr1 | 119623660 | 119647767 | ZNF697 | 0 | - | 119623660 | 119647767 | 0 | 3 | 457,263,77 | 0,2215,24031 | -0.9385525 | 0.0000943 | 0.0596632 |
** Please note: ** there are very few differentially methylated sites on the host cells in this data set. Those peaks are ones that are different between Ctl and Infected group.
Differential expression analysis
The “emerged/lost peak figure (analysis)” before was an incorrect way to analyzing the data. Those emerged/lost peads mostly reflected differentially expressed genes.
deseq2_res <- RADAR::DESeq2(RSV.A549$geneSum, matrix(c("Ctl","Infected","Infected"),ncol = 1) )[1] "without pc ..."
Using input matrix without additional normalization...#write.table(deseq2_res[which(deseq2_res$p.adjust<0.05),], file = "~/RSV/201803/Host_diff_expression.xls",sep = "\t", quote = F,row.names = F)
knitr::kable(deseq2_res[which(deseq2_res$p.adjust<0.1),], format = "pandoc", row.names = F)| GeneID | log2FC | p.adjust | pvalue |
|---|---|---|---|
| A4GALT | 2.57 | 0.0000000 | 0.00e+00 |
| AAAS | -1.23 | 0.0000000 | 0.00e+00 |
| AACS | 0.68 | 0.0011486 | 3.57e-04 |
| AADAC | 1.48 | 0.0000025 | 5.00e-07 |
| AADAT | -1.79 | 0.0038961 | 1.35e-03 |
| AARS | 0.25 | 0.0194316 | 7.99e-03 |
| AARSD1 | -1.13 | 0.0000091 | 2.00e-06 |
| AASDHPPT | -1.59 | 0.0000000 | 0.00e+00 |
| AATF | 0.66 | 0.0001012 | 2.63e-05 |
| ABALON | 2.74 | 0.0903933 | 4.59e-02 |
| ABAT | 3.74 | 0.0005702 | 1.67e-04 |
| ABCA1 | 0.99 | 0.0001283 | 3.39e-05 |
| ABCA2 | -0.54 | 0.0298873 | 1.29e-02 |
| ABCA3 | 0.74 | 0.0040767 | 1.42e-03 |
| ABCB1 | 3.22 | 0.0255717 | 1.09e-02 |
| ABCB10 | -1.32 | 0.0003303 | 9.28e-05 |
| ABCB6 | -0.41 | 0.0688732 | 3.34e-02 |
| ABCB8 | -0.59 | 0.0204123 | 8.44e-03 |
| ABCC1 | 1.80 | 0.0000000 | 0.00e+00 |
| ABCC10 | 1.13 | 0.0016591 | 5.31e-04 |
| ABCC2 | -0.94 | 0.0000000 | 0.00e+00 |
| ABCC3 | 0.85 | 0.0000000 | 0.00e+00 |
| ABCC4 | -1.94 | 0.0000000 | 0.00e+00 |
| ABCC9 | 7.03 | 0.0060309 | 2.19e-03 |
| ABCD1 | 1.63 | 0.0054132 | 1.94e-03 |
| ABCD3 | -2.32 | 0.0000000 | 0.00e+00 |
| ABCD4 | 0.65 | 0.0110672 | 4.30e-03 |
| ABCE1 | -2.17 | 0.0000000 | 0.00e+00 |
| ABCF2 | -0.74 | 0.0000022 | 5.00e-07 |
| ABCG1 | 2.33 | 0.0000000 | 0.00e+00 |
| ABCG2 | -2.72 | 0.0000000 | 0.00e+00 |
| ABHD10 | -1.35 | 0.0000316 | 7.70e-06 |
| ABHD12 | 0.58 | 0.0001026 | 2.67e-05 |
| ABHD14A-ACY1 | -1.07 | 0.0066972 | 2.46e-03 |
| ABHD14B | -0.77 | 0.0359063 | 1.59e-02 |
| ABHD17C | 0.96 | 0.0003533 | 1.00e-04 |
| ABHD4 | 0.88 | 0.0000000 | 0.00e+00 |
| ABHD8 | 1.89 | 0.0000361 | 8.80e-06 |
| ABI1 | 0.79 | 0.0000008 | 2.00e-07 |
| ABI2 | -0.40 | 0.0542863 | 2.54e-02 |
| ABL1 | 1.07 | 0.0000000 | 0.00e+00 |
| ABL2 | 2.49 | 0.0000000 | 0.00e+00 |
| ABLIM1 | 0.66 | 0.0000037 | 8.00e-07 |
| ABLIM2 | 2.38 | 0.0772379 | 3.82e-02 |
| ABLIM3 | 2.29 | 0.0000000 | 0.00e+00 |
| ABR | 0.75 | 0.0000575 | 1.44e-05 |
| ABTB1 | 0.75 | 0.0088158 | 3.34e-03 |
| ABTB2 | 3.47 | 0.0000000 | 0.00e+00 |
| ACAA2 | -0.81 | 0.0010610 | 3.27e-04 |
| ACACA | -1.00 | 0.0000002 | 0.00e+00 |
| ACAD9 | -0.73 | 0.0068287 | 2.51e-03 |
| ACADM | -1.43 | 0.0000782 | 1.99e-05 |
| ACADSB | -1.27 | 0.0002281 | 6.25e-05 |
| ACADVL | 0.59 | 0.0000001 | 0.00e+00 |
| ACAN | 6.75 | 0.0000386 | 9.50e-06 |
| ACAP2 | -0.83 | 0.0088696 | 3.36e-03 |
| ACAP3 | -0.84 | 0.0617375 | 2.94e-02 |
| ACAT1 | -1.79 | 0.0000000 | 0.00e+00 |
| ACAT2 | 0.42 | 0.0130205 | 5.14e-03 |
| ACBD3 | 0.36 | 0.0697827 | 3.39e-02 |
| ACE2 | 5.53 | 0.0810935 | 4.05e-02 |
| ACHE | 6.90 | 0.0000000 | 0.00e+00 |
| ACKR1 | 8.09 | 0.0006350 | 1.88e-04 |
| ACKR3 | 1.69 | 0.0537325 | 2.51e-02 |
| ACLY | -0.27 | 0.0017872 | 5.77e-04 |
| ACO2 | 0.36 | 0.0252187 | 1.07e-02 |
| ACOT13 | -1.26 | 0.0000000 | 0.00e+00 |
| ACOT7 | 1.05 | 0.0000000 | 0.00e+00 |
| ACOT9 | 0.33 | 0.0630650 | 3.01e-02 |
| ACP1 | -1.26 | 0.0000000 | 0.00e+00 |
| ACP2 | 2.69 | 0.0000000 | 0.00e+00 |
| ACP6 | -1.31 | 0.0175211 | 7.14e-03 |
| ACSF3 | 0.62 | 0.0504330 | 2.33e-02 |
| ACSL1 | 0.62 | 0.0173642 | 7.06e-03 |
| ACSL4 | 1.85 | 0.0000000 | 0.00e+00 |
| ACSL5 | 6.06 | 0.0000003 | 1.00e-07 |
| ACSM3 | -1.83 | 0.0009159 | 2.79e-04 |
| ACSS1 | -0.78 | 0.0259347 | 1.10e-02 |
| ACSS2 | 1.92 | 0.0000000 | 0.00e+00 |
| ACTA2 | 0.87 | 0.0984634 | 5.07e-02 |
| ACTB | 0.54 | 0.0000000 | 0.00e+00 |
| ACTL6A | -1.59 | 0.0000000 | 0.00e+00 |
| ACTL8 | -5.32 | 0.0337735 | 1.48e-02 |
| ACTN1 | 1.93 | 0.0000000 | 0.00e+00 |
| ACTN2 | 10.59 | 0.0000014 | 3.00e-07 |
| ACTN4 | 0.80 | 0.0000000 | 0.00e+00 |
| ACTR10 | -0.85 | 0.0007378 | 2.20e-04 |
| ACTR1B | -0.41 | 0.0886523 | 4.49e-02 |
| ACTR2 | -0.85 | 0.0000000 | 0.00e+00 |
| ACTR3B | -1.66 | 0.0129805 | 5.12e-03 |
| ACTR5 | -0.96 | 0.0246075 | 1.04e-02 |
| ACTR6 | -1.12 | 0.0010604 | 3.27e-04 |
| ACVR1 | 1.82 | 0.0000000 | 0.00e+00 |
| ACVR1B | 0.72 | 0.0022361 | 7.41e-04 |
| ACVRL1 | 8.50 | 0.0000000 | 0.00e+00 |
| ACY1 | -1.17 | 0.0040227 | 1.40e-03 |
| ACY3 | 1.75 | 0.0805924 | 4.01e-02 |
| ACYP1 | -1.24 | 0.0384296 | 1.72e-02 |
| ADAM10 | -0.24 | 0.0881778 | 4.46e-02 |
| ADAM12 | 3.86 | 0.0000000 | 0.00e+00 |
| ADAM17 | 0.75 | 0.0000072 | 1.60e-06 |
| ADAM19 | 3.28 | 0.0000000 | 0.00e+00 |
| ADAM28 | 3.32 | 0.0170968 | 6.94e-03 |
| ADAM9 | -1.05 | 0.0000000 | 0.00e+00 |
| ADAMTS10 | 0.85 | 0.0011228 | 3.48e-04 |
| ADAMTS2 | 3.73 | 0.0192650 | 7.92e-03 |
| ADAMTS4 | 10.91 | 0.0000006 | 1.00e-07 |
| ADAMTS9 | 2.86 | 0.0000000 | 0.00e+00 |
| ADAMTSL4 | 1.81 | 0.0101758 | 3.92e-03 |
| ADAP1 | 1.26 | 0.0092804 | 3.53e-03 |
| ADAP2 | 1.14 | 0.0310483 | 1.35e-02 |
| ADAR | 1.72 | 0.0000000 | 0.00e+00 |
| ADCK2 | -0.79 | 0.0459852 | 2.10e-02 |
| ADCK3 | -1.41 | 0.0000817 | 2.09e-05 |
| ADCK4 | 0.43 | 0.0240913 | 1.02e-02 |
| ADCY4 | 5.76 | 0.0612786 | 2.91e-02 |
| ADCY5 | 2.17 | 0.0000057 | 1.20e-06 |
| ADD1 | 0.58 | 0.0000673 | 1.70e-05 |
| ADD2 | -2.03 | 0.0000000 | 0.00e+00 |
| ADGRA3 | -2.62 | 0.0000000 | 0.00e+00 |
| ADGRE1 | 3.05 | 0.0000000 | 0.00e+00 |
| ADGRE2 | 2.51 | 0.0000000 | 0.00e+00 |
| ADGRE5 | 1.83 | 0.0000000 | 0.00e+00 |
| ADGRG1 | 1.76 | 0.0000000 | 0.00e+00 |
| ADGRG6 | -1.16 | 0.0000000 | 0.00e+00 |
| ADGRL2 | -0.76 | 0.0176392 | 7.20e-03 |
| ADH5 | -1.08 | 0.0000000 | 0.00e+00 |
| ADI1 | -0.75 | 0.0019444 | 6.34e-04 |
| ADIPOR2 | 0.27 | 0.0698789 | 3.39e-02 |
| ADK | -0.96 | 0.0017236 | 5.54e-04 |
| ADM | 1.05 | 0.0000000 | 0.00e+00 |
| ADM2 | 4.03 | 0.0000000 | 0.00e+00 |
| ADNP2 | 0.71 | 0.0004738 | 1.37e-04 |
| ADORA2A | 7.29 | 0.0000000 | 0.00e+00 |
| ADORA2B | -1.11 | 0.0249154 | 1.06e-02 |
| ADPRH | 1.43 | 0.0107962 | 4.18e-03 |
| ADPRHL1 | -1.18 | 0.0047197 | 1.67e-03 |
| ADPRHL2 | 0.90 | 0.0029649 | 1.01e-03 |
| ADPRM | 1.00 | 0.0397683 | 1.78e-02 |
| ADRA1B | 1.40 | 0.0003568 | 1.01e-04 |
| ADRA1D | 3.02 | 0.0000000 | 0.00e+00 |
| ADRB2 | 7.51 | 0.0022554 | 7.49e-04 |
| ADRBK1 | 0.47 | 0.0149991 | 6.01e-03 |
| ADRM1 | 0.31 | 0.0255980 | 1.09e-02 |
| ADSL | -1.15 | 0.0000000 | 0.00e+00 |
| ADSSL1 | -1.52 | 0.0076349 | 2.85e-03 |
| AEBP2 | 0.94 | 0.0000743 | 1.89e-05 |
| AEN | 1.49 | 0.0000000 | 0.00e+00 |
| AES | 0.62 | 0.0000000 | 0.00e+00 |
| AFAP1 | 1.48 | 0.0000000 | 0.00e+00 |
| AFAP1L2 | 1.15 | 0.0000000 | 0.00e+00 |
| AFF1 | 1.05 | 0.0000000 | 0.00e+00 |
| AFF3 | 5.69 | 0.0651644 | 3.13e-02 |
| AFF4 | 0.83 | 0.0000000 | 0.00e+00 |
| AFG3L1P | -0.70 | 0.0600549 | 2.85e-02 |
| AFMID | -0.97 | 0.0173120 | 7.03e-03 |
| AFTPH | 0.36 | 0.0694373 | 3.37e-02 |
| AGAP1 | -0.71 | 0.0047360 | 1.68e-03 |
| AGAP3 | 0.50 | 0.0178214 | 7.28e-03 |
| AGBL5 | -1.50 | 0.0000000 | 0.00e+00 |
| AGFG1 | -0.44 | 0.0013400 | 4.21e-04 |
| AGFG2 | 2.46 | 0.0000000 | 0.00e+00 |
| AGGF1 | -0.70 | 0.0072625 | 2.69e-03 |
| AGL | -1.23 | 0.0057547 | 2.08e-03 |
| AGMAT | -2.15 | 0.0000222 | 5.30e-06 |
| AGMO | 1.79 | 0.0957657 | 4.91e-02 |
| AGO2 | 1.16 | 0.0000000 | 0.00e+00 |
| AGPAT2 | -0.46 | 0.0212676 | 8.84e-03 |
| AGPAT4 | 1.40 | 0.0059199 | 2.15e-03 |
| AGPAT5 | -2.52 | 0.0000000 | 0.00e+00 |
| AGPS | -1.08 | 0.0000000 | 0.00e+00 |
| AGRN | 1.82 | 0.0000000 | 0.00e+00 |
| AGTPBP1 | -1.38 | 0.0000236 | 5.60e-06 |
| AGTRAP | 0.99 | 0.0000005 | 1.00e-07 |
| AHCTF1 | -1.27 | 0.0000001 | 0.00e+00 |
| AHCTF1P1 | -1.05 | 0.0003560 | 1.01e-04 |
| AHCY | -0.81 | 0.0000000 | 0.00e+00 |
| AHCYL2 | 4.52 | 0.0000000 | 0.00e+00 |
| AHDC1 | 0.68 | 0.0397458 | 1.78e-02 |
| AHI1 | -1.26 | 0.0019778 | 6.46e-04 |
| AHNAK | 0.39 | 0.0001026 | 2.67e-05 |
| AHNAK2 | -0.30 | 0.0175444 | 7.15e-03 |
| AHR | 1.31 | 0.0000000 | 0.00e+00 |
| AHSA1 | -0.45 | 0.0002091 | 5.69e-05 |
| AIFM1 | -1.41 | 0.0000000 | 0.00e+00 |
| AIFM2 | 1.29 | 0.0000000 | 0.00e+00 |
| AIM1 | 3.10 | 0.0000420 | 1.03e-05 |
| AIMP1 | -0.84 | 0.0003206 | 8.99e-05 |
| AIMP2 | -1.11 | 0.0000006 | 1.00e-07 |
| AIP | 0.71 | 0.0000213 | 5.10e-06 |
| AJUBA | 1.14 | 0.0000000 | 0.00e+00 |
| AK2 | -0.33 | 0.0441407 | 2.01e-02 |
| AK3 | -0.60 | 0.0085506 | 3.23e-03 |
| AK5 | -3.11 | 0.0041295 | 1.44e-03 |
| AK8 | 3.39 | 0.0000235 | 5.60e-06 |
| AKAP1 | -0.39 | 0.0272971 | 1.17e-02 |
| AKAP11 | -1.25 | 0.0000090 | 2.00e-06 |
| AKAP12 | 1.00 | 0.0000000 | 0.00e+00 |
| AKAP13 | 1.44 | 0.0000000 | 0.00e+00 |
| AKAP17A | 1.23 | 0.0000000 | 0.00e+00 |
| AKAP2 | 3.44 | 0.0000000 | 0.00e+00 |
| AKAP8L | 1.25 | 0.0000000 | 0.00e+00 |
| AKAP9 | -0.93 | 0.0003116 | 8.72e-05 |
| AKIP1 | 0.73 | 0.0031132 | 1.06e-03 |
| AKIRIN1 | 0.38 | 0.0660512 | 3.18e-02 |
| AKIRIN2 | 0.74 | 0.0081433 | 3.06e-03 |
| AKNA | 1.89 | 0.0000000 | 0.00e+00 |
| AKR1A1 | -0.51 | 0.0222122 | 9.27e-03 |
| AKR1B1 | 1.33 | 0.0000000 | 0.00e+00 |
| AKR1B10 | -1.26 | 0.0000000 | 0.00e+00 |
| AKR1B15 | -1.13 | 0.0000000 | 0.00e+00 |
| AKR1C1 | -1.26 | 0.0000000 | 0.00e+00 |
| AKR1C2 | -1.46 | 0.0000000 | 0.00e+00 |
| AKR1C3 | -1.35 | 0.0000000 | 0.00e+00 |
| AKR1C4 | -1.45 | 0.0000000 | 0.00e+00 |
| AKR7A2 | -1.60 | 0.0000001 | 0.00e+00 |
| AKT3 | 0.64 | 0.0082026 | 3.08e-03 |
| AKTIP | -0.87 | 0.0725658 | 3.55e-02 |
| ALAD | -0.70 | 0.0383109 | 1.71e-02 |
| ALAS1 | 1.55 | 0.0000000 | 0.00e+00 |
| ALCAM | 0.53 | 0.0000179 | 4.20e-06 |
| ALDH18A1 | -0.91 | 0.0000005 | 1.00e-07 |
| ALDH1A1 | -2.64 | 0.0000000 | 0.00e+00 |
| ALDH1B1 | -1.66 | 0.0000000 | 0.00e+00 |
| ALDH2 | 1.55 | 0.0000000 | 0.00e+00 |
| ALDH3A1 | -2.79 | 0.0000000 | 0.00e+00 |
| ALDH3A2 | -2.28 | 0.0000000 | 0.00e+00 |
| ALDH3B1 | -1.14 | 0.0000000 | 0.00e+00 |
| ALDH4A1 | -1.39 | 0.0000192 | 4.50e-06 |
| ALDH5A1 | -0.85 | 0.0622037 | 2.97e-02 |
| ALDH7A1 | -1.81 | 0.0000000 | 0.00e+00 |
| ALDH9A1 | -0.89 | 0.0000028 | 6.00e-07 |
| ALDOA | 0.21 | 0.0134545 | 5.34e-03 |
| ALDOC | 1.26 | 0.0162148 | 6.55e-03 |
| ALG10 | -1.27 | 0.0744400 | 3.66e-02 |
| ALG10B | -1.48 | 0.0055272 | 1.99e-03 |
| ALG13 | -0.72 | 0.0324356 | 1.42e-02 |
| ALG14 | -2.62 | 0.0000920 | 2.37e-05 |
| ALG6 | -1.44 | 0.0187468 | 7.69e-03 |
| ALG8 | -1.20 | 0.0000044 | 1.00e-06 |
| ALKBH1 | 0.78 | 0.0108453 | 4.20e-03 |
| ALKBH2 | -1.27 | 0.0001242 | 3.27e-05 |
| ALKBH5 | -0.29 | 0.0972509 | 5.00e-02 |
| ALKBH8 | -1.08 | 0.0713871 | 3.48e-02 |
| ALMS1 | -1.26 | 0.0000162 | 3.80e-06 |
| ALOX12B | 2.74 | 0.0009099 | 2.77e-04 |
| ALOX12P2 | 1.07 | 0.0908802 | 4.62e-02 |
| ALOXE3 | 5.52 | 0.0000000 | 0.00e+00 |
| ALPK1 | 0.66 | 0.0620066 | 2.96e-02 |
| ALPK2 | 1.13 | 0.0000000 | 0.00e+00 |
| ALPP | 3.36 | 0.0045501 | 1.61e-03 |
| ALPPL2 | 8.35 | 0.0003561 | 1.01e-04 |
| ALS2CL | 1.19 | 0.0002665 | 7.38e-05 |
| ALYREF | -1.40 | 0.0000000 | 0.00e+00 |
| AMACR | -0.74 | 0.0656335 | 3.15e-02 |
| AMBRA1 | 2.46 | 0.0000000 | 0.00e+00 |
| AMD1 | -1.18 | 0.0000000 | 0.00e+00 |
| AMFR | 0.81 | 0.0000004 | 1.00e-07 |
| AMH | -1.34 | 0.0716367 | 3.50e-02 |
| AMHR2 | 3.61 | 0.0000595 | 1.50e-05 |
| AMIGO2 | -0.52 | 0.0000022 | 5.00e-07 |
| AMMECR1L | 0.48 | 0.0428851 | 1.94e-02 |
| AMOTL1 | 0.78 | 0.0000067 | 1.50e-06 |
| AMOTL2 | 1.65 | 0.0000000 | 0.00e+00 |
| AMPD3 | 4.86 | 0.0000000 | 0.00e+00 |
| AMZ2 | -1.31 | 0.0000000 | 0.00e+00 |
| AMZ2P1 | -1.15 | 0.0110612 | 4.30e-03 |
| ANAPC1 | -1.35 | 0.0000000 | 0.00e+00 |
| ANAPC11 | -1.11 | 0.0000000 | 0.00e+00 |
| ANAPC13 | -0.85 | 0.0042223 | 1.48e-03 |
| ANAPC15 | -1.60 | 0.0000000 | 0.00e+00 |
| ANAPC16 | -0.89 | 0.0000186 | 4.40e-06 |
| ANAPC1P1 | -1.71 | 0.0490104 | 2.25e-02 |
| ANAPC4 | -1.38 | 0.0019695 | 6.42e-04 |
| ANAPC5 | -1.24 | 0.0000000 | 0.00e+00 |
| ANAPC7 | -0.81 | 0.0000394 | 9.70e-06 |
| ANG | 0.76 | 0.0305982 | 1.33e-02 |
| ANGPTL1 | 5.44 | 0.0911055 | 4.64e-02 |
| ANGPTL4 | 3.30 | 0.0000000 | 0.00e+00 |
| ANK3 | -1.13 | 0.0740621 | 3.64e-02 |
| ANKDD1A | -2.24 | 0.0978722 | 5.04e-02 |
| ANKFY1 | 0.31 | 0.0396026 | 1.77e-02 |
| ANKH | -1.18 | 0.0012216 | 3.81e-04 |
| ANKHD1 | 0.57 | 0.0000758 | 1.93e-05 |
| ANKHD1-EIF4EBP3 | 0.57 | 0.0000837 | 2.15e-05 |
| ANKLE2 | 1.63 | 0.0000000 | 0.00e+00 |
| ANKMY2 | -0.70 | 0.0572008 | 2.70e-02 |
| ANKRD1 | 1.90 | 0.0000000 | 0.00e+00 |
| ANKRD10 | 1.13 | 0.0000015 | 3.00e-07 |
| ANKRD11 | 0.41 | 0.0035245 | 1.21e-03 |
| ANKRD13A | 1.64 | 0.0000000 | 0.00e+00 |
| ANKRD13B | -1.05 | 0.0180316 | 7.37e-03 |
| ANKRD13D | 0.88 | 0.0009725 | 2.98e-04 |
| ANKRD18A | -1.00 | 0.0515345 | 2.39e-02 |
| ANKRD2 | -3.50 | 0.0000854 | 2.19e-05 |
| ANKRD20A5P | 1.69 | 0.0619141 | 2.95e-02 |
| ANKRD24 | 1.98 | 0.0569706 | 2.68e-02 |
| ANKRD26 | -0.82 | 0.0886793 | 4.49e-02 |
| ANKRD26P1 | 7.61 | 0.0017949 | 5.79e-04 |
| ANKRD30A | -1.89 | 0.0063251 | 2.31e-03 |
| ANKRD33B | 1.72 | 0.0000000 | 0.00e+00 |
| ANKRD36 | -1.88 | 0.0001533 | 4.10e-05 |
| ANKRD36B | -1.90 | 0.0018842 | 6.12e-04 |
| ANKRD36C | -1.88 | 0.0003649 | 1.04e-04 |
| ANKRD39 | -0.93 | 0.0472980 | 2.17e-02 |
| ANKRD50 | -0.89 | 0.0198710 | 8.20e-03 |
| ANKRD52 | 0.77 | 0.0000000 | 0.00e+00 |
| ANKRD54 | -0.80 | 0.0198075 | 8.17e-03 |
| ANKS1A | 0.88 | 0.0021399 | 7.05e-04 |
| ANKS4B | -2.89 | 0.0000894 | 2.30e-05 |
| ANKZF1 | 1.15 | 0.0000017 | 3.00e-07 |
| ANLN | -2.86 | 0.0000000 | 0.00e+00 |
| ANO6 | 0.48 | 0.0010267 | 3.16e-04 |
| ANO8 | 1.30 | 0.0004538 | 1.31e-04 |
| ANO9 | 4.43 | 0.0000000 | 0.00e+00 |
| ANP32A | -1.46 | 0.0000000 | 0.00e+00 |
| ANP32AP1 | -1.55 | 0.0523357 | 2.43e-02 |
| ANP32B | -0.42 | 0.0012155 | 3.79e-04 |
| ANP32E | -1.44 | 0.0000000 | 0.00e+00 |
| ANPEP | 1.70 | 0.0000061 | 1.30e-06 |
| ANTXR1 | -1.19 | 0.0000000 | 0.00e+00 |
| ANTXR2 | 3.07 | 0.0000039 | 8.00e-07 |
| ANXA11 | 0.46 | 0.0000668 | 1.69e-05 |
| ANXA13 | -3.53 | 0.0000000 | 0.00e+00 |
| ANXA2 | -0.91 | 0.0000000 | 0.00e+00 |
| ANXA2P1 | -0.95 | 0.0000000 | 0.00e+00 |
| ANXA2P2 | -0.90 | 0.0000000 | 0.00e+00 |
| ANXA2P3 | -1.06 | 0.0099426 | 3.82e-03 |
| ANXA4 | -0.53 | 0.0000005 | 1.00e-07 |
| ANXA5 | 0.72 | 0.0000000 | 0.00e+00 |
| ANXA8 | 1.54 | 0.0000000 | 0.00e+00 |
| ANXA8L1 | 1.47 | 0.0000000 | 0.00e+00 |
| ANXA9 | -2.84 | 0.0189077 | 7.77e-03 |
| AP1B1 | 1.19 | 0.0000000 | 0.00e+00 |
| AP1G1 | -0.50 | 0.0018805 | 6.10e-04 |
| AP1M2 | -1.49 | 0.0000023 | 5.00e-07 |
| AP1S1 | -1.19 | 0.0000000 | 0.00e+00 |
| AP1S2 | -1.44 | 0.0083424 | 3.14e-03 |
| AP2A1 | 1.17 | 0.0000000 | 0.00e+00 |
| AP2A2 | 0.84 | 0.0000828 | 2.12e-05 |
| AP2B1 | 0.19 | 0.0857340 | 4.32e-02 |
| AP2M1 | -0.33 | 0.0219763 | 9.16e-03 |
| AP2S1 | -0.55 | 0.0020586 | 6.76e-04 |
| AP3B1 | -0.56 | 0.0047892 | 1.70e-03 |
| AP3B2 | 2.81 | 0.0798152 | 3.97e-02 |
| AP3D1 | 0.78 | 0.0000000 | 0.00e+00 |
| AP3M1 | -1.01 | 0.0000000 | 0.00e+00 |
| AP3M2 | -1.74 | 0.0000010 | 2.00e-07 |
| AP3S1 | -0.74 | 0.0092678 | 3.53e-03 |
| AP3S2 | 0.38 | 0.0116113 | 4.53e-03 |
| AP4B1 | 1.52 | 0.0000001 | 0.00e+00 |
| AP5B1 | 1.25 | 0.0000000 | 0.00e+00 |
| AP5M1 | -0.62 | 0.0423530 | 1.91e-02 |
| AP5Z1 | 1.76 | 0.0000000 | 0.00e+00 |
| APBA3 | 1.94 | 0.0000000 | 0.00e+00 |
| APBB1 | 0.66 | 0.0092132 | 3.51e-03 |
| APBB1IP | 1.61 | 0.0085247 | 3.21e-03 |
| APBB2 | 1.00 | 0.0008084 | 2.44e-04 |
| APC | -1.26 | 0.0000970 | 2.51e-05 |
| APEH | -0.99 | 0.0000000 | 0.00e+00 |
| APEX1 | -1.19 | 0.0000000 | 0.00e+00 |
| API5 | -1.23 | 0.0000000 | 0.00e+00 |
| APIP | -1.04 | 0.0053880 | 1.93e-03 |
| APITD1 | -2.28 | 0.0000000 | 0.00e+00 |
| APITD1-CORT | -2.14 | 0.0000014 | 3.00e-07 |
| APLP1 | 0.70 | 0.0003299 | 9.27e-05 |
| APLP2 | 0.52 | 0.0000008 | 2.00e-07 |
| APMAP | -1.24 | 0.0000000 | 0.00e+00 |
| APOA1BP | -1.11 | 0.0000000 | 0.00e+00 |
| APOBEC3B | -1.15 | 0.0183044 | 7.49e-03 |
| APOBEC3F | 0.93 | 0.0452950 | 2.07e-02 |
| APOBEC3G | 4.52 | 0.0260055 | 1.11e-02 |
| APOBR | 5.44 | 0.0025679 | 8.61e-04 |
| APOC1 | -0.94 | 0.0020823 | 6.85e-04 |
| APOH | -4.13 | 0.0000039 | 8.00e-07 |
| APOL1 | 7.08 | 0.0000000 | 0.00e+00 |
| APOL2 | 4.09 | 0.0000000 | 0.00e+00 |
| APOL3 | 10.43 | 0.0000022 | 4.00e-07 |
| APOL6 | 4.85 | 0.0000000 | 0.00e+00 |
| APOLD1 | 2.52 | 0.0112861 | 4.40e-03 |
| APOOL | -1.11 | 0.0051160 | 1.83e-03 |
| APP | 1.25 | 0.0000000 | 0.00e+00 |
| APPBP2 | 0.49 | 0.0344298 | 1.52e-02 |
| APPL1 | -1.56 | 0.0000006 | 1.00e-07 |
| APRT | -0.95 | 0.0000000 | 0.00e+00 |
| AQP3 | 0.72 | 0.0472395 | 2.17e-02 |
| AQP9 | 8.81 | 0.0001204 | 3.17e-05 |
| AQR | -1.07 | 0.0000000 | 0.00e+00 |
| ARAF | 0.92 | 0.0000001 | 0.00e+00 |
| ARAP1 | 1.54 | 0.0000000 | 0.00e+00 |
| ARAP2 | 2.87 | 0.0000001 | 0.00e+00 |
| ARAP3 | -0.71 | 0.0506424 | 2.34e-02 |
| ARC | 3.61 | 0.0011651 | 3.62e-04 |
| AREL1 | 0.69 | 0.0000297 | 7.20e-06 |
| ARF1 | 0.65 | 0.0000000 | 0.00e+00 |
| ARF3 | 0.30 | 0.0072019 | 2.67e-03 |
| ARF4 | 0.49 | 0.0010783 | 3.33e-04 |
| ARF5 | -0.38 | 0.0549289 | 2.57e-02 |
| ARF6 | 0.61 | 0.0000136 | 3.20e-06 |
| ARFGAP1 | 0.40 | 0.0152511 | 6.12e-03 |
| ARFGAP2 | 0.88 | 0.0000012 | 2.00e-07 |
| ARFGAP3 | 1.68 | 0.0000000 | 0.00e+00 |
| ARFGEF2 | -0.33 | 0.0944208 | 4.84e-02 |
| ARFRP1 | 0.93 | 0.0003484 | 9.84e-05 |
| ARG2 | 4.01 | 0.0001056 | 2.75e-05 |
| ARHGAP1 | 1.09 | 0.0000000 | 0.00e+00 |
| ARHGAP10 | 1.18 | 0.0006200 | 1.83e-04 |
| ARHGAP11A | -3.12 | 0.0000000 | 0.00e+00 |
| ARHGAP11B | -2.77 | 0.0000000 | 0.00e+00 |
| ARHGAP12 | 0.50 | 0.0044932 | 1.59e-03 |
| ARHGAP17 | 1.70 | 0.0000000 | 0.00e+00 |
| ARHGAP18 | -1.82 | 0.0000000 | 0.00e+00 |
| ARHGAP19 | -1.48 | 0.0004823 | 1.40e-04 |
| ARHGAP19-SLIT1 | -1.31 | 0.0686446 | 3.32e-02 |
| ARHGAP21 | 0.58 | 0.0010074 | 3.10e-04 |
| ARHGAP24 | 1.72 | 0.0000003 | 1.00e-07 |
| ARHGAP26 | 0.74 | 0.0000050 | 1.10e-06 |
| ARHGAP29 | -1.67 | 0.0000000 | 0.00e+00 |
| ARHGAP30 | 4.09 | 0.0061385 | 2.24e-03 |
| ARHGAP31 | 3.65 | 0.0000000 | 0.00e+00 |
| ARHGAP32 | 0.60 | 0.0506424 | 2.34e-02 |
| ARHGAP42 | 1.71 | 0.0000000 | 0.00e+00 |
| ARHGAP5-AS1 | 1.10 | 0.0022849 | 7.59e-04 |
| ARHGDIA | 0.59 | 0.0000000 | 0.00e+00 |
| ARHGEF1 | 0.54 | 0.0001282 | 3.39e-05 |
| ARHGEF10L | 2.41 | 0.0000000 | 0.00e+00 |
| ARHGEF11 | 0.72 | 0.0000473 | 1.17e-05 |
| ARHGEF18 | 1.73 | 0.0000000 | 0.00e+00 |
| ARHGEF2 | 1.96 | 0.0000000 | 0.00e+00 |
| ARHGEF26 | -1.59 | 0.0269343 | 1.15e-02 |
| ARHGEF28 | 1.53 | 0.0000000 | 0.00e+00 |
| ARHGEF3 | 1.93 | 0.0000000 | 0.00e+00 |
| ARHGEF38 | 3.91 | 0.0852017 | 4.29e-02 |
| ARHGEF39 | -2.46 | 0.0000000 | 0.00e+00 |
| ARHGEF4 | 3.21 | 0.0000000 | 0.00e+00 |
| ARHGEF40 | 1.23 | 0.0000000 | 0.00e+00 |
| ARHGEF9 | -1.19 | 0.0034407 | 1.18e-03 |
| ARID1A | 0.42 | 0.0229050 | 9.60e-03 |
| ARID2 | -0.90 | 0.0005670 | 1.66e-04 |
| ARID3A | 1.65 | 0.0000002 | 0.00e+00 |
| ARID3B | 2.85 | 0.0000000 | 0.00e+00 |
| ARID4B | 0.46 | 0.0448712 | 2.04e-02 |
| ARID5A | 2.83 | 0.0000000 | 0.00e+00 |
| ARID5B | 3.76 | 0.0000000 | 0.00e+00 |
| ARIH1 | 0.45 | 0.0043976 | 1.55e-03 |
| ARL14 | 1.60 | 0.0989897 | 5.11e-02 |
| ARL16 | -0.69 | 0.0680787 | 3.29e-02 |
| ARL2 | -0.44 | 0.0146944 | 5.87e-03 |
| ARL3 | -1.20 | 0.0000745 | 1.89e-05 |
| ARL4D | 1.59 | 0.0000000 | 0.00e+00 |
| ARL5A | -1.20 | 0.0001085 | 2.83e-05 |
| ARL5B | 0.95 | 0.0004234 | 1.22e-04 |
| ARL6IP1 | -1.71 | 0.0000000 | 0.00e+00 |
| ARL6IP5 | -0.86 | 0.0000041 | 9.00e-07 |
| ARL8A | 1.52 | 0.0000000 | 0.00e+00 |
| ARMC1 | -1.17 | 0.0000001 | 0.00e+00 |
| ARMC5 | 0.91 | 0.0449800 | 2.05e-02 |
| ARMC6 | -1.31 | 0.0000047 | 1.00e-06 |
| ARMC9 | -0.97 | 0.0334637 | 1.47e-02 |
| ARMCX3 | 0.46 | 0.0989663 | 5.10e-02 |
| ARMT1 | -0.83 | 0.0279450 | 1.20e-02 |
| ARNT | 0.76 | 0.0008039 | 2.42e-04 |
| ARNT2 | 0.62 | 0.0034319 | 1.18e-03 |
| ARNTL2 | 2.97 | 0.0000000 | 0.00e+00 |
| ARPC1B | 0.49 | 0.0000169 | 4.00e-06 |
| ARPC4 | 0.25 | 0.0660858 | 3.18e-02 |
| ARPC4-TTLL3 | 0.54 | 0.0019891 | 6.50e-04 |
| ARPC5 | 0.58 | 0.0000030 | 6.00e-07 |
| ARPP19 | -0.32 | 0.0223639 | 9.35e-03 |
| ARRB2 | -1.86 | 0.0000000 | 0.00e+00 |
| ARRDC1 | 0.82 | 0.0022766 | 7.56e-04 |
| ARRDC2 | 2.04 | 0.0000000 | 0.00e+00 |
| ARRDC3 | 0.97 | 0.0000143 | 3.30e-06 |
| ARSA | 2.43 | 0.0000000 | 0.00e+00 |
| ARSB | 1.15 | 0.0000002 | 0.00e+00 |
| ARSD | 1.53 | 0.0000000 | 0.00e+00 |
| ARSE | 0.90 | 0.0000003 | 1.00e-07 |
| ARSJ | 1.03 | 0.0025448 | 8.53e-04 |
| ARV1 | -1.37 | 0.0002453 | 6.75e-05 |
| AS3MT | -2.96 | 0.0000019 | 4.00e-07 |
| ASAH1 | -1.21 | 0.0000000 | 0.00e+00 |
| ASAP1 | 0.64 | 0.0000025 | 5.00e-07 |
| ASAP2 | 0.95 | 0.0000001 | 0.00e+00 |
| ASB11 | 5.61 | 0.0719686 | 3.52e-02 |
| ASB13 | -0.93 | 0.0052680 | 1.89e-03 |
| ASB9 | -2.26 | 0.0000000 | 0.00e+00 |
| ASCC1 | -0.80 | 0.0001625 | 4.37e-05 |
| ASCC2 | 0.91 | 0.0000005 | 1.00e-07 |
| ASCC3 | -1.24 | 0.0000000 | 0.00e+00 |
| ASF1A | -0.65 | 0.0999221 | 5.16e-02 |
| ASF1B | -2.07 | 0.0000000 | 0.00e+00 |
| ASIC1 | -1.46 | 0.0083349 | 3.14e-03 |
| ASMTL | 0.94 | 0.0020942 | 6.89e-04 |
| ASPH | -2.28 | 0.0000000 | 0.00e+00 |
| ASPHD2 | 1.44 | 0.0704866 | 3.43e-02 |
| ASPM | -3.39 | 0.0000000 | 0.00e+00 |
| ASRGL1 | -2.39 | 0.0000000 | 0.00e+00 |
| ASS1 | 4.31 | 0.0000000 | 0.00e+00 |
| ASTN2 | -1.90 | 0.0011533 | 3.58e-04 |
| ASUN | -1.70 | 0.0000000 | 0.00e+00 |
| ASXL1 | 0.75 | 0.0000008 | 2.00e-07 |
| ATAD2 | -2.95 | 0.0000000 | 0.00e+00 |
| ATAD3A | -0.95 | 0.0017433 | 5.61e-04 |
| ATAD5 | -3.23 | 0.0000000 | 0.00e+00 |
| ATE1 | -1.01 | 0.0000086 | 1.90e-06 |
| ATF3 | 4.88 | 0.0000000 | 0.00e+00 |
| ATF4 | 0.56 | 0.0000000 | 0.00e+00 |
| ATF5 | 1.28 | 0.0000000 | 0.00e+00 |
| ATF6 | 0.80 | 0.0000023 | 5.00e-07 |
| ATF6B | 0.98 | 0.0063701 | 2.33e-03 |
| ATF7 | 0.71 | 0.0022925 | 7.62e-04 |
| ATF7IP | 0.57 | 0.0426432 | 1.93e-02 |
| ATG13 | 0.98 | 0.0000000 | 0.00e+00 |
| ATG14 | 1.00 | 0.0009150 | 2.79e-04 |
| ATG2A | 2.54 | 0.0000000 | 0.00e+00 |
| ATG2B | -1.39 | 0.0000024 | 5.00e-07 |
| ATG3 | -0.50 | 0.0456153 | 2.08e-02 |
| ATG4C | -1.77 | 0.0049842 | 1.78e-03 |
| ATG4D | 1.19 | 0.0001393 | 3.70e-05 |
| ATG7 | 1.76 | 0.0000000 | 0.00e+00 |
| ATG9A | 1.16 | 0.0000000 | 0.00e+00 |
| ATHL1 | 0.95 | 0.0021896 | 7.23e-04 |
| ATIC | -0.87 | 0.0000000 | 0.00e+00 |
| ATL2 | -0.62 | 0.0014944 | 4.74e-04 |
| ATL3 | -0.62 | 0.0014531 | 4.60e-04 |
| ATM | -1.60 | 0.0000001 | 0.00e+00 |
| ATN1 | 1.33 | 0.0000000 | 0.00e+00 |
| ATOH8 | 1.31 | 0.0387878 | 1.73e-02 |
| ATP10A | 1.28 | 0.0000028 | 6.00e-07 |
| ATP10D | 1.57 | 0.0003108 | 8.69e-05 |
| ATP11C | -1.05 | 0.0051402 | 1.84e-03 |
| ATP13A1 | 1.33 | 0.0000000 | 0.00e+00 |
| ATP13A2 | 1.33 | 0.0000000 | 0.00e+00 |
| ATP13A3 | -0.92 | 0.0000020 | 4.00e-07 |
| ATP1B1 | 1.31 | 0.0000000 | 0.00e+00 |
| ATP1B3 | 1.53 | 0.0000000 | 0.00e+00 |
| ATP2A1-AS1 | -2.25 | 0.0341413 | 1.50e-02 |
| ATP2A2 | 1.02 | 0.0000000 | 0.00e+00 |
| ATP2B1 | -0.28 | 0.0450821 | 2.06e-02 |
| ATP2B4 | 3.18 | 0.0000000 | 0.00e+00 |
| ATP4A | 12.90 | 0.0000000 | 0.00e+00 |
| ATP5A1 | -1.12 | 0.0000000 | 0.00e+00 |
| ATP5B | -1.06 | 0.0000000 | 0.00e+00 |
| ATP5C1 | -0.85 | 0.0000000 | 0.00e+00 |
| ATP5E | -0.61 | 0.0000072 | 1.60e-06 |
| ATP5F1 | -1.63 | 0.0000000 | 0.00e+00 |
| ATP5G1 | -1.81 | 0.0000000 | 0.00e+00 |
| ATP5G2 | -0.29 | 0.0388034 | 1.73e-02 |
| ATP5G3 | -1.44 | 0.0000000 | 0.00e+00 |
| ATP5H | -0.50 | 0.0018987 | 6.17e-04 |
| ATP5I | -0.77 | 0.0000437 | 1.08e-05 |
| ATP5J | -0.78 | 0.0010787 | 3.33e-04 |
| ATP5J2 | -0.80 | 0.0000000 | 0.00e+00 |
| ATP5L | -0.81 | 0.0000046 | 1.00e-06 |
| ATP5L2 | -1.37 | 0.0793708 | 3.94e-02 |
| ATP5O | -0.96 | 0.0000102 | 2.30e-06 |
| ATP5SL | -0.50 | 0.0035436 | 1.22e-03 |
| ATP6AP1 | 1.21 | 0.0000000 | 0.00e+00 |
| ATP6AP2 | -0.82 | 0.0000053 | 1.10e-06 |
| ATP6V0A1 | 0.92 | 0.0000035 | 8.00e-07 |
| ATP6V0C | 0.90 | 0.0000000 | 0.00e+00 |
| ATP6V0E1 | 0.37 | 0.0107529 | 4.16e-03 |
| ATP6V0E2 | -1.51 | 0.0000001 | 0.00e+00 |
| ATP6V1A | -0.79 | 0.0008609 | 2.61e-04 |
| ATP6V1B2 | -0.73 | 0.0001769 | 4.77e-05 |
| ATP6V1C1 | -0.46 | 0.0354180 | 1.57e-02 |
| ATP6V1C2 | 6.75 | 0.0000000 | 0.00e+00 |
| ATP7A | -1.01 | 0.0055083 | 1.98e-03 |
| ATP8B2 | 0.38 | 0.0702788 | 3.41e-02 |
| ATP9A | 0.39 | 0.0689924 | 3.34e-02 |
| ATP9B | 0.84 | 0.0010463 | 3.23e-04 |
| ATPAF1 | -1.07 | 0.0001144 | 3.00e-05 |
| ATPIF1 | -0.86 | 0.0019891 | 6.50e-04 |
| ATR | -1.77 | 0.0000000 | 0.00e+00 |
| ATRAID | -0.68 | 0.0000099 | 2.20e-06 |
| ATRIP | -1.05 | 0.0292715 | 1.26e-02 |
| ATRNL1 | 2.70 | 0.0981007 | 5.05e-02 |
| ATRX | -0.53 | 0.0103292 | 3.98e-03 |
| ATXN1 | 1.83 | 0.0000000 | 0.00e+00 |
| ATXN1L | 0.65 | 0.0003091 | 8.64e-05 |
| ATXN2 | 0.88 | 0.0000887 | 2.28e-05 |
| ATXN2L | 1.99 | 0.0000000 | 0.00e+00 |
| ATXN7 | 0.77 | 0.0250829 | 1.06e-02 |
| ATXN7L1 | 1.67 | 0.0000107 | 2.40e-06 |
| ATXN7L3 | 0.86 | 0.0000000 | 0.00e+00 |
| AUNIP | -2.19 | 0.0004136 | 1.19e-04 |
| AUP1 | 0.63 | 0.0000000 | 0.00e+00 |
| AURKA | -1.08 | 0.0000000 | 0.00e+00 |
| AURKAIP1 | -0.66 | 0.0000848 | 2.18e-05 |
| AURKB | -2.07 | 0.0000000 | 0.00e+00 |
| AUTS2 | 2.48 | 0.0000000 | 0.00e+00 |
| AVIL | 3.95 | 0.0095090 | 3.63e-03 |
| AVPI1 | 1.25 | 0.0000000 | 0.00e+00 |
| AVPR1A | 4.70 | 0.0173878 | 7.08e-03 |
| AXIN1 | 0.95 | 0.0000000 | 0.00e+00 |
| AXIN2 | 1.73 | 0.0000001 | 0.00e+00 |
| AXL | 1.46 | 0.0000000 | 0.00e+00 |
| AZIN1 | 1.09 | 0.0000000 | 0.00e+00 |
| AZIN2 | 3.05 | 0.0000291 | 7.00e-06 |
| B2M | 3.70 | 0.0000000 | 0.00e+00 |
| B3GALNT2 | -0.99 | 0.0014290 | 4.52e-04 |
| B3GALT5 | 1.50 | 0.0553377 | 2.59e-02 |
| B3GAT3 | 0.46 | 0.0544587 | 2.55e-02 |
| B3GNT2 | 0.59 | 0.0087402 | 3.31e-03 |
| B3GNT3 | 0.98 | 0.0006066 | 1.79e-04 |
| B3GNT5 | 1.55 | 0.0001011 | 2.62e-05 |
| B3GNT7 | 2.18 | 0.0000252 | 6.00e-06 |
| B3GNTL1 | -2.12 | 0.0425915 | 1.93e-02 |
| B4GALNT3 | 2.70 | 0.0049420 | 1.76e-03 |
| B4GALNT4 | -1.87 | 0.0004380 | 1.26e-04 |
| B4GALT1 | 1.60 | 0.0000000 | 0.00e+00 |
| B4GALT2 | -0.82 | 0.0008893 | 2.70e-04 |
| B4GALT3 | 0.83 | 0.0009276 | 2.83e-04 |
| B4GALT4 | -0.93 | 0.0000000 | 0.00e+00 |
| B4GALT5 | 1.74 | 0.0000000 | 0.00e+00 |
| B4GAT1 | -0.99 | 0.0141908 | 5.65e-03 |
| B9D1 | -1.58 | 0.0000099 | 2.20e-06 |
| BAAT | -1.26 | 0.0535417 | 2.50e-02 |
| BACH1 | 1.75 | 0.0000000 | 0.00e+00 |
| BACH2 | 3.29 | 0.0000578 | 1.45e-05 |
| BAD | -0.57 | 0.0368854 | 1.64e-02 |
| BAG2 | -1.65 | 0.0000001 | 0.00e+00 |
| BAG3 | 1.09 | 0.0000000 | 0.00e+00 |
| BAHCC1 | 0.97 | 0.0000041 | 9.00e-07 |
| BAHD1 | 0.65 | 0.0336785 | 1.48e-02 |
| BAIAP2-AS1 | -1.39 | 0.0208589 | 8.65e-03 |
| BAIAP2L1 | 0.34 | 0.0398562 | 1.79e-02 |
| BAIAP2L2 | 1.83 | 0.0000000 | 0.00e+00 |
| BAK1 | 0.72 | 0.0210460 | 8.74e-03 |
| BAMBI | 2.70 | 0.0000000 | 0.00e+00 |
| BANF1 | -0.99 | 0.0000000 | 0.00e+00 |
| BANP | 1.02 | 0.0033453 | 1.15e-03 |
| BARD1 | -2.17 | 0.0000005 | 1.00e-07 |
| BASP1 | 0.51 | 0.0000002 | 0.00e+00 |
| BATF2 | 6.34 | 0.0000000 | 0.00e+00 |
| BATF3 | 1.52 | 0.0000001 | 0.00e+00 |
| BAX | -0.31 | 0.0649980 | 3.12e-02 |
| BAZ1A | 0.75 | 0.0000132 | 3.00e-06 |
| BAZ2A | 2.09 | 0.0000000 | 0.00e+00 |
| BBC3 | 4.98 | 0.0000000 | 0.00e+00 |
| BBIP1 | -0.79 | 0.0388031 | 1.73e-02 |
| BBS12 | 1.26 | 0.0338955 | 1.49e-02 |
| BBS2 | -1.01 | 0.0021081 | 6.94e-04 |
| BBX | -0.42 | 0.0184609 | 7.56e-03 |
| BCAM | 0.86 | 0.0001259 | 3.32e-05 |
| BCAR1 | 2.57 | 0.0000000 | 0.00e+00 |
| BCAR3 | 1.08 | 0.0000000 | 0.00e+00 |
| BCAS1 | -2.95 | 0.0000009 | 2.00e-07 |
| BCAS4 | -1.29 | 0.0001849 | 5.00e-05 |
| BCAT2 | -1.62 | 0.0000000 | 0.00e+00 |
| BCCIP | -1.28 | 0.0000000 | 0.00e+00 |
| BCDIN3D | 0.73 | 0.0979293 | 5.04e-02 |
| BCKDHA | 0.54 | 0.0024616 | 8.23e-04 |
| BCKDHB | -1.38 | 0.0390334 | 1.75e-02 |
| BCKDK | 0.74 | 0.0012303 | 3.84e-04 |
| BCL10 | 2.12 | 0.0000000 | 0.00e+00 |
| BCL2A1 | 5.31 | 0.0000000 | 0.00e+00 |
| BCL2L1 | 0.75 | 0.0000000 | 0.00e+00 |
| BCL2L11 | 2.01 | 0.0000000 | 0.00e+00 |
| BCL2L12 | -0.58 | 0.0260308 | 1.11e-02 |
| BCL2L13 | 1.33 | 0.0000000 | 0.00e+00 |
| BCL2L14 | 5.51 | 0.0022542 | 7.48e-04 |
| BCL2L15 | 4.07 | 0.0000000 | 0.00e+00 |
| BCL2L2 | 0.77 | 0.0001370 | 3.64e-05 |
| BCL3 | 2.21 | 0.0000000 | 0.00e+00 |
| BCL6 | 3.16 | 0.0000000 | 0.00e+00 |
| BCL7A | 1.41 | 0.0000000 | 0.00e+00 |
| BCL7B | 0.73 | 0.0003619 | 1.03e-04 |
| BCL7C | -1.22 | 0.0000286 | 6.90e-06 |
| BCL9L | 1.64 | 0.0000000 | 0.00e+00 |
| BCLAF1 | -1.52 | 0.0000000 | 0.00e+00 |
| BCOR | 0.72 | 0.0001211 | 3.19e-05 |
| BCORL1 | 3.10 | 0.0000000 | 0.00e+00 |
| BCR | 1.32 | 0.0000000 | 0.00e+00 |
| BCS1L | -1.56 | 0.0000002 | 0.00e+00 |
| BCYRN1 | -2.37 | 0.0000001 | 0.00e+00 |
| BDKRB1 | 2.41 | 0.0000025 | 5.00e-07 |
| BDKRB2 | 2.43 | 0.0000000 | 0.00e+00 |
| BDP1 | -0.90 | 0.0186714 | 7.66e-03 |
| BEAN1 | 3.72 | 0.0000000 | 0.00e+00 |
| BECN1 | 0.39 | 0.0156669 | 6.31e-03 |
| BEGAIN | 2.40 | 0.0001910 | 5.18e-05 |
| BEST1 | 3.00 | 0.0097251 | 3.72e-03 |
| BET1L | 0.47 | 0.0231986 | 9.75e-03 |
| BHLHE40 | 1.60 | 0.0000000 | 0.00e+00 |
| BHLHE41 | 5.56 | 0.0000058 | 1.30e-06 |
| BICC1 | -1.07 | 0.0000001 | 0.00e+00 |
| BID | 1.09 | 0.0000000 | 0.00e+00 |
| BIK | 4.92 | 0.0000000 | 0.00e+00 |
| BIN1 | -1.85 | 0.0000000 | 0.00e+00 |
| BIN3 | 0.66 | 0.0096150 | 3.67e-03 |
| BIRC2 | 0.80 | 0.0001521 | 4.07e-05 |
| BIRC3 | 4.20 | 0.0000000 | 0.00e+00 |
| BIRC5 | -3.24 | 0.0000000 | 0.00e+00 |
| BIRC6 | -0.60 | 0.0000750 | 1.91e-05 |
| BISPR | 7.69 | 0.0015118 | 4.80e-04 |
| BIVM | -1.49 | 0.0058096 | 2.10e-03 |
| BLACAT1 | 5.61 | 0.0735043 | 3.61e-02 |
| BLCAP | 0.73 | 0.0012093 | 3.77e-04 |
| BLM | -3.18 | 0.0000000 | 0.00e+00 |
| BLMH | -0.52 | 0.0297227 | 1.28e-02 |
| BLOC1S1 | -0.58 | 0.0115972 | 4.53e-03 |
| BLOC1S3 | 1.30 | 0.0004624 | 1.34e-04 |
| BLOC1S4 | 0.71 | 0.0749372 | 3.68e-02 |
| BLOC1S5-TXNDC5 | -1.03 | 0.0000000 | 0.00e+00 |
| BLOC1S6 | -1.12 | 0.0005213 | 1.52e-04 |
| BMF | 1.86 | 0.0009662 | 2.96e-04 |
| BMP1 | 1.65 | 0.0000000 | 0.00e+00 |
| BMP2 | 7.72 | 0.0000010 | 2.00e-07 |
| BMP6 | 2.71 | 0.0000000 | 0.00e+00 |
| BMP7 | 1.97 | 0.0052665 | 1.89e-03 |
| BMP8B | -1.06 | 0.0757049 | 3.73e-02 |
| BMPR1A | -0.89 | 0.0096936 | 3.71e-03 |
| BNIP3L | 0.41 | 0.0606222 | 2.88e-02 |
| BOC | 1.41 | 0.0341057 | 1.50e-02 |
| BOD1L1 | -0.57 | 0.0060226 | 2.19e-03 |
| BOLA1 | -0.74 | 0.0968441 | 4.97e-02 |
| BOLA3 | -1.53 | 0.0000005 | 1.00e-07 |
| BOP1 | -1.69 | 0.0000000 | 0.00e+00 |
| BORA | -1.41 | 0.0045278 | 1.60e-03 |
| BPGM | 1.76 | 0.0000000 | 0.00e+00 |
| BPTF | -0.63 | 0.0020270 | 6.64e-04 |
| BRAF | 1.41 | 0.0010915 | 3.38e-04 |
| BRCA1 | -3.11 | 0.0000000 | 0.00e+00 |
| BRCA2 | -1.74 | 0.0001119 | 2.93e-05 |
| BRCC3 | -1.10 | 0.0002845 | 7.91e-05 |
| BRD1 | 1.00 | 0.0001375 | 3.65e-05 |
| BRD3 | 0.69 | 0.0009103 | 2.77e-04 |
| BRD4 | 2.00 | 0.0000000 | 0.00e+00 |
| BRD8 | -0.36 | 0.0825036 | 4.13e-02 |
| BRF2 | 0.87 | 0.0187038 | 7.67e-03 |
| BRI3 | 0.68 | 0.0000357 | 8.70e-06 |
| BRI3BP | -1.41 | 0.0000784 | 2.00e-05 |
| BRIP1 | -1.50 | 0.0000952 | 2.46e-05 |
| BRIX1 | -1.05 | 0.0000291 | 7.00e-06 |
| BRMS1 | -0.62 | 0.0016910 | 5.43e-04 |
| BRMS1L | -1.27 | 0.0065429 | 2.40e-03 |
| BRPF1 | 0.64 | 0.0060584 | 2.20e-03 |
| BRPF3 | 1.10 | 0.0000088 | 2.00e-06 |
| BRSK1 | 1.98 | 0.0000006 | 1.00e-07 |
| BSCL2 | -1.49 | 0.0020541 | 6.74e-04 |
| BSDC1 | 1.26 | 0.0000000 | 0.00e+00 |
| BSG | 0.82 | 0.0000000 | 0.00e+00 |
| BST1 | -1.48 | 0.0031322 | 1.07e-03 |
| BST2 | 8.97 | 0.0000000 | 0.00e+00 |
| BTBD1 | -0.42 | 0.0232294 | 9.76e-03 |
| BTBD11 | -1.21 | 0.0000015 | 3.00e-07 |
| BTBD3 | -1.01 | 0.0006147 | 1.81e-04 |
| BTBD6 | -0.66 | 0.0280119 | 1.20e-02 |
| BTBD7 | -0.91 | 0.0654076 | 3.14e-02 |
| BTBD9 | 1.62 | 0.0000069 | 1.50e-06 |
| BTF3L4 | -0.70 | 0.0377814 | 1.68e-02 |
| BTG1 | 2.60 | 0.0000000 | 0.00e+00 |
| BTG2 | 1.26 | 0.0000844 | 2.16e-05 |
| BTG3 | 1.65 | 0.0000000 | 0.00e+00 |
| BTN2A1 | 2.32 | 0.0000000 | 0.00e+00 |
| BTN2A2 | 3.72 | 0.0000000 | 0.00e+00 |
| BTN2A3P | 2.78 | 0.0000001 | 0.00e+00 |
| BTN3A1 | 2.62 | 0.0000000 | 0.00e+00 |
| BTN3A2 | 1.74 | 0.0000000 | 0.00e+00 |
| BTN3A3 | 3.46 | 0.0000000 | 0.00e+00 |
| BTRC | 0.53 | 0.0917306 | 4.68e-02 |
| BUB1 | -2.64 | 0.0000000 | 0.00e+00 |
| BUB1B | -3.85 | 0.0000000 | 0.00e+00 |
| BUB3 | -1.16 | 0.0000000 | 0.00e+00 |
| BVES | 1.44 | 0.0000253 | 6.00e-06 |
| BZW1 | -0.58 | 0.0000007 | 1.00e-07 |
| BZW2 | -1.05 | 0.0000000 | 0.00e+00 |
| C10orf10 | 5.90 | 0.0000000 | 0.00e+00 |
| C10orf11 | 4.51 | 0.0000503 | 1.25e-05 |
| C10orf2 | -0.52 | 0.0548320 | 2.56e-02 |
| C10orf32-ASMT | -1.41 | 0.0013373 | 4.20e-04 |
| C10orf54 | 1.24 | 0.0000000 | 0.00e+00 |
| C10orf55 | 2.76 | 0.0065790 | 2.41e-03 |
| C10orf91 | -4.32 | 0.0532516 | 2.48e-02 |
| C11orf1 | -1.22 | 0.0400850 | 1.80e-02 |
| C11orf31 | -1.14 | 0.0000049 | 1.10e-06 |
| C11orf58 | -0.33 | 0.0366800 | 1.63e-02 |
| C11orf68 | 1.07 | 0.0000529 | 1.32e-05 |
| C11orf70 | -1.41 | 0.0863409 | 4.35e-02 |
| C11orf71 | -1.36 | 0.0396101 | 1.77e-02 |
| C11orf84 | 0.63 | 0.0010202 | 3.14e-04 |
| C11orf96 | 7.19 | 0.0000000 | 0.00e+00 |
| C12orf10 | -0.68 | 0.0022246 | 7.37e-04 |
| C12orf29 | -0.89 | 0.0124816 | 4.91e-03 |
| C12orf4 | -0.87 | 0.0181435 | 7.42e-03 |
| C12orf45 | 0.63 | 0.0768382 | 3.80e-02 |
| C12orf56 | 6.83 | 0.0089857 | 3.41e-03 |
| C12orf57 | 0.33 | 0.0548300 | 2.56e-02 |
| C12orf66 | -0.80 | 0.0810496 | 4.04e-02 |
| C12orf75 | -0.35 | 0.0964468 | 4.95e-02 |
| C14orf105 | -2.72 | 0.0034241 | 1.18e-03 |
| C14orf119 | 0.34 | 0.0806217 | 4.02e-02 |
| C14orf132 | -2.41 | 0.0003233 | 9.07e-05 |
| C14orf142 | -1.52 | 0.0227523 | 9.53e-03 |
| C14orf2 | -1.00 | 0.0000000 | 0.00e+00 |
| C14orf80 | -0.93 | 0.0018210 | 5.89e-04 |
| C15orf26 | 5.97 | 0.0428851 | 1.94e-02 |
| C15orf38-AP3S2 | 0.39 | 0.0080316 | 3.01e-03 |
| C15orf39 | 2.84 | 0.0000000 | 0.00e+00 |
| C15orf41 | -0.93 | 0.0751844 | 3.70e-02 |
| C15orf48 | 7.17 | 0.0000000 | 0.00e+00 |
| C16orf13 | -0.86 | 0.0050297 | 1.80e-03 |
| C16orf45 | 1.83 | 0.0000000 | 0.00e+00 |
| C16orf52 | 0.85 | 0.0557065 | 2.61e-02 |
| C16orf58 | 0.46 | 0.0059868 | 2.17e-03 |
| C16orf59 | -1.87 | 0.0000068 | 1.50e-06 |
| C16orf72 | 0.45 | 0.0660333 | 3.18e-02 |
| C16orf74 | -2.40 | 0.0007677 | 2.30e-04 |
| C16orf87 | 0.85 | 0.0671597 | 3.24e-02 |
| C16orf93 | -1.75 | 0.0829323 | 4.15e-02 |
| C17orf104 | -2.62 | 0.0000002 | 0.00e+00 |
| C17orf51 | -0.59 | 0.0854366 | 4.30e-02 |
| C17orf59 | 0.68 | 0.0556105 | 2.61e-02 |
| C17orf62 | 0.72 | 0.0000126 | 2.90e-06 |
| C17orf67 | 1.18 | 0.0497074 | 2.29e-02 |
| C17orf75 | -0.53 | 0.0935357 | 4.79e-02 |
| C17orf80 | -0.86 | 0.0031539 | 1.08e-03 |
| C17orf85 | -0.49 | 0.0255259 | 1.09e-02 |
| C17orf89 | -1.65 | 0.0000000 | 0.00e+00 |
| C17orf96 | 2.15 | 0.0000000 | 0.00e+00 |
| C18orf54 | -3.94 | 0.0001614 | 4.34e-05 |
| C19orf12 | 2.36 | 0.0000000 | 0.00e+00 |
| C19orf43 | -0.39 | 0.0271651 | 1.16e-02 |
| C19orf47 | 0.47 | 0.0878814 | 4.44e-02 |
| C19orf48 | -1.23 | 0.0000000 | 0.00e+00 |
| C19orf54 | 0.87 | 0.0108662 | 4.21e-03 |
| C19orf66 | 2.76 | 0.0000000 | 0.00e+00 |
| C19orf70 | -1.11 | 0.0003048 | 8.51e-05 |
| C1GALT1C1 | -0.67 | 0.0186710 | 7.65e-03 |
| C1GALT1C1L | -2.58 | 0.0756914 | 3.73e-02 |
| C1orf106 | 0.98 | 0.0000000 | 0.00e+00 |
| C1orf109 | -0.94 | 0.0134545 | 5.34e-03 |
| C1orf112 | -2.10 | 0.0000000 | 0.00e+00 |
| C1orf116 | 0.69 | 0.0791304 | 3.92e-02 |
| C1orf123 | -0.79 | 0.0275595 | 1.18e-02 |
| C1orf198 | 0.46 | 0.0398081 | 1.79e-02 |
| C1orf21 | -1.87 | 0.0070663 | 2.61e-03 |
| C1orf216 | -1.09 | 0.0220170 | 9.18e-03 |
| C1orf228 | 2.70 | 0.0628466 | 3.00e-02 |
| C1orf233 | -1.89 | 0.0751642 | 3.70e-02 |
| C1orf27 | -0.71 | 0.0215407 | 8.96e-03 |
| C1QBP | -1.27 | 0.0000000 | 0.00e+00 |
| C1QL1 | 1.47 | 0.0000000 | 0.00e+00 |
| C1QTNF1 | 5.41 | 0.0000000 | 0.00e+00 |
| C1QTNF3-AMACR | -0.79 | 0.0596059 | 2.82e-02 |
| C1QTNF6 | -0.57 | 0.0362062 | 1.61e-02 |
| C1R | 5.04 | 0.0000000 | 0.00e+00 |
| C1RL | 2.10 | 0.0000000 | 0.00e+00 |
| C1RL-AS1 | 0.69 | 0.0135274 | 5.37e-03 |
| C1S | 3.96 | 0.0000000 | 0.00e+00 |
| C20orf141 | 5.94 | 0.0000006 | 1.00e-07 |
| C20orf194 | 0.76 | 0.0195807 | 8.07e-03 |
| C20orf27 | -1.72 | 0.0000000 | 0.00e+00 |
| C21orf58 | -3.01 | 0.0000003 | 1.00e-07 |
| C21orf59 | -0.81 | 0.0057207 | 2.07e-03 |
| C21orf91 | 1.08 | 0.0625997 | 2.99e-02 |
| C22orf29 | -0.76 | 0.0851805 | 4.29e-02 |
| C2CD4A | 5.60 | 0.0016555 | 5.30e-04 |
| C2CD5 | -1.38 | 0.0000009 | 2.00e-07 |
| C2orf44 | -0.99 | 0.0016675 | 5.34e-04 |
| C2orf48 | -3.00 | 0.0396910 | 1.78e-02 |
| C2orf69 | -1.33 | 0.0014334 | 4.54e-04 |
| C2orf72 | -1.32 | 0.0031021 | 1.06e-03 |
| C2orf74 | -1.05 | 0.0708334 | 3.45e-02 |
| C3 | 6.63 | 0.0000000 | 0.00e+00 |
| C3AR1 | 6.14 | 0.0313259 | 1.36e-02 |
| C3orf14 | -2.26 | 0.0000222 | 5.30e-06 |
| C3orf17 | 0.43 | 0.0635907 | 3.04e-02 |
| C3orf18 | -1.54 | 0.0602210 | 2.86e-02 |
| C3orf52 | 1.77 | 0.0000013 | 3.00e-07 |
| C4orf19 | 5.90 | 0.0492013 | 2.27e-02 |
| C4orf27 | -1.18 | 0.0085970 | 3.25e-03 |
| C4orf32 | 1.20 | 0.0450061 | 2.05e-02 |
| C4orf46 | -2.26 | 0.0000225 | 5.30e-06 |
| C5 | -3.20 | 0.0000000 | 0.00e+00 |
| C5orf15 | 1.41 | 0.0000000 | 0.00e+00 |
| C5orf24 | -0.79 | 0.0009812 | 3.01e-04 |
| C5orf30 | -1.12 | 0.0021543 | 7.11e-04 |
| C5orf34 | -1.75 | 0.0054010 | 1.94e-03 |
| C5orf42 | -1.26 | 0.0008645 | 2.62e-04 |
| C5orf45 | -1.59 | 0.0311640 | 1.35e-02 |
| C5orf51 | -0.63 | 0.0327447 | 1.43e-02 |
| C5orf56 | 4.76 | 0.0000094 | 2.10e-06 |
| C6orf1 | 0.99 | 0.0052125 | 1.87e-03 |
| C6orf106 | 1.34 | 0.0000000 | 0.00e+00 |
| C6orf132 | 3.06 | 0.0000001 | 0.00e+00 |
| C6orf141 | 0.77 | 0.0955321 | 4.90e-02 |
| C6orf223 | 3.22 | 0.0250436 | 1.06e-02 |
| C6orf48 | 1.82 | 0.0000030 | 6.00e-07 |
| C6orf58 | 7.59 | 0.0018648 | 6.05e-04 |
| C6orf62 | 0.88 | 0.0000171 | 4.00e-06 |
| C7orf26 | 0.95 | 0.0004508 | 1.30e-04 |
| C7orf31 | 1.21 | 0.0792688 | 3.93e-02 |
| C7orf43 | 0.93 | 0.0059143 | 2.14e-03 |
| C7orf50 | -1.54 | 0.0000000 | 0.00e+00 |
| C7orf55 | -0.84 | 0.0639825 | 3.06e-02 |
| C7orf55-LUC7L2 | -0.46 | 0.0250406 | 1.06e-02 |
| C7orf73 | -0.77 | 0.0103688 | 4.00e-03 |
| C8orf33 | -0.66 | 0.0006027 | 1.77e-04 |
| C8orf4 | 1.54 | 0.0000000 | 0.00e+00 |
| C8orf46 | 4.25 | 0.0463868 | 2.12e-02 |
| C8orf59 | -0.57 | 0.0297464 | 1.29e-02 |
| C8orf76 | 0.56 | 0.0695098 | 3.37e-02 |
| C9orf142 | -0.86 | 0.0036784 | 1.27e-03 |
| C9orf16 | 2.61 | 0.0000000 | 0.00e+00 |
| C9orf3 | -0.97 | 0.0000000 | 0.00e+00 |
| C9orf78 | -0.51 | 0.0253592 | 1.08e-02 |
| CA12 | 0.60 | 0.0000000 | 0.00e+00 |
| CA2 | 3.46 | 0.0000000 | 0.00e+00 |
| CA4 | 5.19 | 0.0054571 | 1.96e-03 |
| CA8 | 2.86 | 0.0000000 | 0.00e+00 |
| CA9 | 2.19 | 0.0412534 | 1.86e-02 |
| CAB39 | 1.04 | 0.0000000 | 0.00e+00 |
| CACNA1A | 3.12 | 0.0332280 | 1.45e-02 |
| CACNA1G | -2.46 | 0.0000525 | 1.31e-05 |
| CACNA1I | 3.09 | 0.0152511 | 6.12e-03 |
| CACNA2D1 | -0.80 | 0.0045606 | 1.61e-03 |
| CACNA2D3 | 5.97 | 0.0423108 | 1.91e-02 |
| CACNB1 | 2.09 | 0.0000000 | 0.00e+00 |
| CACNG4 | -2.25 | 0.0017363 | 5.59e-04 |
| CACNG6 | 1.61 | 0.0000000 | 0.00e+00 |
| CACNG8 | 0.99 | 0.0054161 | 1.95e-03 |
| CACYBP | -1.07 | 0.0000000 | 0.00e+00 |
| CAD | -1.31 | 0.0000000 | 0.00e+00 |
| CADM3 | 7.00 | 0.0064268 | 2.35e-03 |
| CADPS2 | 2.62 | 0.0120168 | 4.70e-03 |
| CALB1 | 4.21 | 0.0494635 | 2.28e-02 |
| CALB2 | -2.51 | 0.0000002 | 0.00e+00 |
| CALCOCO1 | 1.65 | 0.0000000 | 0.00e+00 |
| CALCOCO2 | 0.80 | 0.0000000 | 0.00e+00 |
| CALM1 | -0.63 | 0.0000000 | 0.00e+00 |
| CALM2 | -0.91 | 0.0000000 | 0.00e+00 |
| CALM3 | -0.41 | 0.0003194 | 8.95e-05 |
| CALML4 | -1.54 | 0.0025843 | 8.67e-04 |
| CALR | 0.67 | 0.0000000 | 0.00e+00 |
| CALU | -0.77 | 0.0000000 | 0.00e+00 |
| CALY | 3.97 | 0.0010907 | 3.37e-04 |
| CAMK1D | 1.71 | 0.0000000 | 0.00e+00 |
| CAMK1G | 8.48 | 0.0002622 | 7.25e-05 |
| CAMK2D | -0.41 | 0.0704066 | 3.42e-02 |
| CAMK2G | -0.64 | 0.0167603 | 6.79e-03 |
| CAMKK1 | -2.59 | 0.0000000 | 0.00e+00 |
| CAMKK2 | 0.41 | 0.0449104 | 2.05e-02 |
| CAMSAP3 | 1.44 | 0.0044491 | 1.57e-03 |
| CAMTA2 | 1.28 | 0.0000000 | 0.00e+00 |
| CAND1 | -0.62 | 0.0000018 | 4.00e-07 |
| CANT1 | 1.29 | 0.0000000 | 0.00e+00 |
| CANX | -0.23 | 0.0113064 | 4.41e-03 |
| CAP1 | 0.31 | 0.0047241 | 1.68e-03 |
| CAPG | 2.33 | 0.0000000 | 0.00e+00 |
| CAPN1 | 0.94 | 0.0000000 | 0.00e+00 |
| CAPN15 | 0.73 | 0.0004450 | 1.28e-04 |
| CAPN2 | -0.20 | 0.0795183 | 3.95e-02 |
| CAPN3 | 2.77 | 0.0838975 | 4.22e-02 |
| CAPN5 | 0.93 | 0.0497446 | 2.30e-02 |
| CAPN7 | -1.12 | 0.0000543 | 1.36e-05 |
| CAPRIN1 | -0.59 | 0.0000001 | 0.00e+00 |
| CAPRIN2 | 0.55 | 0.0463834 | 2.12e-02 |
| CAPZA1 | -0.53 | 0.0008074 | 2.43e-04 |
| CAPZA2 | -0.78 | 0.0000055 | 1.20e-06 |
| CARD11 | 5.10 | 0.0000000 | 0.00e+00 |
| CARD14 | 4.97 | 0.0001465 | 3.91e-05 |
| CARD16 | 6.14 | 0.0313259 | 1.36e-02 |
| CARD6 | 1.44 | 0.0002670 | 7.40e-05 |
| CARHSP1 | 0.86 | 0.0000000 | 0.00e+00 |
| CARNMT1 | -1.89 | 0.0000045 | 1.00e-06 |
| CARS | 1.63 | 0.0000000 | 0.00e+00 |
| CARS2 | -1.76 | 0.0000000 | 0.00e+00 |
| CASC10 | -0.83 | 0.0245147 | 1.04e-02 |
| CASC3 | 0.50 | 0.0008308 | 2.51e-04 |
| CASC5 | -3.54 | 0.0000000 | 0.00e+00 |
| CASD1 | -1.43 | 0.0007956 | 2.39e-04 |
| CASK | -0.83 | 0.0008353 | 2.52e-04 |
| CASKIN2 | 1.22 | 0.0000004 | 1.00e-07 |
| CASP1 | 4.56 | 0.0000378 | 9.20e-06 |
| CASP10 | 2.68 | 0.0000000 | 0.00e+00 |
| CASP4 | 3.35 | 0.0000000 | 0.00e+00 |
| CASP6 | -1.35 | 0.0018440 | 5.97e-04 |
| CASP7 | 1.83 | 0.0000000 | 0.00e+00 |
| CASP8AP2 | -1.52 | 0.0000496 | 1.24e-05 |
| CAST | 0.22 | 0.0632904 | 3.02e-02 |
| CASZ1 | 3.03 | 0.0000000 | 0.00e+00 |
| CAT | -2.05 | 0.0000000 | 0.00e+00 |
| CATSPER1 | 2.44 | 0.0262728 | 1.12e-02 |
| CBFA2T2 | 0.88 | 0.0088027 | 3.33e-03 |
| CBL | 0.46 | 0.0711988 | 3.47e-02 |
| CBLL1 | 1.04 | 0.0000021 | 4.00e-07 |
| CBLN2 | -4.00 | 0.0334242 | 1.46e-02 |
| CBLN3 | 2.54 | 0.0000002 | 0.00e+00 |
| CBR1 | -1.11 | 0.0000000 | 0.00e+00 |
| CBR3 | 1.28 | 0.0000064 | 1.40e-06 |
| CBS | 1.61 | 0.0000000 | 0.00e+00 |
| CBWD1 | -0.75 | 0.0336731 | 1.48e-02 |
| CBWD5 | -0.59 | 0.0554699 | 2.60e-02 |
| CBWD6 | -0.83 | 0.0338839 | 1.49e-02 |
| CBX1 | -1.19 | 0.0000000 | 0.00e+00 |
| CBX2 | -1.75 | 0.0000000 | 0.00e+00 |
| CBX3 | -1.53 | 0.0000000 | 0.00e+00 |
| CBX4 | 1.05 | 0.0000000 | 0.00e+00 |
| CBX5 | -1.78 | 0.0000000 | 0.00e+00 |
| CBX6 | 0.84 | 0.0000002 | 0.00e+00 |
| CBX8 | -0.83 | 0.0270641 | 1.16e-02 |
| CBY1 | -0.86 | 0.0998977 | 5.16e-02 |
| CC2D1A | 1.34 | 0.0000000 | 0.00e+00 |
| CC2D1B | 1.10 | 0.0000001 | 0.00e+00 |
| CC2D2A | -2.05 | 0.0000447 | 1.11e-05 |
| CCAR1 | -1.33 | 0.0000000 | 0.00e+00 |
| CCAT1 | 1.43 | 0.0871482 | 4.40e-02 |
| CCBE1 | 2.72 | 0.0076998 | 2.87e-03 |
| CCBL1 | -1.07 | 0.0253202 | 1.08e-02 |
| CCDC101 | 1.10 | 0.0016101 | 5.14e-04 |
| CCDC107 | 0.70 | 0.0483578 | 2.22e-02 |
| CCDC113 | -1.76 | 0.0165738 | 6.71e-03 |
| CCDC117 | 0.68 | 0.0096165 | 3.67e-03 |
| CCDC121 | -1.78 | 0.0287073 | 1.24e-02 |
| CCDC130 | 1.51 | 0.0000004 | 1.00e-07 |
| CCDC138 | -2.92 | 0.0000216 | 5.10e-06 |
| CCDC14 | -1.74 | 0.0000000 | 0.00e+00 |
| CCDC144A | -1.49 | 0.0605152 | 2.87e-02 |
| CCDC144B | -1.14 | 0.0364585 | 1.62e-02 |
| CCDC146 | 1.67 | 0.0502611 | 2.32e-02 |
| CCDC149 | 1.55 | 0.0230090 | 9.65e-03 |
| CCDC15 | -2.08 | 0.0902875 | 4.59e-02 |
| CCDC150 | -2.73 | 0.0002416 | 6.64e-05 |
| CCDC174 | 1.58 | 0.0000000 | 0.00e+00 |
| CCDC18 | -3.23 | 0.0000061 | 1.30e-06 |
| CCDC22 | 0.81 | 0.0035634 | 1.23e-03 |
| CCDC25 | -0.95 | 0.0000162 | 3.80e-06 |
| CCDC3 | -3.14 | 0.0829550 | 4.16e-02 |
| CCDC34 | -2.98 | 0.0000000 | 0.00e+00 |
| CCDC40 | -1.74 | 0.0459591 | 2.10e-02 |
| CCDC47 | -0.33 | 0.0300597 | 1.30e-02 |
| CCDC50 | 1.09 | 0.0000001 | 0.00e+00 |
| CCDC51 | -0.92 | 0.0066164 | 2.43e-03 |
| CCDC6 | 1.07 | 0.0000000 | 0.00e+00 |
| CCDC64 | 1.39 | 0.0000028 | 6.00e-07 |
| CCDC68 | 0.62 | 0.0798676 | 3.97e-02 |
| CCDC69 | 2.09 | 0.0000000 | 0.00e+00 |
| CCDC71L | 2.23 | 0.0000000 | 0.00e+00 |
| CCDC74A | -2.66 | 0.0000391 | 9.60e-06 |
| CCDC74B | -2.48 | 0.0033254 | 1.14e-03 |
| CCDC77 | -1.32 | 0.0002135 | 5.83e-05 |
| CCDC78 | -2.45 | 0.0872792 | 4.41e-02 |
| CCDC85B | -2.32 | 0.0000000 | 0.00e+00 |
| CCDC86 | -0.42 | 0.0296138 | 1.28e-02 |
| CCDC88A | -0.49 | 0.0188326 | 7.73e-03 |
| CCDC9 | 1.51 | 0.0000000 | 0.00e+00 |
| CCDC92 | 1.49 | 0.0003250 | 9.13e-05 |
| CCDC94 | 1.10 | 0.0000345 | 8.40e-06 |
| CCDC97 | 1.32 | 0.0000000 | 0.00e+00 |
| CCK | 2.82 | 0.0022542 | 7.48e-04 |
| CCL15 | 5.90 | 0.0466313 | 2.14e-02 |
| CCL15-CCL14 | 5.76 | 0.0578985 | 2.73e-02 |
| CCL17 | 11.75 | 0.0000001 | 0.00e+00 |
| CCL19 | 5.61 | 0.0719686 | 3.52e-02 |
| CCL2 | 5.45 | 0.0000000 | 0.00e+00 |
| CCL20 | 10.20 | 0.0000000 | 0.00e+00 |
| CCL22 | 10.52 | 0.0000017 | 4.00e-07 |
| CCL28 | 4.80 | 0.0000072 | 1.60e-06 |
| CCL3 | 11.83 | 0.0000000 | 0.00e+00 |
| CCL3L1 | 12.01 | 0.0000000 | 0.00e+00 |
| CCL3L3 | 8.74 | 0.0001440 | 3.84e-05 |
| CCL4 | 9.14 | 0.0000551 | 1.38e-05 |
| CCL4L1 | 9.69 | 0.0000141 | 3.30e-06 |
| CCL4L2 | 9.72 | 0.0000130 | 3.00e-06 |
| CCL5 | 10.58 | 0.0000000 | 0.00e+00 |
| CCM2 | 0.69 | 0.0008453 | 2.56e-04 |
| CCNA2 | -3.60 | 0.0000000 | 0.00e+00 |
| CCNB1 | -3.05 | 0.0000000 | 0.00e+00 |
| CCNB1IP1 | 0.50 | 0.0767624 | 3.79e-02 |
| CCNB2 | -2.61 | 0.0000000 | 0.00e+00 |
| CCNC | -0.90 | 0.0016026 | 5.12e-04 |
| CCND3 | -0.69 | 0.0000000 | 0.00e+00 |
| CCNE2 | -3.05 | 0.0000517 | 1.29e-05 |
| CCNF | -1.94 | 0.0000000 | 0.00e+00 |
| CCNG1 | 0.38 | 0.0367126 | 1.63e-02 |
| CCNG2 | 1.70 | 0.0000697 | 1.77e-05 |
| CCNH | -0.74 | 0.0219451 | 9.15e-03 |
| CCNI | 1.23 | 0.0000000 | 0.00e+00 |
| CCNK | 1.83 | 0.0000000 | 0.00e+00 |
| CCNL1 | 0.95 | 0.0000005 | 1.00e-07 |
| CCP110 | -1.23 | 0.0014587 | 4.62e-04 |
| CCR7 | 4.52 | 0.0000000 | 0.00e+00 |
| CCRL2 | 5.44 | 0.0911055 | 4.64e-02 |
| CCSAP | -1.42 | 0.0012702 | 3.98e-04 |
| CCT2 | -1.12 | 0.0000000 | 0.00e+00 |
| CCT3 | -0.86 | 0.0000000 | 0.00e+00 |
| CCT4 | -1.02 | 0.0000000 | 0.00e+00 |
| CCT5 | -1.26 | 0.0000000 | 0.00e+00 |
| CCT6A | -1.30 | 0.0000000 | 0.00e+00 |
| CCT7 | -1.20 | 0.0000000 | 0.00e+00 |
| CCT8 | -0.99 | 0.0000000 | 0.00e+00 |
| CD109 | -0.72 | 0.0000286 | 6.90e-06 |
| CD14 | 2.59 | 0.0000000 | 0.00e+00 |
| CD163L1 | 3.68 | 0.0041716 | 1.46e-03 |
| CD164 | -0.44 | 0.0080205 | 3.01e-03 |
| CD177 | 6.25 | 0.0258134 | 1.10e-02 |
| CD24 | -0.51 | 0.0001032 | 2.68e-05 |
| CD274 | 7.06 | 0.0000000 | 0.00e+00 |
| CD276 | 1.94 | 0.0000000 | 0.00e+00 |
| CD2BP2 | 0.55 | 0.0001000 | 2.59e-05 |
| CD320 | -0.76 | 0.0032821 | 1.12e-03 |
| CD34 | 3.95 | 0.0807166 | 4.02e-02 |
| CD38 | 3.25 | 0.0000000 | 0.00e+00 |
| CD3EAP | -2.39 | 0.0000000 | 0.00e+00 |
| CD40 | 3.35 | 0.0000000 | 0.00e+00 |
| CD44 | 2.60 | 0.0000000 | 0.00e+00 |
| CD46 | -0.33 | 0.0157352 | 6.34e-03 |
| CD47 | 1.69 | 0.0000000 | 0.00e+00 |
| CD55 | 2.96 | 0.0000000 | 0.00e+00 |
| CD58 | 0.72 | 0.0989469 | 5.10e-02 |
| CD59 | 0.50 | 0.0000289 | 7.00e-06 |
| CD63 | 0.34 | 0.0010385 | 3.20e-04 |
| CD68 | 4.21 | 0.0000000 | 0.00e+00 |
| CD70 | 2.44 | 0.0000000 | 0.00e+00 |
| CD74 | 9.18 | 0.0000513 | 1.28e-05 |
| CD80 | 5.44 | 0.0911055 | 4.64e-02 |
| CD81 | 1.01 | 0.0000000 | 0.00e+00 |
| CD82 | 4.56 | 0.0000000 | 0.00e+00 |
| CD83 | 3.71 | 0.0000000 | 0.00e+00 |
| CD86 | 6.20 | 0.0294725 | 1.27e-02 |
| CD9 | 0.88 | 0.0000000 | 0.00e+00 |
| CD99 | 0.72 | 0.0000039 | 8.00e-07 |
| CD99L2 | -0.63 | 0.0000448 | 1.11e-05 |
| CD99P1 | 1.70 | 0.0240065 | 1.01e-02 |
| CDA | 2.04 | 0.0000000 | 0.00e+00 |
| CDC123 | -1.24 | 0.0000000 | 0.00e+00 |
| CDC14A | 1.56 | 0.0012762 | 4.00e-04 |
| CDC14B | 0.86 | 0.0059179 | 2.15e-03 |
| CDC16 | -0.77 | 0.0004491 | 1.30e-04 |
| CDC20 | -3.04 | 0.0000000 | 0.00e+00 |
| CDC23 | -1.00 | 0.0000039 | 8.00e-07 |
| CDC25A | -2.07 | 0.0000001 | 0.00e+00 |
| CDC25B | 0.69 | 0.0000000 | 0.00e+00 |
| CDC25C | -2.06 | 0.0000000 | 0.00e+00 |
| CDC27 | -0.71 | 0.0000015 | 3.00e-07 |
| CDC37 | 0.38 | 0.0016229 | 5.19e-04 |
| CDC37L1 | 2.45 | 0.0000000 | 0.00e+00 |
| CDC42 | -0.34 | 0.0233167 | 9.81e-03 |
| CDC42BPB | 0.51 | 0.0000193 | 4.50e-06 |
| CDC42EP1 | 2.35 | 0.0000000 | 0.00e+00 |
| CDC42EP2 | 3.25 | 0.0000000 | 0.00e+00 |
| CDC42EP3 | 0.72 | 0.0092836 | 3.54e-03 |
| CDC42EP4 | 0.86 | 0.0000000 | 0.00e+00 |
| CDC42EP5 | 7.03 | 0.0060020 | 2.18e-03 |
| CDC42SE1 | 1.50 | 0.0000000 | 0.00e+00 |
| CDC42SE2 | 0.78 | 0.0000074 | 1.60e-06 |
| CDC45 | -3.02 | 0.0000000 | 0.00e+00 |
| CDC6 | -3.00 | 0.0000000 | 0.00e+00 |
| CDC7 | -2.26 | 0.0000016 | 3.00e-07 |
| CDC73 | -0.84 | 0.0003445 | 9.71e-05 |
| CDCA2 | -2.70 | 0.0000000 | 0.00e+00 |
| CDCA3 | -3.34 | 0.0000000 | 0.00e+00 |
| CDCA4 | -0.80 | 0.0009307 | 2.84e-04 |
| CDCA5 | -2.46 | 0.0000000 | 0.00e+00 |
| CDCA7 | -2.80 | 0.0000000 | 0.00e+00 |
| CDCA7L | -4.70 | 0.0000000 | 0.00e+00 |
| CDCA8 | -2.33 | 0.0000000 | 0.00e+00 |
| CDCP1 | 5.15 | 0.0000000 | 0.00e+00 |
| CDH1 | 0.91 | 0.0000000 | 0.00e+00 |
| CDH11 | 9.06 | 0.0000658 | 1.66e-05 |
| CDH17 | -1.95 | 0.0000000 | 0.00e+00 |
| CDH23 | 1.98 | 0.0018199 | 5.88e-04 |
| CDH24 | -1.27 | 0.0412082 | 1.86e-02 |
| CDH3 | 5.09 | 0.0000000 | 0.00e+00 |
| CDH4 | 1.75 | 0.0000251 | 6.00e-06 |
| CDH5 | 7.20 | 0.0043348 | 1.52e-03 |
| CDHR2 | 7.40 | 0.0028243 | 9.54e-04 |
| CDIPT | 0.35 | 0.0532116 | 2.48e-02 |
| CDK1 | -2.89 | 0.0000000 | 0.00e+00 |
| CDK11B | 0.60 | 0.0068861 | 2.54e-03 |
| CDK13 | 0.49 | 0.0486781 | 2.24e-02 |
| CDK16 | 0.52 | 0.0000323 | 7.80e-06 |
| CDK17 | 0.69 | 0.0068568 | 2.52e-03 |
| CDK18 | 4.13 | 0.0000000 | 0.00e+00 |
| CDK19 | -1.37 | 0.0019794 | 6.46e-04 |
| CDK2 | -1.44 | 0.0000000 | 0.00e+00 |
| CDK2AP1 | -0.92 | 0.0000009 | 2.00e-07 |
| CDK2AP2 | 0.87 | 0.0000083 | 1.80e-06 |
| CDK4 | -1.31 | 0.0000000 | 0.00e+00 |
| CDK5 | -1.01 | 0.0101574 | 3.91e-03 |
| CDK5R2 | 5.10 | 0.0000000 | 0.00e+00 |
| CDK5RAP2 | -0.59 | 0.0034584 | 1.19e-03 |
| CDK5RAP3 | -0.37 | 0.0407415 | 1.83e-02 |
| CDK7 | 1.27 | 0.0000017 | 3.00e-07 |
| CDK9 | 1.04 | 0.0000000 | 0.00e+00 |
| CDKL2 | -3.10 | 0.0447793 | 2.04e-02 |
| CDKL5 | 0.98 | 0.0079463 | 2.98e-03 |
| CDKN1A | 2.73 | 0.0000000 | 0.00e+00 |
| CDKN1B | 1.14 | 0.0000000 | 0.00e+00 |
| CDKN2AIP | 0.73 | 0.0310413 | 1.35e-02 |
| CDKN2AIPNL | -1.30 | 0.0000073 | 1.60e-06 |
| CDKN3 | -3.15 | 0.0000000 | 0.00e+00 |
| CDRT4 | 1.37 | 0.0002648 | 7.33e-05 |
| CDS2 | 0.32 | 0.0328399 | 1.43e-02 |
| CDT1 | -1.24 | 0.0001861 | 5.04e-05 |
| CDV3 | 0.62 | 0.0000016 | 3.00e-07 |
| CDYL | 0.57 | 0.0154466 | 6.21e-03 |
| CEACAM1 | 5.91 | 0.0000000 | 0.00e+00 |
| CEACAM19 | 2.40 | 0.0000000 | 0.00e+00 |
| CEACAM3 | 2.42 | 0.0000017 | 3.00e-07 |
| CEACAM5 | 3.79 | 0.0000000 | 0.00e+00 |
| CEACAM6 | 1.46 | 0.0000000 | 0.00e+00 |
| CEACAM7 | 2.67 | 0.0015839 | 5.05e-04 |
| CEACAM8 | 2.03 | 0.0061797 | 2.25e-03 |
| CEBPA | 2.19 | 0.0000000 | 0.00e+00 |
| CEBPB | 1.98 | 0.0000000 | 0.00e+00 |
| CEBPB-AS1 | 3.27 | 0.0000311 | 7.50e-06 |
| CEBPD | 3.25 | 0.0000000 | 0.00e+00 |
| CEBPG | 0.89 | 0.0000000 | 0.00e+00 |
| CEBPZ | -0.99 | 0.0000000 | 0.00e+00 |
| CEBPZOS | -0.71 | 0.0092941 | 3.54e-03 |
| CECR5 | -0.97 | 0.0006789 | 2.02e-04 |
| CELF1 | 0.69 | 0.0000001 | 0.00e+00 |
| CELSR1 | 0.38 | 0.0132492 | 5.24e-03 |
| CEMIP | 4.76 | 0.0000000 | 0.00e+00 |
| CENPA | -3.30 | 0.0000000 | 0.00e+00 |
| CENPC | -1.27 | 0.0129501 | 5.11e-03 |
| CENPE | -3.45 | 0.0000000 | 0.00e+00 |
| CENPF | -3.76 | 0.0000000 | 0.00e+00 |
| CENPH | -2.21 | 0.0000000 | 0.00e+00 |
| CENPI | -2.53 | 0.0000000 | 0.00e+00 |
| CENPJ | -1.86 | 0.0000001 | 0.00e+00 |
| CENPK | -3.17 | 0.0000000 | 0.00e+00 |
| CENPL | -1.05 | 0.0264787 | 1.13e-02 |
| CENPM | -2.99 | 0.0000000 | 0.00e+00 |
| CENPN | -1.81 | 0.0000000 | 0.00e+00 |
| CENPO | -2.09 | 0.0000000 | 0.00e+00 |
| CENPP | -1.62 | 0.0149991 | 6.01e-03 |
| CENPQ | -1.28 | 0.0395513 | 1.77e-02 |
| CENPU | -3.25 | 0.0000000 | 0.00e+00 |
| CENPV | -0.72 | 0.0016772 | 5.38e-04 |
| CENPW | -2.33 | 0.0000005 | 1.00e-07 |
| CEP126 | -1.66 | 0.0089025 | 3.37e-03 |
| CEP128 | -2.40 | 0.0000691 | 1.75e-05 |
| CEP131 | -0.85 | 0.0787549 | 3.90e-02 |
| CEP152 | -2.32 | 0.0000005 | 1.00e-07 |
| CEP170 | 0.53 | 0.0381059 | 1.70e-02 |
| CEP170B | 0.69 | 0.0001681 | 4.53e-05 |
| CEP192 | -1.67 | 0.0000002 | 0.00e+00 |
| CEP250 | -1.18 | 0.0000093 | 2.10e-06 |
| CEP290 | -1.63 | 0.0000012 | 2.00e-07 |
| CEP295 | -1.54 | 0.0000503 | 1.25e-05 |
| CEP350 | -0.45 | 0.0483421 | 2.22e-02 |
| CEP44 | -1.22 | 0.0126654 | 4.99e-03 |
| CEP55 | -2.56 | 0.0000000 | 0.00e+00 |
| CEP57L1 | -1.81 | 0.0171064 | 6.95e-03 |
| CEP63 | -1.04 | 0.0325481 | 1.42e-02 |
| CEP70 | -1.43 | 0.0230233 | 9.66e-03 |
| CEP78 | -2.79 | 0.0000000 | 0.00e+00 |
| CEP83 | -2.19 | 0.0002562 | 7.07e-05 |
| CEP85 | -1.00 | 0.0006677 | 1.98e-04 |
| CEP97 | -1.23 | 0.0022139 | 7.32e-04 |
| CEPT1 | 1.08 | 0.0016811 | 5.39e-04 |
| CERK | -1.31 | 0.0000017 | 4.00e-07 |
| CERS2 | 0.23 | 0.0638247 | 3.05e-02 |
| CERS4 | 1.32 | 0.0082390 | 3.10e-03 |
| CERS5 | 0.79 | 0.0014280 | 4.52e-04 |
| CERS6 | -1.18 | 0.0000001 | 0.00e+00 |
| CES2 | 0.56 | 0.0041770 | 1.46e-03 |
| CETN2 | -1.01 | 0.0001501 | 4.01e-05 |
| CETN3 | -1.28 | 0.0056171 | 2.03e-03 |
| CFAP44 | -2.02 | 0.0089264 | 3.39e-03 |
| CFAP45 | 3.13 | 0.0874170 | 4.41e-02 |
| CFAP57 | 2.07 | 0.0055208 | 1.99e-03 |
| CFAP97 | -1.18 | 0.0001102 | 2.88e-05 |
| CFB | 9.93 | 0.0000078 | 1.70e-06 |
| CFD | 1.91 | 0.0000000 | 0.00e+00 |
| CFH | 1.11 | 0.0000000 | 0.00e+00 |
| CFL2 | 0.42 | 0.0125754 | 4.95e-03 |
| CFLAR | 1.94 | 0.0000000 | 0.00e+00 |
| CGN | 1.72 | 0.0000022 | 5.00e-07 |
| CGNL1 | 1.91 | 0.0000143 | 3.30e-06 |
| CGREF1 | -1.12 | 0.0535417 | 2.50e-02 |
| CH17-360D5.1 | -2.35 | 0.0000000 | 0.00e+00 |
| CH25H | 4.18 | 0.0003532 | 9.99e-05 |
| CHAC1 | 4.12 | 0.0000000 | 0.00e+00 |
| CHAC2 | -1.93 | 0.0001093 | 2.85e-05 |
| CHAF1A | -1.84 | 0.0000000 | 0.00e+00 |
| CHAF1B | -2.12 | 0.0000000 | 0.00e+00 |
| CHCHD2 | -0.55 | 0.0000013 | 3.00e-07 |
| CHCHD3 | -0.72 | 0.0001088 | 2.84e-05 |
| CHCHD4 | -0.85 | 0.0042475 | 1.49e-03 |
| CHCHD7 | -1.24 | 0.0225432 | 9.43e-03 |
| CHD1 | -0.55 | 0.0097859 | 3.75e-03 |
| CHD1L | -1.01 | 0.0000007 | 1.00e-07 |
| CHD2 | 1.59 | 0.0000000 | 0.00e+00 |
| CHD8 | 0.68 | 0.0000177 | 4.20e-06 |
| CHD9 | -0.54 | 0.0169593 | 6.88e-03 |
| CHDH | -1.35 | 0.0530267 | 2.47e-02 |
| CHEK1 | -1.81 | 0.0000000 | 0.00e+00 |
| CHEK2 | 1.28 | 0.0000015 | 3.00e-07 |
| CHERP | 0.86 | 0.0000371 | 9.10e-06 |
| CHFR | 1.55 | 0.0000000 | 0.00e+00 |
| CHGA | 3.59 | 0.0000002 | 0.00e+00 |
| CHGB | 1.05 | 0.0003230 | 9.06e-05 |
| CHI3L2 | 9.34 | 0.0000340 | 8.30e-06 |
| CHIC1 | -0.96 | 0.0603642 | 2.87e-02 |
| CHIC2 | 2.16 | 0.0000122 | 2.80e-06 |
| CHKB | 0.75 | 0.0263552 | 1.12e-02 |
| CHKB-CPT1B | 0.97 | 0.0000124 | 2.80e-06 |
| CHM | -1.19 | 0.0004927 | 1.43e-04 |
| CHML | -2.95 | 0.0000000 | 0.00e+00 |
| CHMP1A | 0.31 | 0.0499976 | 2.31e-02 |
| CHMP3 | -0.59 | 0.0000220 | 5.20e-06 |
| CHMP4B | 1.37 | 0.0000000 | 0.00e+00 |
| CHMP5 | 0.47 | 0.0047042 | 1.67e-03 |
| CHN2 | 6.03 | 0.0377181 | 1.68e-02 |
| CHORDC1 | -1.25 | 0.0000000 | 0.00e+00 |
| CHP1 | 0.28 | 0.0440886 | 2.00e-02 |
| CHPF | 1.57 | 0.0000000 | 0.00e+00 |
| CHPF2 | 1.33 | 0.0000000 | 0.00e+00 |
| CHPT1 | -1.09 | 0.0000004 | 1.00e-07 |
| CHRM4 | 4.43 | 0.0319475 | 1.39e-02 |
| CHRNA5 | -1.40 | 0.0000104 | 2.40e-06 |
| CHST1 | 3.55 | 0.0003850 | 1.10e-04 |
| CHST10 | -0.96 | 0.0077833 | 2.91e-03 |
| CHST11 | 1.84 | 0.0000000 | 0.00e+00 |
| CHST15 | 2.29 | 0.0000000 | 0.00e+00 |
| CHST2 | 5.71 | 0.0000000 | 0.00e+00 |
| CHST3 | 0.76 | 0.0005418 | 1.58e-04 |
| CHST4 | 4.36 | 0.0367833 | 1.63e-02 |
| CHST7 | 1.15 | 0.0006594 | 1.95e-04 |
| CHTF18 | -1.72 | 0.0000028 | 6.00e-07 |
| CHUK | -0.60 | 0.0110368 | 4.29e-03 |
| CHURC1 | -0.60 | 0.0055190 | 1.99e-03 |
| CHURC1-FNTB | -0.61 | 0.0012262 | 3.83e-04 |
| CIAO1 | -0.73 | 0.0000112 | 2.50e-06 |
| CIB1 | 0.63 | 0.0000029 | 6.00e-07 |
| CIC | 2.12 | 0.0000000 | 0.00e+00 |
| CIDECP | 1.77 | 0.0001190 | 3.13e-05 |
| CINP | -0.61 | 0.0564623 | 2.65e-02 |
| CIPC | -0.89 | 0.0205878 | 8.52e-03 |
| CIR1 | 0.89 | 0.0023837 | 7.95e-04 |
| CIRH1A | -0.64 | 0.0031369 | 1.07e-03 |
| CISD1 | -0.59 | 0.0639421 | 3.06e-02 |
| CIT | -1.96 | 0.0000000 | 0.00e+00 |
| CITED2 | 4.79 | 0.0000000 | 0.00e+00 |
| CITED4 | 3.25 | 0.0000000 | 0.00e+00 |
| CIZ1 | 0.53 | 0.0005500 | 1.61e-04 |
| CKAP2 | -2.44 | 0.0000000 | 0.00e+00 |
| CKAP2L | -2.89 | 0.0000000 | 0.00e+00 |
| CKAP4 | 1.50 | 0.0000000 | 0.00e+00 |
| CKAP5 | -0.87 | 0.0000000 | 0.00e+00 |
| CKB | 2.03 | 0.0000000 | 0.00e+00 |
| CKLF | -1.02 | 0.0061323 | 2.23e-03 |
| CKLF-CMTM1 | -0.94 | 0.0333831 | 1.46e-02 |
| CKMT1A | -2.65 | 0.0046851 | 1.66e-03 |
| CKMT1B | -2.83 | 0.0021280 | 7.01e-04 |
| CKS1B | -1.84 | 0.0000000 | 0.00e+00 |
| CKS2 | -1.98 | 0.0000000 | 0.00e+00 |
| CLASP2 | -0.79 | 0.0002605 | 7.19e-05 |
| CLASRP | 0.46 | 0.0236138 | 9.95e-03 |
| CLCC1 | -1.07 | 0.0002199 | 6.01e-05 |
| CLCF1 | 1.99 | 0.0000000 | 0.00e+00 |
| CLCN3 | -0.71 | 0.0024546 | 8.20e-04 |
| CLCN4 | -1.06 | 0.0983085 | 5.06e-02 |
| CLCN6 | 1.58 | 0.0000000 | 0.00e+00 |
| CLCN7 | 1.38 | 0.0000000 | 0.00e+00 |
| CLDN1 | 2.08 | 0.0000000 | 0.00e+00 |
| CLDN12 | 1.34 | 0.0000000 | 0.00e+00 |
| CLDN14 | 7.14 | 0.0047358 | 1.68e-03 |
| CLDN4 | 5.25 | 0.0000000 | 0.00e+00 |
| CLEC17A | 5.27 | 0.0042524 | 1.49e-03 |
| CLEC2B | 3.34 | 0.0000034 | 7.00e-07 |
| CLEC2D | -1.63 | 0.0000000 | 0.00e+00 |
| CLEC4E | 12.05 | 0.0000000 | 0.00e+00 |
| CLIC4 | 1.43 | 0.0000000 | 0.00e+00 |
| CLINT1 | 0.64 | 0.0000001 | 0.00e+00 |
| CLIP2 | 2.75 | 0.0000000 | 0.00e+00 |
| CLK3 | 1.47 | 0.0000000 | 0.00e+00 |
| CLN3 | 1.27 | 0.0000000 | 0.00e+00 |
| CLN6 | -0.84 | 0.0000718 | 1.82e-05 |
| CLNS1A | -0.57 | 0.0038094 | 1.32e-03 |
| CLOCK | -0.96 | 0.0113383 | 4.42e-03 |
| CLPTM1 | 0.91 | 0.0000000 | 0.00e+00 |
| CLSPN | -3.11 | 0.0000000 | 0.00e+00 |
| CLSTN1 | 2.13 | 0.0000000 | 0.00e+00 |
| CLSTN3 | 1.26 | 0.0000000 | 0.00e+00 |
| CLTA | 0.31 | 0.0156522 | 6.30e-03 |
| CLTB | 0.55 | 0.0015689 | 5.00e-04 |
| CLTC | -0.31 | 0.0012372 | 3.87e-04 |
| CLU | 1.52 | 0.0000000 | 0.00e+00 |
| CLUH | 1.05 | 0.0000000 | 0.00e+00 |
| CLUHP3 | 1.04 | 0.0606388 | 2.88e-02 |
| CMBL | -1.93 | 0.0000018 | 4.00e-07 |
| CMC2 | -0.74 | 0.0139960 | 5.57e-03 |
| CMC4 | -0.98 | 0.0447920 | 2.04e-02 |
| CMIP | 1.45 | 0.0000000 | 0.00e+00 |
| CMPK2 | 9.75 | 0.0000000 | 0.00e+00 |
| CMSS1 | -1.51 | 0.0000002 | 0.00e+00 |
| CMTM3 | 1.07 | 0.0000000 | 0.00e+00 |
| CMTM4 | -1.94 | 0.0000000 | 0.00e+00 |
| CMTR1 | 0.98 | 0.0000001 | 0.00e+00 |
| CMTR2 | -0.57 | 0.0844140 | 4.24e-02 |
| CNBP | -0.25 | 0.0361935 | 1.60e-02 |
| CNIH1 | -0.36 | 0.0547812 | 2.56e-02 |
| CNKSR2 | -1.05 | 0.0122090 | 4.79e-03 |
| CNKSR3 | 2.65 | 0.0000000 | 0.00e+00 |
| CNN2 | 0.71 | 0.0000086 | 1.90e-06 |
| CNNM2 | 1.35 | 0.0000537 | 1.34e-05 |
| CNNM4 | 0.87 | 0.0001419 | 3.78e-05 |
| CNOT1 | -0.34 | 0.0072336 | 2.68e-03 |
| CNOT3 | 1.67 | 0.0791847 | 3.93e-02 |
| CNOT4 | 1.38 | 0.0000000 | 0.00e+00 |
| CNOT6 | 0.39 | 0.0764584 | 3.77e-02 |
| CNOT7 | -1.37 | 0.0000000 | 0.00e+00 |
| CNP | 0.93 | 0.0000000 | 0.00e+00 |
| CNPPD1 | 0.48 | 0.0436035 | 1.98e-02 |
| CNPY2 | -0.42 | 0.0271612 | 1.16e-02 |
| CNPY3 | 0.46 | 0.0301916 | 1.31e-02 |
| CNPY4 | -1.44 | 0.0302519 | 1.31e-02 |
| CNTN1 | -3.36 | 0.0000000 | 0.00e+00 |
| CNTNAP3 | -0.86 | 0.0016015 | 5.11e-04 |
| CNTNAP3B | -0.58 | 0.0165164 | 6.68e-03 |
| CNTNAP3P2 | -0.70 | 0.0283415 | 1.22e-02 |
| CNTRL | -1.40 | 0.0001194 | 3.14e-05 |
| COA1 | -0.91 | 0.0000137 | 3.20e-06 |
| COA3 | -0.92 | 0.0000205 | 4.80e-06 |
| COA4 | -0.61 | 0.0004438 | 1.28e-04 |
| COA5 | -0.84 | 0.0657247 | 3.16e-02 |
| COA6 | -0.48 | 0.0758741 | 3.74e-02 |
| COA7 | -0.91 | 0.0145362 | 5.80e-03 |
| COG1 | 0.81 | 0.0007669 | 2.30e-04 |
| COG2 | -0.48 | 0.0814750 | 4.07e-02 |
| COG4 | -0.42 | 0.0207553 | 8.60e-03 |
| COG6 | -0.77 | 0.0891887 | 4.52e-02 |
| COG7 | 0.62 | 0.0185744 | 7.61e-03 |
| COG8 | -0.43 | 0.0693957 | 3.37e-02 |
| COIL | -0.57 | 0.0361281 | 1.60e-02 |
| COL11A1 | -1.88 | 0.0034223 | 1.18e-03 |
| COL12A1 | 1.93 | 0.0000000 | 0.00e+00 |
| COL13A1 | 8.14 | 0.0005598 | 1.64e-04 |
| COL15A1 | 5.51 | 0.0022822 | 7.58e-04 |
| COL16A1 | 1.28 | 0.0073945 | 2.75e-03 |
| COL17A1 | 3.85 | 0.0000000 | 0.00e+00 |
| COL18A1 | 0.35 | 0.0222910 | 9.31e-03 |
| COL1A1 | 1.06 | 0.0625465 | 2.99e-02 |
| COL21A1 | -3.05 | 0.0475486 | 2.18e-02 |
| COL22A1 | 10.69 | 0.0000011 | 2.00e-07 |
| COL26A1 | -2.26 | 0.0098968 | 3.80e-03 |
| COL27A1 | 1.36 | 0.0000000 | 0.00e+00 |
| COL4A1 | 2.51 | 0.0000000 | 0.00e+00 |
| COL4A2 | 2.74 | 0.0000000 | 0.00e+00 |
| COL4A3BP | 0.67 | 0.0072625 | 2.69e-03 |
| COL5A1 | 2.64 | 0.0000000 | 0.00e+00 |
| COL5A2 | 0.32 | 0.0016718 | 5.36e-04 |
| COL5A3 | 4.93 | 0.0000000 | 0.00e+00 |
| COL6A1 | 1.29 | 0.0000386 | 9.50e-06 |
| COL6A2 | 2.81 | 0.0000000 | 0.00e+00 |
| COL7A1 | 2.07 | 0.0000000 | 0.00e+00 |
| COL9A2 | 5.46 | 0.0000000 | 0.00e+00 |
| COLGALT1 | 0.38 | 0.0060838 | 2.21e-03 |
| COMMD10 | -0.88 | 0.0212101 | 8.81e-03 |
| COMMD4 | -1.05 | 0.0000000 | 0.00e+00 |
| COMMD6 | -0.69 | 0.0178123 | 7.27e-03 |
| COMMD8 | -1.46 | 0.0069969 | 2.58e-03 |
| COMMD9 | -0.76 | 0.0122913 | 4.83e-03 |
| COMT | 0.80 | 0.0000052 | 1.10e-06 |
| COPA | 0.54 | 0.0000027 | 6.00e-07 |
| COPB1 | -0.44 | 0.0006753 | 2.00e-04 |
| COPG1 | 1.24 | 0.0000000 | 0.00e+00 |
| COPG2 | -0.68 | 0.0151993 | 6.10e-03 |
| COPRS | -0.90 | 0.0001266 | 3.34e-05 |
| COPS2 | -1.11 | 0.0000001 | 0.00e+00 |
| COPS3 | -1.09 | 0.0000000 | 0.00e+00 |
| COPS4 | -1.08 | 0.0002459 | 6.77e-05 |
| COPS5 | -0.48 | 0.0092464 | 3.52e-03 |
| COPS6 | -0.73 | 0.0000033 | 7.00e-07 |
| COPS7A | -1.19 | 0.0000000 | 0.00e+00 |
| COPS8 | -1.17 | 0.0000022 | 5.00e-07 |
| COPZ1 | -0.83 | 0.0000000 | 0.00e+00 |
| COPZ2 | -1.51 | 0.0378795 | 1.69e-02 |
| COQ10B | 0.79 | 0.0019033 | 6.19e-04 |
| COQ2 | -1.65 | 0.0000101 | 2.30e-06 |
| COQ3 | -1.61 | 0.0061797 | 2.25e-03 |
| COQ4 | -0.64 | 0.0692695 | 3.36e-02 |
| COQ5 | -0.44 | 0.0943406 | 4.83e-02 |
| CORO1B | 1.35 | 0.0000000 | 0.00e+00 |
| CORO2B | 2.86 | 0.0322318 | 1.41e-02 |
| CORO6 | 1.20 | 0.0175363 | 7.15e-03 |
| CORO7 | 1.70 | 0.0000072 | 1.60e-06 |
| CORO7-PAM16 | 0.78 | 0.0140590 | 5.60e-03 |
| COTL1 | -0.36 | 0.0014890 | 4.73e-04 |
| COX11 | -1.56 | 0.0000000 | 0.00e+00 |
| COX14 | -0.54 | 0.0442069 | 2.01e-02 |
| COX15 | -0.63 | 0.0035918 | 1.24e-03 |
| COX16 | -1.04 | 0.0001372 | 3.64e-05 |
| COX18 | -1.11 | 0.0568928 | 2.68e-02 |
| COX20 | -0.91 | 0.0015568 | 4.96e-04 |
| COX4I1 | -0.50 | 0.0000338 | 8.20e-06 |
| COX5B | -0.46 | 0.0018607 | 6.03e-04 |
| COX6A1 | -0.60 | 0.0000281 | 6.80e-06 |
| COX6B1 | -0.35 | 0.0007684 | 2.30e-04 |
| COX6C | -0.99 | 0.0000000 | 0.00e+00 |
| COX7A2 | -0.33 | 0.0292194 | 1.26e-02 |
| COX7A2L | -0.29 | 0.0715409 | 3.49e-02 |
| COX7B | -0.71 | 0.0000178 | 4.20e-06 |
| COX7C | -0.63 | 0.0000354 | 8.60e-06 |
| COX8A | -0.60 | 0.0000130 | 3.00e-06 |
| CP | 2.54 | 0.0000000 | 0.00e+00 |
| CPA4 | -3.06 | 0.0799928 | 3.98e-02 |
| CPAMD8 | 8.92 | 0.0000933 | 2.41e-05 |
| CPE | 2.32 | 0.0898677 | 4.56e-02 |
| CPEB2 | 0.88 | 0.0001149 | 3.01e-05 |
| CPEB3 | 2.24 | 0.0147928 | 5.92e-03 |
| CPEB4 | 0.99 | 0.0000001 | 0.00e+00 |
| CPLX2 | -2.42 | 0.0000000 | 0.00e+00 |
| CPM | 1.93 | 0.0000000 | 0.00e+00 |
| CPNE3 | -0.82 | 0.0000516 | 1.29e-05 |
| CPNE4 | 1.07 | 0.0750825 | 3.69e-02 |
| CPNE5 | 7.22 | 0.0040291 | 1.41e-03 |
| CPNE7 | -1.69 | 0.0000000 | 0.00e+00 |
| CPNE8 | 0.73 | 0.0096659 | 3.70e-03 |
| CPPED1 | -0.45 | 0.0251244 | 1.07e-02 |
| CPSF2 | -0.77 | 0.0000236 | 5.60e-06 |
| CPSF3 | -1.08 | 0.0000000 | 0.00e+00 |
| CPSF6 | -0.94 | 0.0000009 | 2.00e-07 |
| CPT1B | 1.06 | 0.0002101 | 5.72e-05 |
| CPTP | 0.72 | 0.0662283 | 3.19e-02 |
| CPVL | 6.44 | 0.0198508 | 8.19e-03 |
| CPXM1 | 11.64 | 0.0000001 | 0.00e+00 |
| CR1L | 3.86 | 0.0917429 | 4.68e-02 |
| CRABP2 | 2.62 | 0.0000001 | 0.00e+00 |
| CRAMP1L | 1.12 | 0.0000019 | 4.00e-07 |
| CRAT | 1.16 | 0.0000000 | 0.00e+00 |
| CRB2 | 7.40 | 0.0028076 | 9.49e-04 |
| CREB3 | 0.77 | 0.0000024 | 5.00e-07 |
| CREB3L1 | 3.80 | 0.0000000 | 0.00e+00 |
| CREB3L2 | 0.88 | 0.0000062 | 1.40e-06 |
| CREB3L3 | 5.09 | 0.0070227 | 2.59e-03 |
| CREB3L4 | -0.89 | 0.0673986 | 3.25e-02 |
| CREB5 | 4.63 | 0.0006976 | 2.07e-04 |
| CREBBP | 1.09 | 0.0000000 | 0.00e+00 |
| CREBRF | 2.47 | 0.0000000 | 0.00e+00 |
| CREBZF | -0.46 | 0.0080505 | 3.02e-03 |
| CREG2 | 2.72 | 0.0144891 | 5.78e-03 |
| CRELD1 | 1.16 | 0.0007593 | 2.27e-04 |
| CREM | 0.65 | 0.0057172 | 2.06e-03 |
| CRIM1 | 0.29 | 0.0067762 | 2.49e-03 |
| CRIP1 | -1.92 | 0.0000000 | 0.00e+00 |
| CRIP2 | -1.98 | 0.0000001 | 0.00e+00 |
| CRIPT | -0.75 | 0.0297144 | 1.28e-02 |
| CRISPLD1 | 1.70 | 0.0173424 | 7.05e-03 |
| CRISPLD2 | 4.38 | 0.0000000 | 0.00e+00 |
| CRKL | 1.10 | 0.0000000 | 0.00e+00 |
| CRLF2 | 5.40 | 0.0000000 | 0.00e+00 |
| CRLS1 | -0.48 | 0.0486699 | 2.24e-02 |
| CRNDE | -2.04 | 0.0001534 | 4.11e-05 |
| CRNKL1 | -0.66 | 0.0023278 | 7.75e-04 |
| CROT | -1.75 | 0.0020371 | 6.68e-04 |
| CRTAC1 | 8.19 | 0.0000002 | 0.00e+00 |
| CRTC2 | 1.64 | 0.0000000 | 0.00e+00 |
| CRTC3 | 0.86 | 0.0000139 | 3.20e-06 |
| CRY2 | 0.77 | 0.0767892 | 3.79e-02 |
| CRYAB | 2.26 | 0.0168462 | 6.82e-03 |
| CRYBG3 | -0.69 | 0.0226182 | 9.47e-03 |
| CRYZ | -1.85 | 0.0000000 | 0.00e+00 |
| CS | -0.27 | 0.0715548 | 3.49e-02 |
| CSAG3 | 7.32 | 0.0000048 | 1.00e-06 |
| CSAG4 | 5.83 | 0.0541189 | 2.53e-02 |
| CSDE1 | -0.88 | 0.0000000 | 0.00e+00 |
| CSE1L | -2.50 | 0.0000000 | 0.00e+00 |
| CSF1 | 5.24 | 0.0000000 | 0.00e+00 |
| CSF2 | 9.33 | 0.0000348 | 8.50e-06 |
| CSF2RA | 2.63 | 0.0000002 | 0.00e+00 |
| CSF3 | 10.73 | 0.0000000 | 0.00e+00 |
| CSGALNACT2 | 0.98 | 0.0010008 | 3.08e-04 |
| CSK | 0.73 | 0.0000864 | 2.22e-05 |
| CSNK1A1 | 0.56 | 0.0000118 | 2.70e-06 |
| CSNK1D | 0.93 | 0.0000000 | 0.00e+00 |
| CSNK1E | 0.84 | 0.0000010 | 2.00e-07 |
| CSNK1G1 | 0.64 | 0.0020576 | 6.75e-04 |
| CSNK1G2 | 0.98 | 0.0000000 | 0.00e+00 |
| CSNK2A1 | -0.42 | 0.0068379 | 2.52e-03 |
| CSNK2A2 | 0.71 | 0.0056321 | 2.03e-03 |
| CSNK2A3 | -0.47 | 0.0362899 | 1.61e-02 |
| CSPG5 | -3.27 | 0.0459420 | 2.10e-02 |
| CSPP1 | -0.90 | 0.0278727 | 1.20e-02 |
| CSRNP1 | 3.79 | 0.0000000 | 0.00e+00 |
| CSRNP2 | 1.76 | 0.0000000 | 0.00e+00 |
| CSRP1 | 0.83 | 0.0000000 | 0.00e+00 |
| CST1 | 4.38 | 0.0000000 | 0.00e+00 |
| CST2 | 2.96 | 0.0018855 | 6.12e-04 |
| CST3 | 1.81 | 0.0000000 | 0.00e+00 |
| CST4 | 3.59 | 0.0000804 | 2.05e-05 |
| CSTA | 1.81 | 0.0815883 | 4.08e-02 |
| CSTB | 1.36 | 0.0000000 | 0.00e+00 |
| CSTF1 | -0.86 | 0.0002730 | 7.57e-05 |
| CSTF3 | -1.98 | 0.0000000 | 0.00e+00 |
| CTC1 | 0.99 | 0.0000092 | 2.10e-06 |
| CTCF | -0.48 | 0.0223639 | 9.35e-03 |
| CTDNEP1 | 0.42 | 0.0022846 | 7.59e-04 |
| CTDP1 | 0.78 | 0.0077970 | 2.92e-03 |
| CTDSP2 | 1.59 | 0.0000000 | 0.00e+00 |
| CTDSPL | -2.53 | 0.0000000 | 0.00e+00 |
| CTDSPL2 | -0.86 | 0.0114575 | 4.47e-03 |
| CTGF | 4.27 | 0.0000000 | 0.00e+00 |
| CTIF | 1.65 | 0.0000000 | 0.00e+00 |
| CTNNA1 | 0.86 | 0.0000000 | 0.00e+00 |
| CTNNAL1 | -1.20 | 0.0000003 | 1.00e-07 |
| CTNNB1 | 0.55 | 0.0000004 | 1.00e-07 |
| CTNNBL1 | 0.76 | 0.0000061 | 1.30e-06 |
| CTNND1 | 0.78 | 0.0000000 | 0.00e+00 |
| CTPS1 | -0.40 | 0.0074852 | 2.78e-03 |
| CTR9 | -0.75 | 0.0001309 | 3.47e-05 |
| CTSA | 0.87 | 0.0000000 | 0.00e+00 |
| CTSB | 1.16 | 0.0000000 | 0.00e+00 |
| CTSC | 0.77 | 0.0000074 | 1.60e-06 |
| CTSD | 1.00 | 0.0000000 | 0.00e+00 |
| CTSE | 9.34 | 0.0000339 | 8.30e-06 |
| CTSH | -1.18 | 0.0038350 | 1.33e-03 |
| CTSL | 1.93 | 0.0000000 | 0.00e+00 |
| CTSO | 2.17 | 0.0678982 | 3.28e-02 |
| CTSS | 4.27 | 0.0000000 | 0.00e+00 |
| CTSV | -2.34 | 0.0000088 | 2.00e-06 |
| CTTNBP2NL | 2.13 | 0.0000000 | 0.00e+00 |
| CTU1 | 0.71 | 0.0847872 | 4.26e-02 |
| CTU2 | -0.93 | 0.0021663 | 7.15e-04 |
| CTXN1 | -2.84 | 0.0139778 | 5.56e-03 |
| CUEDC1 | 0.36 | 0.0232810 | 9.78e-03 |
| CUEDC2 | 0.44 | 0.0529603 | 2.47e-02 |
| CUL1 | -0.62 | 0.0003141 | 8.79e-05 |
| CUL2 | -1.18 | 0.0000063 | 1.40e-06 |
| CUL3 | -1.43 | 0.0000000 | 0.00e+00 |
| CUL4B | -0.86 | 0.0000060 | 1.30e-06 |
| CUL5 | -1.65 | 0.0000005 | 1.00e-07 |
| CUTA | -0.54 | 0.0903743 | 4.59e-02 |
| CUTC | -1.58 | 0.0000000 | 0.00e+00 |
| CUX1 | 0.43 | 0.0063184 | 2.31e-03 |
| CWC27 | -1.28 | 0.0008628 | 2.62e-04 |
| CX3CL1 | 7.28 | 0.0000000 | 0.00e+00 |
| CXCL1 | 7.66 | 0.0000000 | 0.00e+00 |
| CXCL10 | 11.85 | 0.0000000 | 0.00e+00 |
| CXCL11 | 12.03 | 0.0000000 | 0.00e+00 |
| CXCL16 | 0.74 | 0.0120739 | 4.73e-03 |
| CXCL2 | 5.28 | 0.0000000 | 0.00e+00 |
| CXCL3 | 5.87 | 0.0000000 | 0.00e+00 |
| CXCL5 | 3.36 | 0.0000000 | 0.00e+00 |
| CXCL6 | 5.83 | 0.0530340 | 2.47e-02 |
| CXCL8 | 8.88 | 0.0000000 | 0.00e+00 |
| CXCR4 | 4.49 | 0.0000582 | 1.46e-05 |
| CXorf38 | 0.70 | 0.0106576 | 4.12e-03 |
| CXorf40B | 0.56 | 0.0400980 | 1.80e-02 |
| CXorf56 | -0.56 | 0.0935407 | 4.79e-02 |
| CXXC5 | -1.04 | 0.0091299 | 3.47e-03 |
| CYB561 | -0.75 | 0.0171383 | 6.96e-03 |
| CYB561A3 | -1.43 | 0.0000001 | 0.00e+00 |
| CYB561D1 | 0.98 | 0.0132407 | 5.24e-03 |
| CYB561D2 | 1.72 | 0.0000000 | 0.00e+00 |
| CYB5A | -1.08 | 0.0000072 | 1.60e-06 |
| CYB5B | -0.86 | 0.0000000 | 0.00e+00 |
| CYB5D1 | -3.12 | 0.0000000 | 0.00e+00 |
| CYB5D2 | -0.76 | 0.0722944 | 3.54e-02 |
| CYB5R1 | 0.65 | 0.0583863 | 2.76e-02 |
| CYB5R2 | 1.72 | 0.0071700 | 2.65e-03 |
| CYB5R3 | 1.18 | 0.0000000 | 0.00e+00 |
| CYB5RL | -1.92 | 0.0141311 | 5.63e-03 |
| CYBA | 1.44 | 0.0000000 | 0.00e+00 |
| CYBRD1 | -1.09 | 0.0000086 | 1.90e-06 |
| CYC1 | -1.04 | 0.0000000 | 0.00e+00 |
| CYCS | -1.03 | 0.0000000 | 0.00e+00 |
| CYFIP2 | -1.47 | 0.0000003 | 0.00e+00 |
| CYLD | 2.44 | 0.0000000 | 0.00e+00 |
| CYMP | 5.69 | 0.0651644 | 3.13e-02 |
| CYP1B1 | 1.09 | 0.0000000 | 0.00e+00 |
| CYP21A1P | 6.09 | 0.0350409 | 1.55e-02 |
| CYP24A1 | -0.65 | 0.0000000 | 0.00e+00 |
| CYP26A1 | -2.81 | 0.0692364 | 3.36e-02 |
| CYP27B1 | 2.97 | 0.0000003 | 1.00e-07 |
| CYP27C1 | 2.17 | 0.0022436 | 7.44e-04 |
| CYP2S1 | 3.30 | 0.0000000 | 0.00e+00 |
| CYP2U1 | -1.85 | 0.0532816 | 2.48e-02 |
| CYP3A5 | 3.17 | 0.0000000 | 0.00e+00 |
| CYP4F12 | 1.23 | 0.0117829 | 4.61e-03 |
| CYP4F22 | 6.61 | 0.0133613 | 5.29e-03 |
| CYP4F3 | -1.14 | 0.0007275 | 2.17e-04 |
| CYP51A1 | 1.17 | 0.0000000 | 0.00e+00 |
| CYR61 | 1.89 | 0.0000000 | 0.00e+00 |
| CYSTM1 | 0.94 | 0.0000000 | 0.00e+00 |
| CYTH1 | 1.73 | 0.0000000 | 0.00e+00 |
| CYTH2 | 1.03 | 0.0000000 | 0.00e+00 |
| CYTH3 | 0.49 | 0.0080936 | 3.04e-03 |
| CYTH4 | 7.51 | 0.0000023 | 5.00e-07 |
| D2HGDH | -1.07 | 0.0173259 | 7.04e-03 |
| DAB2 | 0.81 | 0.0050520 | 1.80e-03 |
| DAB2IP | 2.73 | 0.0000000 | 0.00e+00 |
| DACT1 | 6.09 | 0.0343344 | 1.51e-02 |
| DAD1 | -1.13 | 0.0000000 | 0.00e+00 |
| DAG1 | 0.62 | 0.0000001 | 0.00e+00 |
| DAGLB | 1.08 | 0.0000426 | 1.05e-05 |
| DALRD3 | -0.68 | 0.0800582 | 3.99e-02 |
| DANCR | -3.12 | 0.0000000 | 0.00e+00 |
| DAP | 1.25 | 0.0000000 | 0.00e+00 |
| DAPK2 | 1.47 | 0.0009521 | 2.92e-04 |
| DAPK3 | 2.81 | 0.0000000 | 0.00e+00 |
| DAPP1 | 7.37 | 0.0029508 | 1.00e-03 |
| DARS | -0.51 | 0.0023902 | 7.97e-04 |
| DARS2 | -2.08 | 0.0000000 | 0.00e+00 |
| DAXX | 1.29 | 0.0005246 | 1.53e-04 |
| DAZAP1 | -0.55 | 0.0002495 | 6.87e-05 |
| DBF4 | -1.40 | 0.0000057 | 1.20e-06 |
| DBF4B | -0.74 | 0.0106066 | 4.10e-03 |
| DBI | -0.37 | 0.0149415 | 5.99e-03 |
| DBN1 | 0.63 | 0.0000694 | 1.76e-05 |
| DBNL | 0.36 | 0.0359573 | 1.59e-02 |
| DBP | -1.97 | 0.0415157 | 1.87e-02 |
| DCAF10 | 0.75 | 0.0069307 | 2.55e-03 |
| DCAF11 | -0.56 | 0.0216896 | 9.03e-03 |
| DCAF12 | 0.55 | 0.0018418 | 5.96e-04 |
| DCAF13 | -2.05 | 0.0000000 | 0.00e+00 |
| DCAF17 | -1.03 | 0.0041121 | 1.44e-03 |
| DCAF4 | -1.62 | 0.0000833 | 2.13e-05 |
| DCAF5 | 1.25 | 0.0000000 | 0.00e+00 |
| DCAF6 | -0.56 | 0.0040103 | 1.40e-03 |
| DCAF7 | 0.25 | 0.0976751 | 5.02e-02 |
| DCAF8 | 0.30 | 0.0682574 | 3.30e-02 |
| DCBLD1 | 0.82 | 0.0144479 | 5.76e-03 |
| DCBLD2 | -0.36 | 0.0007082 | 2.11e-04 |
| DCDC2 | -1.38 | 0.0000017 | 3.00e-07 |
| DCHS1 | 3.08 | 0.0009783 | 3.00e-04 |
| DCK | -1.50 | 0.0000357 | 8.70e-06 |
| DCLK2 | 1.00 | 0.0307080 | 1.33e-02 |
| DCLK3 | 8.24 | 0.0004914 | 1.43e-04 |
| DCLRE1A | -1.53 | 0.0000010 | 2.00e-07 |
| DCLRE1B | 1.30 | 0.0000086 | 1.90e-06 |
| DCP1A | 1.34 | 0.0000000 | 0.00e+00 |
| DCP1B | 1.85 | 0.0000000 | 0.00e+00 |
| DCTN2 | 0.27 | 0.0941631 | 4.82e-02 |
| DCTN3 | -0.73 | 0.0050811 | 1.82e-03 |
| DCTPP1 | -1.43 | 0.0000000 | 0.00e+00 |
| DCUN1D3 | 1.40 | 0.0001110 | 2.90e-05 |
| DCUN1D4 | -0.95 | 0.0076176 | 2.84e-03 |
| DCUN1D5 | -0.53 | 0.0329473 | 1.44e-02 |
| DCXR | -1.25 | 0.0000097 | 2.20e-06 |
| DDA1 | 0.65 | 0.0005731 | 1.68e-04 |
| DDB1 | -0.73 | 0.0000000 | 0.00e+00 |
| DDB2 | -1.06 | 0.0000004 | 1.00e-07 |
| DDC | -4.48 | 0.0000000 | 0.00e+00 |
| DDHD2 | -0.52 | 0.0447489 | 2.04e-02 |
| DDIAS | -2.78 | 0.0007707 | 2.31e-04 |
| DDIT3 | 4.05 | 0.0000000 | 0.00e+00 |
| DDIT4 | 0.50 | 0.0000113 | 2.60e-06 |
| DDR1 | 0.86 | 0.0117536 | 4.59e-03 |
| DDRGK1 | 0.96 | 0.0000000 | 0.00e+00 |
| DDX1 | -0.78 | 0.0000000 | 0.00e+00 |
| DDX12P | -1.11 | 0.0002019 | 5.48e-05 |
| DDX18 | -1.55 | 0.0000000 | 0.00e+00 |
| DDX21 | -0.33 | 0.0027245 | 9.19e-04 |
| DDX23 | -0.28 | 0.0215147 | 8.95e-03 |
| DDX27 | 0.28 | 0.0912586 | 4.65e-02 |
| DDX39A | -0.86 | 0.0000003 | 1.00e-07 |
| DDX3X | -0.91 | 0.0000000 | 0.00e+00 |
| DDX3Y | -0.92 | 0.0000021 | 4.00e-07 |
| DDX41 | -0.58 | 0.0031570 | 1.08e-03 |
| DDX42 | -0.27 | 0.0617298 | 2.94e-02 |
| DDX46 | -1.55 | 0.0000000 | 0.00e+00 |
| DDX47 | 0.30 | 0.0800582 | 3.99e-02 |
| DDX49 | -0.73 | 0.0049100 | 1.75e-03 |
| DDX51 | -0.62 | 0.0116739 | 4.56e-03 |
| DDX55 | -0.61 | 0.0078049 | 2.92e-03 |
| DDX56 | -0.55 | 0.0000964 | 2.49e-05 |
| DDX58 | 6.97 | 0.0000000 | 0.00e+00 |
| DDX6 | -0.68 | 0.0001988 | 5.39e-05 |
| DDX60 | 4.86 | 0.0000000 | 0.00e+00 |
| DDX60L | 2.28 | 0.0000000 | 0.00e+00 |
| DEAF1 | 0.52 | 0.0933257 | 4.77e-02 |
| DECR2 | -1.15 | 0.0330824 | 1.45e-02 |
| DEDD | 1.28 | 0.0000000 | 0.00e+00 |
| DEDD2 | 1.77 | 0.0000000 | 0.00e+00 |
| DEFB1 | 2.22 | 0.0078044 | 2.92e-03 |
| DEFB4A | 5.61 | 0.0717731 | 3.50e-02 |
| DEGS1 | 0.38 | 0.0163584 | 6.61e-03 |
| DEK | -1.39 | 0.0000000 | 0.00e+00 |
| DENND1A | 1.71 | 0.0000000 | 0.00e+00 |
| DENND2D | 3.39 | 0.0000000 | 0.00e+00 |
| DENND3 | 2.01 | 0.0000000 | 0.00e+00 |
| DENND4A | 1.74 | 0.0000000 | 0.00e+00 |
| DENND4B | 0.49 | 0.0172476 | 7.01e-03 |
| DENND4C | 0.52 | 0.0054529 | 1.96e-03 |
| DENND5A | 1.81 | 0.0000000 | 0.00e+00 |
| DENND5B | -0.87 | 0.0000114 | 2.60e-06 |
| DENR | -0.67 | 0.0003515 | 9.94e-05 |
| DEPDC1 | -3.24 | 0.0000000 | 0.00e+00 |
| DEPDC1B | -3.70 | 0.0000000 | 0.00e+00 |
| DERA | -1.53 | 0.0000000 | 0.00e+00 |
| DESI2 | -0.77 | 0.0010150 | 3.12e-04 |
| DEXI | -0.72 | 0.0569001 | 2.68e-02 |
| DFFA | -0.78 | 0.0103490 | 3.99e-03 |
| DGCR14 | 0.80 | 0.0080348 | 3.01e-03 |
| DGCR2 | 0.60 | 0.0085976 | 3.25e-03 |
| DGCR5 | -1.43 | 0.0799487 | 3.98e-02 |
| DGCR6L | -1.15 | 0.0029475 | 9.99e-04 |
| DGKA | 1.12 | 0.0001218 | 3.20e-05 |
| DGKH | 0.69 | 0.0367529 | 1.63e-02 |
| DGKZ | 0.51 | 0.0871109 | 4.40e-02 |
| DHCR24 | 1.39 | 0.0000000 | 0.00e+00 |
| DHCR7 | 1.31 | 0.0000000 | 0.00e+00 |
| DHFR | -3.81 | 0.0000000 | 0.00e+00 |
| DHFRL1 | -1.63 | 0.0010640 | 3.28e-04 |
| DHODH | -2.32 | 0.0000011 | 2.00e-07 |
| DHRS11 | -1.66 | 0.0702976 | 3.42e-02 |
| DHRS13 | 1.01 | 0.0098706 | 3.78e-03 |
| DHRS3 | 1.57 | 0.0000000 | 0.00e+00 |
| DHRS4 | -1.20 | 0.0006299 | 1.86e-04 |
| DHRS4-AS1 | -0.87 | 0.0520550 | 2.42e-02 |
| DHRS4L2 | -1.44 | 0.0003082 | 8.61e-05 |
| DHRS7 | 1.03 | 0.0000000 | 0.00e+00 |
| DHRSX | 1.06 | 0.0000057 | 1.20e-06 |
| DHX15 | -1.76 | 0.0000000 | 0.00e+00 |
| DHX29 | -1.00 | 0.0000002 | 0.00e+00 |
| DHX30 | 0.33 | 0.0342530 | 1.51e-02 |
| DHX33 | -0.76 | 0.0001432 | 3.82e-05 |
| DHX36 | -0.58 | 0.0112368 | 4.37e-03 |
| DHX37 | 0.33 | 0.0686086 | 3.32e-02 |
| DHX40 | -0.87 | 0.0000061 | 1.30e-06 |
| DHX57 | -1.12 | 0.0000079 | 1.80e-06 |
| DHX58 | 8.87 | 0.0000000 | 0.00e+00 |
| DHX8 | 0.71 | 0.0000003 | 0.00e+00 |
| DHX9 | -0.98 | 0.0000000 | 0.00e+00 |
| DIAPH1 | 1.25 | 0.0000000 | 0.00e+00 |
| DIAPH2 | 1.02 | 0.0016603 | 5.32e-04 |
| DIAPH3 | -2.61 | 0.0000000 | 0.00e+00 |
| DICER1 | -0.57 | 0.0096971 | 3.71e-03 |
| DIMT1 | -0.75 | 0.0133067 | 5.27e-03 |
| DIP2B | 1.06 | 0.0000000 | 0.00e+00 |
| DIS3 | -0.95 | 0.0001534 | 4.11e-05 |
| DIS3L | -1.02 | 0.0000002 | 0.00e+00 |
| DISC1 | 2.48 | 0.0352662 | 1.56e-02 |
| DIXDC1 | -1.79 | 0.0009380 | 2.87e-04 |
| DKC1 | -1.06 | 0.0000000 | 0.00e+00 |
| DKFZP434I0714 | -1.81 | 0.0807166 | 4.02e-02 |
| DKK1 | -1.23 | 0.0000000 | 0.00e+00 |
| DLAT | -0.81 | 0.0003599 | 1.02e-04 |
| DLD | -1.06 | 0.0000000 | 0.00e+00 |
| DLEU1 | -3.66 | 0.0000001 | 0.00e+00 |
| DLG1 | -1.21 | 0.0000623 | 1.57e-05 |
| DLG3 | 1.06 | 0.0035433 | 1.22e-03 |
| DLG4 | 0.73 | 0.0886793 | 4.49e-02 |
| DLGAP4 | 2.56 | 0.0000000 | 0.00e+00 |
| DLGAP5 | -3.39 | 0.0000000 | 0.00e+00 |
| DLL4 | 5.85 | 0.0000000 | 0.00e+00 |
| DMAP1 | 0.95 | 0.0005814 | 1.71e-04 |
| DMKN | 1.32 | 0.0003488 | 9.85e-05 |
| DMTN | -0.45 | 0.0402570 | 1.81e-02 |
| DMWD | 1.10 | 0.0000001 | 0.00e+00 |
| DMXL2 | -0.90 | 0.0000083 | 1.80e-06 |
| DNA2 | -3.38 | 0.0000000 | 0.00e+00 |
| DNAAF1 | 6.09 | 0.0342207 | 1.51e-02 |
| DNAAF5 | -0.84 | 0.0000595 | 1.50e-05 |
| DNAH14 | -1.12 | 0.0392600 | 1.76e-02 |
| DNAH17 | 5.00 | 0.0000000 | 0.00e+00 |
| DNAJB1 | 0.27 | 0.0306442 | 1.33e-02 |
| DNAJB12 | 0.75 | 0.0000207 | 4.90e-06 |
| DNAJB14 | -1.14 | 0.0047981 | 1.71e-03 |
| DNAJB2 | 1.61 | 0.0000000 | 0.00e+00 |
| DNAJB5 | 3.77 | 0.0000000 | 0.00e+00 |
| DNAJB6 | 0.40 | 0.0223709 | 9.35e-03 |
| DNAJB9 | 1.76 | 0.0000000 | 0.00e+00 |
| DNAJC1 | 2.23 | 0.0000000 | 0.00e+00 |
| DNAJC10 | -0.93 | 0.0000000 | 0.00e+00 |
| DNAJC11 | -0.62 | 0.0059143 | 2.14e-03 |
| DNAJC12 | 1.10 | 0.0000007 | 1.00e-07 |
| DNAJC13 | -1.28 | 0.0000000 | 0.00e+00 |
| DNAJC15 | 3.67 | 0.0231254 | 9.71e-03 |
| DNAJC2 | -0.63 | 0.0113985 | 4.44e-03 |
| DNAJC22 | -0.76 | 0.0005109 | 1.49e-04 |
| DNAJC24 | -0.79 | 0.0906683 | 4.61e-02 |
| DNAJC25-GNG10 | -1.19 | 0.0003621 | 1.03e-04 |
| DNAJC5 | 1.31 | 0.0000000 | 0.00e+00 |
| DNAJC8 | -0.34 | 0.0887707 | 4.50e-02 |
| DNAJC9 | -1.98 | 0.0000000 | 0.00e+00 |
| DNASE2 | 0.57 | 0.0058451 | 2.12e-03 |
| DNER | 2.36 | 0.0099089 | 3.80e-03 |
| DNHD1 | 0.96 | 0.0036637 | 1.27e-03 |
| DNM1 | 0.28 | 0.0485453 | 2.23e-02 |
| DNM1L | -0.54 | 0.0009347 | 2.86e-04 |
| DNM2 | 0.55 | 0.0000962 | 2.49e-05 |
| DNMBP | 2.09 | 0.0000000 | 0.00e+00 |
| DNMT1 | -0.55 | 0.0007902 | 2.37e-04 |
| DNMT3A | 0.62 | 0.0387483 | 1.73e-02 |
| DNMT3B | -0.85 | 0.0226498 | 9.48e-03 |
| DNPEP | 0.71 | 0.0001584 | 4.25e-05 |
| DNTTIP1 | 1.07 | 0.0000003 | 1.00e-07 |
| DNTTIP2 | -0.36 | 0.0864706 | 4.36e-02 |
| DOC2B | 1.83 | 0.0134188 | 5.32e-03 |
| DOCK1 | -0.86 | 0.0000052 | 1.10e-06 |
| DOCK4 | 2.53 | 0.0000000 | 0.00e+00 |
| DOCK6 | 1.02 | 0.0002996 | 8.35e-05 |
| DOCK7 | -1.14 | 0.0001374 | 3.65e-05 |
| DOK4 | -0.76 | 0.0000001 | 0.00e+00 |
| DONSON | -1.55 | 0.0000961 | 2.48e-05 |
| DOPEY2 | 0.87 | 0.0307987 | 1.34e-02 |
| DOT1L | 2.24 | 0.0000000 | 0.00e+00 |
| DPCD | -1.72 | 0.0016294 | 5.21e-04 |
| DPEP1 | -2.36 | 0.0550818 | 2.58e-02 |
| DPF2 | 1.14 | 0.0000000 | 0.00e+00 |
| DPH1 | 0.57 | 0.0001966 | 5.33e-05 |
| DPH2 | -0.91 | 0.0004816 | 1.40e-04 |
| DPH5 | -1.15 | 0.0013933 | 4.40e-04 |
| DPH6 | -1.54 | 0.0166499 | 6.74e-03 |
| DPM1 | -0.85 | 0.0010907 | 3.37e-04 |
| DPP4 | 2.20 | 0.0223639 | 9.35e-03 |
| DPP9 | 0.64 | 0.0000070 | 1.60e-06 |
| DPY19L1 | -1.21 | 0.0000000 | 0.00e+00 |
| DPY19L3 | -1.77 | 0.0000028 | 6.00e-07 |
| DPY19L4 | -1.05 | 0.0006591 | 1.95e-04 |
| DPY30 | -0.80 | 0.0051077 | 1.83e-03 |
| DPYD | 1.56 | 0.0016857 | 5.41e-04 |
| DPYSL3 | -1.05 | 0.0085982 | 3.25e-03 |
| DPYSL5 | -4.28 | 0.0000000 | 0.00e+00 |
| DRAM1 | 1.87 | 0.0000000 | 0.00e+00 |
| DRAM2 | 0.90 | 0.0008263 | 2.50e-04 |
| DROSHA | -0.66 | 0.0024340 | 8.13e-04 |
| DSC2 | 1.47 | 0.0006046 | 1.78e-04 |
| DSCC1 | -4.01 | 0.0000000 | 0.00e+00 |
| DSE | 1.87 | 0.0000000 | 0.00e+00 |
| DSN1 | -2.38 | 0.0000000 | 0.00e+00 |
| DSP | 0.76 | 0.0000000 | 0.00e+00 |
| DST | -0.94 | 0.0000000 | 0.00e+00 |
| DSTN | -0.38 | 0.0003360 | 9.45e-05 |
| DSTNP2 | -0.69 | 0.0586659 | 2.77e-02 |
| DTD1 | -0.97 | 0.0000004 | 1.00e-07 |
| DTD2 | -1.27 | 0.0007380 | 2.20e-04 |
| DTL | -2.62 | 0.0000000 | 0.00e+00 |
| DTWD1 | -2.00 | 0.0055172 | 1.98e-03 |
| DTX2 | 2.71 | 0.0000000 | 0.00e+00 |
| DTX2P1-UPK3BP1-PMS2P11 | 1.98 | 0.0000000 | 0.00e+00 |
| DTX3 | 0.95 | 0.0088346 | 3.35e-03 |
| DTX3L | 3.79 | 0.0000000 | 0.00e+00 |
| DTX4 | 2.03 | 0.0000000 | 0.00e+00 |
| DTYMK | -2.90 | 0.0000000 | 0.00e+00 |
| DUOX2 | 10.74 | 0.0000010 | 2.00e-07 |
| DUOXA1 | 3.21 | 0.0750936 | 3.69e-02 |
| DUOXA2 | 6.88 | 0.0000288 | 6.90e-06 |
| DUSP1 | 2.04 | 0.0000000 | 0.00e+00 |
| DUSP10 | 4.37 | 0.0000000 | 0.00e+00 |
| DUSP11 | 0.71 | 0.0073259 | 2.72e-03 |
| DUSP14 | 0.83 | 0.0086461 | 3.27e-03 |
| DUSP15 | 2.74 | 0.0903933 | 4.59e-02 |
| DUSP16 | 1.67 | 0.0000000 | 0.00e+00 |
| DUSP19 | -1.88 | 0.0724628 | 3.55e-02 |
| DUSP2 | 3.21 | 0.0123485 | 4.85e-03 |
| DUSP22 | 0.79 | 0.0173551 | 7.06e-03 |
| DUSP3 | -0.48 | 0.0018311 | 5.93e-04 |
| DUSP4 | 1.33 | 0.0000000 | 0.00e+00 |
| DUSP5 | 2.73 | 0.0000000 | 0.00e+00 |
| DUSP6 | 4.14 | 0.0000000 | 0.00e+00 |
| DUSP8 | 3.81 | 0.0000003 | 0.00e+00 |
| DUT | -1.53 | 0.0000000 | 0.00e+00 |
| DUXAP10 | -0.86 | 0.0362062 | 1.61e-02 |
| DVL1 | 0.67 | 0.0003018 | 8.41e-05 |
| DVL2 | 0.56 | 0.0022210 | 7.35e-04 |
| DVL3 | 0.72 | 0.0004878 | 1.42e-04 |
| DYNC1H1 | 0.66 | 0.0000000 | 0.00e+00 |
| DYNC1I2 | -0.50 | 0.0112601 | 4.38e-03 |
| DYNC2H1 | -2.84 | 0.0000000 | 0.00e+00 |
| DYNC2LI1 | -0.86 | 0.0878814 | 4.44e-02 |
| DYNLL1 | -0.68 | 0.0000001 | 0.00e+00 |
| DYNLRB1 | 1.15 | 0.0000000 | 0.00e+00 |
| DYRK1A | 0.65 | 0.0121747 | 4.78e-03 |
| DYRK1B | 1.92 | 0.0000000 | 0.00e+00 |
| DYRK2 | 0.73 | 0.0024502 | 8.19e-04 |
| DYSF | 4.38 | 0.0000000 | 0.00e+00 |
| DYX1C1 | -2.72 | 0.0071583 | 2.65e-03 |
| DZIP1L | 1.12 | 0.0349714 | 1.54e-02 |
| DZIP3 | -1.31 | 0.0002259 | 6.19e-05 |
| E2F1 | -3.42 | 0.0000000 | 0.00e+00 |
| E2F2 | -3.32 | 0.0000000 | 0.00e+00 |
| E2F4 | 0.87 | 0.0000000 | 0.00e+00 |
| E2F7 | -2.32 | 0.0000002 | 0.00e+00 |
| E2F8 | -2.62 | 0.0000001 | 0.00e+00 |
| EAF1 | 1.11 | 0.0000001 | 0.00e+00 |
| EAPP | 0.59 | 0.0157029 | 6.33e-03 |
| EARS2 | -1.07 | 0.0000043 | 9.00e-07 |
| EBF1 | 1.72 | 0.0059522 | 2.16e-03 |
| EBF4 | 3.96 | 0.0000000 | 0.00e+00 |
| EBI3 | 6.50 | 0.0000000 | 0.00e+00 |
| EBNA1BP2 | -1.79 | 0.0000000 | 0.00e+00 |
| EBP | 0.39 | 0.0189622 | 7.79e-03 |
| EBPL | -1.95 | 0.0000000 | 0.00e+00 |
| ECD | -0.42 | 0.0867425 | 4.38e-02 |
| ECE1 | 2.03 | 0.0000000 | 0.00e+00 |
| ECE2 | -1.36 | 0.0132069 | 5.22e-03 |
| ECEL1 | 3.91 | 0.0002335 | 6.41e-05 |
| ECHDC1 | -0.97 | 0.0015068 | 4.79e-04 |
| ECHS1 | -0.80 | 0.0000011 | 2.00e-07 |
| ECI1 | -0.55 | 0.0118661 | 4.64e-03 |
| ECI2 | -0.69 | 0.0109841 | 4.27e-03 |
| ECM1 | 4.13 | 0.0000000 | 0.00e+00 |
| ECSCR | 5.61 | 0.0735043 | 3.61e-02 |
| ECT2 | -1.25 | 0.0000000 | 0.00e+00 |
| EDA2R | -0.84 | 0.0342652 | 1.51e-02 |
| EDAR | 4.29 | 0.0443644 | 2.02e-02 |
| EDC4 | 0.66 | 0.0001400 | 3.72e-05 |
| EDF1 | 0.46 | 0.0000344 | 8.40e-06 |
| EDN1 | 0.82 | 0.0000002 | 0.00e+00 |
| EDNRA | 3.07 | 0.0149581 | 5.99e-03 |
| EDRF1 | -0.72 | 0.0250785 | 1.06e-02 |
| EEA1 | -0.91 | 0.0424233 | 1.92e-02 |
| EEF1A1 | -0.23 | 0.0041938 | 1.47e-03 |
| EEF1A2 | 0.94 | 0.0000001 | 0.00e+00 |
| EEF1B2 | -0.69 | 0.0000000 | 0.00e+00 |
| EEF1E1 | -1.55 | 0.0000250 | 6.00e-06 |
| EEF1E1-BLOC1S5 | -0.77 | 0.0205611 | 8.51e-03 |
| EEF1G | -0.29 | 0.0006533 | 1.93e-04 |
| EEF2 | 0.89 | 0.0000000 | 0.00e+00 |
| EEF2K | -0.67 | 0.0221215 | 9.23e-03 |
| EFCAB11 | -2.61 | 0.0000090 | 2.00e-06 |
| EFCAB5 | 4.61 | 0.0211942 | 8.80e-03 |
| EFEMP1 | -3.70 | 0.0000000 | 0.00e+00 |
| EFEMP2 | 2.17 | 0.0680787 | 3.29e-02 |
| EFHC1 | -2.18 | 0.0000009 | 2.00e-07 |
| EFHD1 | 1.57 | 0.0110424 | 4.29e-03 |
| EFNA1 | 5.13 | 0.0000000 | 0.00e+00 |
| EFNA3 | 1.64 | 0.0005565 | 1.63e-04 |
| EFNA5 | 1.20 | 0.0000061 | 1.30e-06 |
| EFNB1 | 3.14 | 0.0000000 | 0.00e+00 |
| EFNB2 | 3.04 | 0.0000000 | 0.00e+00 |
| EFR3B | 1.59 | 0.0000000 | 0.00e+00 |
| EFTUD1 | 0.91 | 0.0000002 | 0.00e+00 |
| EGFLAM | 5.61 | 0.0724836 | 3.55e-02 |
| EGFR | 1.14 | 0.0000000 | 0.00e+00 |
| EGLN1 | 0.45 | 0.0002979 | 8.30e-05 |
| EGLN2 | 1.08 | 0.0000000 | 0.00e+00 |
| EGLN3 | 1.60 | 0.0000001 | 0.00e+00 |
| EGOT | 6.07 | 0.0003930 | 1.12e-04 |
| EGR1 | 7.11 | 0.0000000 | 0.00e+00 |
| EGR2 | 4.54 | 0.0010114 | 3.11e-04 |
| EGR3 | 5.93 | 0.0000006 | 1.00e-07 |
| EGR4 | 8.47 | 0.0002721 | 7.54e-05 |
| EHBP1 | -1.38 | 0.0000000 | 0.00e+00 |
| EHBP1L1 | 1.78 | 0.0000000 | 0.00e+00 |
| EHD1 | 2.36 | 0.0000000 | 0.00e+00 |
| EHD2 | 1.18 | 0.0000000 | 0.00e+00 |
| EHD4 | 2.70 | 0.0000000 | 0.00e+00 |
| EHHADH | -0.96 | 0.0864104 | 4.36e-02 |
| EI24 | -0.59 | 0.0001437 | 3.83e-05 |
| EID1 | -0.97 | 0.0000000 | 0.00e+00 |
| EID2 | -0.45 | 0.0951235 | 4.88e-02 |
| EID2B | -1.28 | 0.0664830 | 3.20e-02 |
| EIF1 | 0.75 | 0.0000000 | 0.00e+00 |
| EIF1AX | -1.22 | 0.0000000 | 0.00e+00 |
| EIF1AY | -0.70 | 0.0512602 | 2.37e-02 |
| EIF2A | -0.37 | 0.0351039 | 1.55e-02 |
| EIF2AK1 | -0.26 | 0.0818011 | 4.09e-02 |
| EIF2AK2 | 1.41 | 0.0000000 | 0.00e+00 |
| EIF2B1 | -0.69 | 0.0016046 | 5.12e-04 |
| EIF2B3 | -1.03 | 0.0014906 | 4.73e-04 |
| EIF2D | -0.61 | 0.0052040 | 1.86e-03 |
| EIF2S1 | -0.66 | 0.0000045 | 1.00e-06 |
| EIF3A | -0.80 | 0.0000000 | 0.00e+00 |
| EIF3B | -0.55 | 0.0000003 | 0.00e+00 |
| EIF3D | -0.33 | 0.0075195 | 2.80e-03 |
| EIF3E | -1.08 | 0.0000000 | 0.00e+00 |
| EIF3F | 0.29 | 0.0943761 | 4.83e-02 |
| EIF3G | 0.55 | 0.0000723 | 1.84e-05 |
| EIF3I | -0.57 | 0.0000140 | 3.20e-06 |
| EIF3K | -0.70 | 0.0000000 | 0.00e+00 |
| EIF3L | -0.93 | 0.0000000 | 0.00e+00 |
| EIF3M | 0.24 | 0.0767604 | 3.79e-02 |
| EIF4A1 | -0.66 | 0.0000000 | 0.00e+00 |
| EIF4A2 | 0.37 | 0.0109644 | 4.26e-03 |
| EIF4B | -1.24 | 0.0000000 | 0.00e+00 |
| EIF4E | -0.88 | 0.0117843 | 4.61e-03 |
| EIF4E3 | -1.03 | 0.0561343 | 2.64e-02 |
| EIF4EBP1 | -0.57 | 0.0026557 | 8.94e-04 |
| EIF4EBP2 | 0.45 | 0.0348149 | 1.54e-02 |
| EIF4G2 | -0.71 | 0.0000000 | 0.00e+00 |
| EIF4H | -0.63 | 0.0000000 | 0.00e+00 |
| EIF5 | 0.20 | 0.0792116 | 3.93e-02 |
| EIF5A | -0.78 | 0.0000000 | 0.00e+00 |
| EIF5AL1 | -0.96 | 0.0000000 | 0.00e+00 |
| EIF5B | -0.96 | 0.0000000 | 0.00e+00 |
| ELAC2 | -0.75 | 0.0000000 | 0.00e+00 |
| ELAVL1 | -0.68 | 0.0000872 | 2.24e-05 |
| ELF1 | 1.13 | 0.0000000 | 0.00e+00 |
| ELF3 | 2.14 | 0.0000000 | 0.00e+00 |
| ELF4 | 2.42 | 0.0000000 | 0.00e+00 |
| ELFN1 | 3.14 | 0.0314682 | 1.37e-02 |
| ELK1 | 0.56 | 0.0096030 | 3.67e-03 |
| ELK3 | 2.07 | 0.0000000 | 0.00e+00 |
| ELL | 2.56 | 0.0000000 | 0.00e+00 |
| ELL2 | 1.92 | 0.0000000 | 0.00e+00 |
| ELMO2 | 0.42 | 0.0604105 | 2.87e-02 |
| ELMOD2 | -1.48 | 0.0002803 | 7.78e-05 |
| ELMSAN1 | 0.83 | 0.0003227 | 9.05e-05 |
| ELOVL1 | 0.61 | 0.0000407 | 1.00e-05 |
| ELOVL3 | 3.54 | 0.0017641 | 5.68e-04 |
| ELOVL7 | 1.82 | 0.0000000 | 0.00e+00 |
| ELP3 | -0.39 | 0.0812630 | 4.06e-02 |
| ELP6 | -1.28 | 0.0013679 | 4.31e-04 |
| EMB | -1.14 | 0.0388722 | 1.74e-02 |
| EMC2 | -0.85 | 0.0377649 | 1.68e-02 |
| EMC6 | -0.37 | 0.0791600 | 3.93e-02 |
| EMC7 | 0.52 | 0.0004406 | 1.27e-04 |
| EMC9 | -0.97 | 0.0021402 | 7.05e-04 |
| EME1 | -3.21 | 0.0000000 | 0.00e+00 |
| EMG1 | -1.14 | 0.0000020 | 4.00e-07 |
| EMILIN2 | 1.22 | 0.0252516 | 1.07e-02 |
| EML1 | -0.64 | 0.0204748 | 8.47e-03 |
| EML2 | 2.07 | 0.0000000 | 0.00e+00 |
| EML4 | -0.81 | 0.0000000 | 0.00e+00 |
| EML6 | -2.03 | 0.0756838 | 3.73e-02 |
| EMP1 | 2.03 | 0.0000000 | 0.00e+00 |
| EMP3 | 0.76 | 0.0000000 | 0.00e+00 |
| ENAH | -0.40 | 0.0180659 | 7.38e-03 |
| ENC1 | 0.91 | 0.0000076 | 1.70e-06 |
| ENDOG | -1.17 | 0.0049333 | 1.76e-03 |
| ENGASE | 0.63 | 0.0066813 | 2.45e-03 |
| ENKUR | 4.16 | 0.0004489 | 1.30e-04 |
| ENO1 | -0.92 | 0.0000000 | 0.00e+00 |
| ENOPH1 | -1.75 | 0.0000000 | 0.00e+00 |
| ENOSF1 | -1.74 | 0.0000000 | 0.00e+00 |
| ENTHD2 | 0.76 | 0.0121030 | 4.74e-03 |
| ENTPD6 | 0.35 | 0.0138835 | 5.52e-03 |
| ENY2 | -0.54 | 0.0130175 | 5.14e-03 |
| EP300 | 1.39 | 0.0000000 | 0.00e+00 |
| EP400 | 0.42 | 0.0087538 | 3.31e-03 |
| EPAS1 | -0.23 | 0.0514906 | 2.39e-02 |
| EPB41 | 0.45 | 0.0903986 | 4.60e-02 |
| EPB41L4A-AS1 | 1.55 | 0.0000001 | 0.00e+00 |
| EPB41L5 | -0.94 | 0.0029421 | 9.96e-04 |
| EPDR1 | -0.36 | 0.0431345 | 1.95e-02 |
| EPG5 | 0.52 | 0.0652068 | 3.13e-02 |
| EPHA2 | 2.51 | 0.0000000 | 0.00e+00 |
| EPHA4 | 3.12 | 0.0047401 | 1.68e-03 |
| EPHB2 | 2.13 | 0.0000000 | 0.00e+00 |
| EPHX1 | -0.94 | 0.0000000 | 0.00e+00 |
| EPHX2 | -2.05 | 0.0000437 | 1.08e-05 |
| EPM2AIP1 | -1.14 | 0.0003997 | 1.14e-04 |
| EPN1 | 1.09 | 0.0000000 | 0.00e+00 |
| EPN2 | 0.73 | 0.0000546 | 1.37e-05 |
| EPPK1 | 2.10 | 0.0267065 | 1.14e-02 |
| EPRS | -0.88 | 0.0000000 | 0.00e+00 |
| EPS15L1 | 0.71 | 0.0105073 | 4.06e-03 |
| EPS8 | 1.13 | 0.0000000 | 0.00e+00 |
| EPS8L3 | 3.46 | 0.0000000 | 0.00e+00 |
| EPSTI1 | 7.02 | 0.0000151 | 3.50e-06 |
| EPT1 | -0.48 | 0.0095782 | 3.66e-03 |
| ERAP1 | 1.69 | 0.0000000 | 0.00e+00 |
| ERAP2 | 2.81 | 0.0000000 | 0.00e+00 |
| ERC1 | 0.38 | 0.0821821 | 4.11e-02 |
| ERCC6L | -3.97 | 0.0000000 | 0.00e+00 |
| ERCC6L2 | -0.63 | 0.0943149 | 4.83e-02 |
| ERCC8 | -0.74 | 0.0849193 | 4.27e-02 |
| EREG | 1.09 | 0.0000000 | 0.00e+00 |
| ERF | 0.66 | 0.0000000 | 0.00e+00 |
| ERGIC1 | -0.49 | 0.0000102 | 2.30e-06 |
| ERGIC2 | -1.07 | 0.0002431 | 6.69e-05 |
| ERGIC3 | 0.40 | 0.0015356 | 4.89e-04 |
| ERH | -1.19 | 0.0000000 | 0.00e+00 |
| ERI1 | -0.65 | 0.0166462 | 6.74e-03 |
| ERI2 | -1.26 | 0.0040511 | 1.41e-03 |
| ERI3 | -1.07 | 0.0000044 | 9.00e-07 |
| ERICH1 | 0.59 | 0.0708789 | 3.45e-02 |
| ERLEC1 | 0.29 | 0.0910093 | 4.63e-02 |
| ERLIN1 | -0.87 | 0.0000135 | 3.10e-06 |
| ERLIN2 | -0.81 | 0.0005960 | 1.75e-04 |
| ERMARD | -2.02 | 0.0293447 | 1.26e-02 |
| ERMP1 | -1.06 | 0.0000004 | 1.00e-07 |
| ERN1 | 2.70 | 0.0000000 | 0.00e+00 |
| ERO1A | 1.03 | 0.0000000 | 0.00e+00 |
| ERP44 | 1.38 | 0.0000000 | 0.00e+00 |
| ERRFI1 | 0.43 | 0.0007988 | 2.40e-04 |
| ESCO2 | -2.74 | 0.0000000 | 0.00e+00 |
| ESD | -1.23 | 0.0000017 | 4.00e-07 |
| ESF1 | -0.74 | 0.0101633 | 3.91e-03 |
| ESPL1 | -1.03 | 0.0000083 | 1.90e-06 |
| ESRRA | 1.24 | 0.0000000 | 0.00e+00 |
| ESRRB | 3.95 | 0.0783729 | 3.88e-02 |
| ESYT1 | -0.56 | 0.0032500 | 1.11e-03 |
| ESYT2 | 0.76 | 0.0000000 | 0.00e+00 |
| ETAA1 | -1.03 | 0.0105223 | 4.06e-03 |
| ETF1 | -0.89 | 0.0000000 | 0.00e+00 |
| ETFA | -0.62 | 0.0000013 | 3.00e-07 |
| ETFB | -0.90 | 0.0000001 | 0.00e+00 |
| ETFDH | -1.05 | 0.0693234 | 3.36e-02 |
| ETHE1 | 1.64 | 0.0000000 | 0.00e+00 |
| ETS1 | 2.63 | 0.0000000 | 0.00e+00 |
| ETS2 | 1.66 | 0.0000000 | 0.00e+00 |
| ETV3 | 3.24 | 0.0000000 | 0.00e+00 |
| ETV3L | 3.81 | 0.0000100 | 2.20e-06 |
| ETV4 | 0.51 | 0.0029868 | 1.01e-03 |
| ETV5 | 1.32 | 0.0000000 | 0.00e+00 |
| ETV6 | 2.09 | 0.0000000 | 0.00e+00 |
| ETV7 | 6.65 | 0.0000551 | 1.38e-05 |
| EVA1B | 1.40 | 0.0018206 | 5.89e-04 |
| EVC | -0.69 | 0.0323059 | 1.41e-02 |
| EVC2 | 1.58 | 0.0014815 | 4.70e-04 |
| EVI5L | 1.02 | 0.0838891 | 4.21e-02 |
| EVX1 | 7.65 | 0.0016470 | 5.27e-04 |
| EWSR1 | 0.39 | 0.0007394 | 2.21e-04 |
| EXO1 | -3.27 | 0.0000000 | 0.00e+00 |
| EXOC3 | 0.51 | 0.0148583 | 5.95e-03 |
| EXOC3L1 | 5.44 | 0.0918945 | 4.69e-02 |
| EXOC3L2 | 2.30 | 0.0735918 | 3.61e-02 |
| EXOC3L4 | 2.29 | 0.0000101 | 2.30e-06 |
| EXOC4 | -0.54 | 0.0055469 | 2.00e-03 |
| EXOC5 | -0.95 | 0.0000388 | 9.50e-06 |
| EXOC7 | 0.68 | 0.0000001 | 0.00e+00 |
| EXOSC2 | -1.49 | 0.0000000 | 0.00e+00 |
| EXOSC3 | -0.75 | 0.0077255 | 2.88e-03 |
| EXOSC5 | -1.63 | 0.0000000 | 0.00e+00 |
| EXOSC7 | -0.77 | 0.0097700 | 3.74e-03 |
| EXOSC8 | -1.11 | 0.0010690 | 3.30e-04 |
| EXOSC9 | -0.84 | 0.0031159 | 1.06e-03 |
| EXT1 | 2.42 | 0.0000000 | 0.00e+00 |
| EXTL2 | -1.19 | 0.0154585 | 6.21e-03 |
| EYA3 | 0.61 | 0.0547981 | 2.56e-02 |
| EYA4 | -0.55 | 0.0751923 | 3.70e-02 |
| EZH2 | 0.63 | 0.0005305 | 1.55e-04 |
| F11R | 1.78 | 0.0000000 | 0.00e+00 |
| F12 | -1.63 | 0.0045971 | 1.63e-03 |
| F2RL1 | 1.47 | 0.0000000 | 0.00e+00 |
| F2RL2 | -1.36 | 0.0012703 | 3.98e-04 |
| F3 | 3.19 | 0.0000000 | 0.00e+00 |
| FA2H | 2.67 | 0.0000000 | 0.00e+00 |
| FAAH | 2.68 | 0.0304007 | 1.32e-02 |
| FAAH2 | 2.46 | 0.0062857 | 2.30e-03 |
| FAAP24 | -1.43 | 0.0006370 | 1.88e-04 |
| FABP5 | -1.84 | 0.0000002 | 0.00e+00 |
| FADD | 0.68 | 0.0000305 | 7.40e-06 |
| FADS1 | -0.38 | 0.0002946 | 8.21e-05 |
| FADS2 | -0.47 | 0.0000003 | 1.00e-07 |
| FADS3 | 1.24 | 0.0000001 | 0.00e+00 |
| FAH | -1.18 | 0.0002305 | 6.32e-05 |
| FAHD1 | -0.97 | 0.0002543 | 7.01e-05 |
| FAM101B | -2.34 | 0.0000000 | 0.00e+00 |
| FAM102A | 1.90 | 0.0000000 | 0.00e+00 |
| FAM104B | -2.29 | 0.0000518 | 1.29e-05 |
| FAM109A | 1.50 | 0.0000871 | 2.24e-05 |
| FAM110B | 2.91 | 0.0618224 | 2.95e-02 |
| FAM111B | -3.69 | 0.0000000 | 0.00e+00 |
| FAM114A1 | 0.57 | 0.0207352 | 8.59e-03 |
| FAM114A2 | -0.93 | 0.0219763 | 9.16e-03 |
| FAM117A | 3.82 | 0.0000000 | 0.00e+00 |
| FAM118A | -0.81 | 0.0856211 | 4.31e-02 |
| FAM120A | -0.60 | 0.0000590 | 1.48e-05 |
| FAM120C | -0.88 | 0.0944751 | 4.84e-02 |
| FAM124A | 1.38 | 0.0863386 | 4.35e-02 |
| FAM126A | -0.66 | 0.0656799 | 3.16e-02 |
| FAM129A | 2.43 | 0.0000000 | 0.00e+00 |
| FAM129B | 1.06 | 0.0000000 | 0.00e+00 |
| FAM134A | 0.41 | 0.0128140 | 5.05e-03 |
| FAM134C | 0.46 | 0.0166024 | 6.72e-03 |
| FAM136A | -1.22 | 0.0000000 | 0.00e+00 |
| FAM160A1 | 4.25 | 0.0463868 | 2.12e-02 |
| FAM160A2 | 0.79 | 0.0062040 | 2.26e-03 |
| FAM160B2 | 1.38 | 0.0000000 | 0.00e+00 |
| FAM161A | -1.79 | 0.0000410 | 1.01e-05 |
| FAM167A | 0.84 | 0.0030586 | 1.04e-03 |
| FAM168B | -1.18 | 0.0000000 | 0.00e+00 |
| FAM169A | -1.36 | 0.0015746 | 5.02e-04 |
| FAM171A1 | -1.36 | 0.0001255 | 3.31e-05 |
| FAM171B | -1.45 | 0.0199181 | 8.22e-03 |
| FAM172A | -2.65 | 0.0000007 | 1.00e-07 |
| FAM173A | -1.01 | 0.0033635 | 1.15e-03 |
| FAM173B | -2.94 | 0.0000063 | 1.40e-06 |
| FAM175A | -1.37 | 0.0124046 | 4.87e-03 |
| FAM177A1 | 0.61 | 0.0038092 | 1.32e-03 |
| FAM189A1 | 2.46 | 0.0615600 | 2.93e-02 |
| FAM189B | 0.81 | 0.0368854 | 1.64e-02 |
| FAM192A | -0.57 | 0.0013623 | 4.29e-04 |
| FAM193A | 1.75 | 0.0000000 | 0.00e+00 |
| FAM193B | 0.57 | 0.0452950 | 2.07e-02 |
| FAM195A | -0.96 | 0.0313269 | 1.36e-02 |
| FAM199X | -0.63 | 0.0023808 | 7.93e-04 |
| FAM201A | -1.65 | 0.0443071 | 2.01e-02 |
| FAM206A | -0.73 | 0.0649862 | 3.12e-02 |
| FAM208A | -1.03 | 0.0000077 | 1.70e-06 |
| FAM208B | -0.74 | 0.0000154 | 3.60e-06 |
| FAM20A | 6.03 | 0.0377181 | 1.68e-02 |
| FAM20B | -1.39 | 0.0000000 | 0.00e+00 |
| FAM210B | -1.16 | 0.0065209 | 2.39e-03 |
| FAM214B | 2.23 | 0.0000000 | 0.00e+00 |
| FAM216A | -2.41 | 0.0000115 | 2.60e-06 |
| FAM217B | -1.78 | 0.0000828 | 2.12e-05 |
| FAM219A | 1.51 | 0.0000000 | 0.00e+00 |
| FAM21A | 0.68 | 0.0011372 | 3.53e-04 |
| FAM21C | 0.58 | 0.0033115 | 1.13e-03 |
| FAM222A | 1.94 | 0.0000031 | 7.00e-07 |
| FAM222B | 1.91 | 0.0000000 | 0.00e+00 |
| FAM224A | -1.84 | 0.0676821 | 3.27e-02 |
| FAM224B | -1.75 | 0.0973445 | 5.00e-02 |
| FAM227A | -2.08 | 0.0876242 | 4.43e-02 |
| FAM26E | 3.67 | 0.0000321 | 7.80e-06 |
| FAM35A | -1.36 | 0.0000134 | 3.10e-06 |
| FAM35BP | -1.24 | 0.0296492 | 1.28e-02 |
| FAM35DP | -1.16 | 0.0835900 | 4.20e-02 |
| FAM3A | 0.44 | 0.0685050 | 3.32e-02 |
| FAM43A | 4.32 | 0.0000000 | 0.00e+00 |
| FAM46A | 2.23 | 0.0000000 | 0.00e+00 |
| FAM46B | 3.89 | 0.0000000 | 0.00e+00 |
| FAM49B | -0.58 | 0.0018814 | 6.11e-04 |
| FAM50A | 0.51 | 0.0019393 | 6.32e-04 |
| FAM53B | 0.66 | 0.0226208 | 9.47e-03 |
| FAM53C | 1.51 | 0.0000000 | 0.00e+00 |
| FAM58A | -0.64 | 0.0587869 | 2.78e-02 |
| FAM60A | 0.79 | 0.0004255 | 1.22e-04 |
| FAM64A | -3.58 | 0.0000000 | 0.00e+00 |
| FAM65A | 1.50 | 0.0000000 | 0.00e+00 |
| FAM65B | 8.14 | 0.0005637 | 1.65e-04 |
| FAM69B | -1.34 | 0.0019141 | 6.23e-04 |
| FAM71E1 | 2.19 | 0.0639247 | 3.06e-02 |
| FAM72A | -2.72 | 0.0000011 | 2.00e-07 |
| FAM72B | -2.33 | 0.0000003 | 1.00e-07 |
| FAM72C | -2.49 | 0.0000002 | 0.00e+00 |
| FAM72D | -2.42 | 0.0000015 | 3.00e-07 |
| FAM73A | -1.22 | 0.0033073 | 1.13e-03 |
| FAM81A | -3.19 | 0.0000000 | 0.00e+00 |
| FAM83A | 3.54 | 0.0000000 | 0.00e+00 |
| FAM83D | -2.46 | 0.0000000 | 0.00e+00 |
| FAM83G | 1.97 | 0.0000000 | 0.00e+00 |
| FAM83H | 0.96 | 0.0000011 | 2.00e-07 |
| FAM84B | 1.12 | 0.0005666 | 1.66e-04 |
| FAM86C2P | -1.19 | 0.0928514 | 4.74e-02 |
| FAM8A1 | 1.14 | 0.0004267 | 1.23e-04 |
| FAM91A1 | -0.69 | 0.0022865 | 7.60e-04 |
| FAM92A1 | -1.29 | 0.0210804 | 8.75e-03 |
| FAM96A | -0.88 | 0.0000034 | 7.00e-07 |
| FAM98A | -0.64 | 0.0019177 | 6.25e-04 |
| FAM98B | -1.06 | 0.0033572 | 1.15e-03 |
| FAN1 | -1.45 | 0.0006897 | 2.05e-04 |
| FANCA | -2.05 | 0.0000000 | 0.00e+00 |
| FANCB | -3.89 | 0.0000019 | 4.00e-07 |
| FANCC | -1.17 | 0.0001588 | 4.27e-05 |
| FANCD2 | -2.65 | 0.0000000 | 0.00e+00 |
| FANCG | -1.83 | 0.0000000 | 0.00e+00 |
| FANCI | -2.22 | 0.0000000 | 0.00e+00 |
| FANCL | -1.21 | 0.0011816 | 3.68e-04 |
| FANCM | -3.16 | 0.0000000 | 0.00e+00 |
| FAP | 5.53 | 0.0806408 | 4.02e-02 |
| FAR1 | -1.89 | 0.0000000 | 0.00e+00 |
| FAR2P2 | -3.06 | 0.0799928 | 3.98e-02 |
| FARP2 | -0.82 | 0.0614942 | 2.93e-02 |
| FARSB | -2.43 | 0.0000000 | 0.00e+00 |
| FAS | 1.39 | 0.0000849 | 2.18e-05 |
| FASN | 1.42 | 0.0000000 | 0.00e+00 |
| FASTK | -0.88 | 0.0000759 | 1.93e-05 |
| FASTKD1 | -2.45 | 0.0000000 | 0.00e+00 |
| FASTKD2 | -1.24 | 0.0000006 | 1.00e-07 |
| FASTKD3 | -0.92 | 0.0343092 | 1.51e-02 |
| FAT3 | -2.36 | 0.0205414 | 8.50e-03 |
| FAXC | -1.86 | 0.0436265 | 1.98e-02 |
| FAXDC2 | 4.17 | 0.0000373 | 9.10e-06 |
| FBF1 | -1.21 | 0.0035845 | 1.24e-03 |
| FBL | -1.07 | 0.0000000 | 0.00e+00 |
| FBLIM1 | 1.13 | 0.0000029 | 6.00e-07 |
| FBLN5 | 3.14 | 0.0000000 | 0.00e+00 |
| FBN1 | 2.17 | 0.0000000 | 0.00e+00 |
| FBN2 | -3.05 | 0.0000000 | 0.00e+00 |
| FBRS | 2.11 | 0.0000000 | 0.00e+00 |
| FBRSL1 | 1.16 | 0.0000933 | 2.41e-05 |
| FBXL12 | 0.95 | 0.0144443 | 5.76e-03 |
| FBXL18 | 0.66 | 0.0100363 | 3.86e-03 |
| FBXL19 | 0.93 | 0.0000550 | 1.38e-05 |
| FBXL20 | 0.93 | 0.0007592 | 2.27e-04 |
| FBXL5 | 0.67 | 0.0043358 | 1.52e-03 |
| FBXO10 | 1.16 | 0.0010865 | 3.36e-04 |
| FBXO11 | 0.85 | 0.0000018 | 4.00e-07 |
| FBXO17 | -0.91 | 0.0000002 | 0.00e+00 |
| FBXO18 | 0.36 | 0.0694777 | 3.37e-02 |
| FBXO2 | 3.74 | 0.0000000 | 0.00e+00 |
| FBXO21 | -0.56 | 0.0036784 | 1.27e-03 |
| FBXO22 | -0.94 | 0.0000042 | 9.00e-07 |
| FBXO25 | -0.76 | 0.0917306 | 4.68e-02 |
| FBXO3 | -1.26 | 0.0010829 | 3.35e-04 |
| FBXO30 | -0.59 | 0.0798110 | 3.97e-02 |
| FBXO32 | 5.10 | 0.0000000 | 0.00e+00 |
| FBXO42 | 0.72 | 0.0246691 | 1.04e-02 |
| FBXO43 | -3.73 | 0.0000186 | 4.40e-06 |
| FBXO45 | -0.92 | 0.0220574 | 9.20e-03 |
| FBXO46 | 2.24 | 0.0000000 | 0.00e+00 |
| FBXO5 | -1.73 | 0.0000018 | 4.00e-07 |
| FBXO6 | 1.72 | 0.0000000 | 0.00e+00 |
| FBXO8 | 0.79 | 0.0344991 | 1.52e-02 |
| FBXW2 | -0.53 | 0.0095193 | 3.63e-03 |
| FBXW4 | 1.15 | 0.0000006 | 1.00e-07 |
| FBXW5 | 1.05 | 0.0000000 | 0.00e+00 |
| FBXW9 | -1.10 | 0.0604702 | 2.87e-02 |
| FCAMR | 6.53 | 0.0161724 | 6.54e-03 |
| FCF1 | -0.65 | 0.0048975 | 1.74e-03 |
| FCGBP | 1.24 | 0.0000002 | 0.00e+00 |
| FCHO2 | 1.00 | 0.0000643 | 1.62e-05 |
| FCHSD1 | 1.67 | 0.0000000 | 0.00e+00 |
| FCRLA | 6.30 | 0.0236212 | 9.95e-03 |
| FDFT1 | 0.91 | 0.0000000 | 0.00e+00 |
| FDPS | 0.85 | 0.0000000 | 0.00e+00 |
| FDXACB1 | -2.11 | 0.0722215 | 3.53e-02 |
| FDXR | -1.63 | 0.0000000 | 0.00e+00 |
| FECH | -1.41 | 0.0000000 | 0.00e+00 |
| FEM1B | 0.46 | 0.0185403 | 7.59e-03 |
| FEM1C | 1.21 | 0.0000237 | 5.60e-06 |
| FEN1 | -2.04 | 0.0000000 | 0.00e+00 |
| FER | -0.99 | 0.0103974 | 4.01e-03 |
| FERMT1 | 1.93 | 0.0000000 | 0.00e+00 |
| FERMT2 | 1.11 | 0.0000000 | 0.00e+00 |
| FEZ1 | 5.17 | 0.0000000 | 0.00e+00 |
| FFAR2 | 9.50 | 0.0000228 | 5.40e-06 |
| FGA | 2.88 | 0.0000000 | 0.00e+00 |
| FGB | 0.64 | 0.0000047 | 1.00e-06 |
| FGD1 | 0.83 | 0.0321172 | 1.40e-02 |
| FGD5-AS1 | -1.06 | 0.0000000 | 0.00e+00 |
| FGD6 | 1.53 | 0.0000000 | 0.00e+00 |
| FGF18 | 6.25 | 0.0002175 | 5.95e-05 |
| FGF2 | 1.67 | 0.0000000 | 0.00e+00 |
| FGFBP3 | -2.19 | 0.0426749 | 1.93e-02 |
| FGFR1OP | -0.93 | 0.0558122 | 2.62e-02 |
| FGFR3 | -3.31 | 0.0000004 | 1.00e-07 |
| FGFR4 | -0.93 | 0.0004003 | 1.15e-04 |
| FGG | 1.58 | 0.0000000 | 0.00e+00 |
| FGL1 | 0.65 | 0.0924883 | 4.72e-02 |
| FH | -1.55 | 0.0000000 | 0.00e+00 |
| FHDC1 | 3.80 | 0.0000000 | 0.00e+00 |
| FHL2 | 1.32 | 0.0000000 | 0.00e+00 |
| FHL3 | 2.14 | 0.0003391 | 9.55e-05 |
| FHOD3 | 2.66 | 0.0000000 | 0.00e+00 |
| FIBP | -0.93 | 0.0001082 | 2.82e-05 |
| FICD | 1.66 | 0.0001776 | 4.80e-05 |
| FIG4 | -1.16 | 0.0350983 | 1.55e-02 |
| FIGNL1 | -2.63 | 0.0000000 | 0.00e+00 |
| FILIP1 | -0.79 | 0.0083122 | 3.13e-03 |
| FILIP1L | 4.78 | 0.0003490 | 9.86e-05 |
| FIS1 | -0.55 | 0.0103648 | 4.00e-03 |
| FIZ1 | 0.86 | 0.0045752 | 1.62e-03 |
| FJX1 | 0.95 | 0.0009492 | 2.91e-04 |
| FKBP11 | 0.95 | 0.0024366 | 8.14e-04 |
| FKBP15 | 1.02 | 0.0000000 | 0.00e+00 |
| FKBP1A | -0.44 | 0.0004171 | 1.20e-04 |
| FKBP2 | 0.57 | 0.0172679 | 7.02e-03 |
| FKBP3 | -0.49 | 0.0057233 | 2.07e-03 |
| FKBP4 | -1.27 | 0.0000000 | 0.00e+00 |
| FKBP5 | -1.54 | 0.0000005 | 1.00e-07 |
| FKBP7 | -2.78 | 0.0000845 | 2.17e-05 |
| FKBP9 | -0.79 | 0.0000158 | 3.70e-06 |
| FKBP9P1 | -0.65 | 0.0911055 | 4.64e-02 |
| FKRP | 1.06 | 0.0000174 | 4.10e-06 |
| FKTN | -1.17 | 0.0011260 | 3.49e-04 |
| FLCN | 1.40 | 0.0000000 | 0.00e+00 |
| FLII | 1.03 | 0.0000000 | 0.00e+00 |
| FLJ13224 | 5.61 | 0.0750936 | 3.69e-02 |
| FLJ32255 | 1.79 | 0.0022095 | 7.31e-04 |
| FLJ42627 | 1.14 | 0.0698148 | 3.39e-02 |
| FLNA | 1.15 | 0.0000000 | 0.00e+00 |
| FLNB | 0.40 | 0.0000038 | 8.00e-07 |
| FLOT2 | 0.45 | 0.0011244 | 3.48e-04 |
| FLRT3 | 0.86 | 0.0000118 | 2.70e-06 |
| FLT3LG | 3.39 | 0.0002899 | 8.07e-05 |
| FLT4 | 1.13 | 0.0004456 | 1.29e-04 |
| FLVCR1 | 0.53 | 0.0215681 | 8.97e-03 |
| FLYWCH1 | 0.78 | 0.0001292 | 3.42e-05 |
| FMNL1 | 1.62 | 0.0000000 | 0.00e+00 |
| FMNL2 | 1.01 | 0.0000000 | 0.00e+00 |
| FMNL3 | 2.24 | 0.0000000 | 0.00e+00 |
| FMR1 | 0.48 | 0.0555108 | 2.60e-02 |
| FN1 | 1.83 | 0.0000000 | 0.00e+00 |
| FN3KRP | -1.32 | 0.0000000 | 0.00e+00 |
| FNBP1 | 1.28 | 0.0000029 | 6.00e-07 |
| FNBP1L | -0.80 | 0.0026409 | 8.88e-04 |
| FNBP4 | 0.42 | 0.0409824 | 1.84e-02 |
| FNDC3A | 1.81 | 0.0000000 | 0.00e+00 |
| FNDC3B | 1.14 | 0.0000000 | 0.00e+00 |
| FNDC4 | 1.77 | 0.0064401 | 2.36e-03 |
| FNTB | -0.76 | 0.0003611 | 1.02e-04 |
| FOCAD | -0.69 | 0.0061797 | 2.25e-03 |
| FOPNL | -1.87 | 0.0000000 | 0.00e+00 |
| FOS | 2.37 | 0.0000000 | 0.00e+00 |
| FOSB | 7.68 | 0.0000000 | 0.00e+00 |
| FOSL1 | 3.45 | 0.0000000 | 0.00e+00 |
| FOSL2 | 2.24 | 0.0000000 | 0.00e+00 |
| FOXA1 | 1.44 | 0.0000000 | 0.00e+00 |
| FOXA2 | 2.16 | 0.0000000 | 0.00e+00 |
| FOXC1 | 1.40 | 0.0000006 | 1.00e-07 |
| FOXC2 | 3.19 | 0.0090880 | 3.45e-03 |
| FOXD1 | 1.87 | 0.0000007 | 1.00e-07 |
| FOXD2 | 1.55 | 0.0894289 | 4.54e-02 |
| FOXF1 | 5.90 | 0.0466313 | 2.14e-02 |
| FOXF2 | 1.03 | 0.0569996 | 2.69e-02 |
| FOXJ2 | 1.26 | 0.0000001 | 0.00e+00 |
| FOXK1 | 0.49 | 0.0083337 | 3.14e-03 |
| FOXK2 | 0.53 | 0.0002680 | 7.43e-05 |
| FOXM1 | -1.54 | 0.0000000 | 0.00e+00 |
| FOXN2 | 0.69 | 0.0111316 | 4.33e-03 |
| FOXO1 | 2.42 | 0.0000000 | 0.00e+00 |
| FOXO3 | 1.86 | 0.0000000 | 0.00e+00 |
| FOXO3B | 0.91 | 0.0074267 | 2.76e-03 |
| FOXO4 | 1.71 | 0.0002004 | 5.44e-05 |
| FOXP1 | 2.27 | 0.0000000 | 0.00e+00 |
| FOXP4 | 1.85 | 0.0000000 | 0.00e+00 |
| FOXQ1 | 5.17 | 0.0000000 | 0.00e+00 |
| FOXRED1 | -1.61 | 0.0000014 | 3.00e-07 |
| FOXRED2 | -2.04 | 0.0000001 | 0.00e+00 |
| FPGS | 0.96 | 0.0000000 | 0.00e+00 |
| FPGT | -1.42 | 0.0026155 | 8.78e-04 |
| FRAS1 | -1.39 | 0.0001429 | 3.81e-05 |
| FRK | 1.11 | 0.0008471 | 2.56e-04 |
| FRMD1 | 5.69 | 0.0662364 | 3.19e-02 |
| FRMD4A | 1.05 | 0.0491948 | 2.26e-02 |
| FRMD4B | 1.17 | 0.0155803 | 6.26e-03 |
| FRMD5 | 2.18 | 0.0013942 | 4.40e-04 |
| FRMD6 | 2.48 | 0.0000000 | 0.00e+00 |
| FRMD8 | 1.71 | 0.0000000 | 0.00e+00 |
| FRS2 | 0.54 | 0.0897723 | 4.56e-02 |
| FSCN1 | 0.66 | 0.0000003 | 1.00e-07 |
| FSD1 | -2.00 | 0.0056496 | 2.04e-03 |
| FSD1L | 2.32 | 0.0000000 | 0.00e+00 |
| FST | 2.41 | 0.0000084 | 1.90e-06 |
| FSTL1 | 2.65 | 0.0000000 | 0.00e+00 |
| FSTL3 | 1.45 | 0.0000000 | 0.00e+00 |
| FSTL4 | 2.01 | 0.0000008 | 2.00e-07 |
| FTH1 | 0.85 | 0.0000000 | 0.00e+00 |
| FTH1P3 | 0.89 | 0.0001473 | 3.93e-05 |
| FTL | -2.46 | 0.0000000 | 0.00e+00 |
| FTSJ2 | -0.58 | 0.0125601 | 4.94e-03 |
| FTSJ3 | -0.30 | 0.0501491 | 2.32e-02 |
| FUCA1 | 0.85 | 0.0299426 | 1.30e-02 |
| FUK | 0.79 | 0.0342814 | 1.51e-02 |
| FURIN | 2.14 | 0.0000000 | 0.00e+00 |
| FUT1 | 3.32 | 0.0586487 | 2.77e-02 |
| FUT2 | 6.57 | 0.0145416 | 5.81e-03 |
| FUT4 | 1.31 | 0.0018914 | 6.15e-04 |
| FUT8 | 0.85 | 0.0015171 | 4.82e-04 |
| FUZ | -0.86 | 0.0666913 | 3.22e-02 |
| FXN | -1.36 | 0.0059385 | 2.15e-03 |
| FXR1 | -0.54 | 0.0011934 | 3.72e-04 |
| FXR2 | 1.00 | 0.0000000 | 0.00e+00 |
| FXYD3 | 2.77 | 0.0873916 | 4.41e-02 |
| FXYD5 | 1.81 | 0.0000000 | 0.00e+00 |
| FYCO1 | 0.51 | 0.0786240 | 3.90e-02 |
| FYN | 2.06 | 0.0000315 | 7.60e-06 |
| FZD2 | -1.29 | 0.0000001 | 0.00e+00 |
| FZD3 | -1.01 | 0.0170469 | 6.92e-03 |
| FZD4 | 2.61 | 0.0000000 | 0.00e+00 |
| FZD5 | 0.73 | 0.0049997 | 1.78e-03 |
| FZD6 | -0.62 | 0.0386387 | 1.73e-02 |
| FZD7 | 1.98 | 0.0000000 | 0.00e+00 |
| FZD8 | 1.90 | 0.0000000 | 0.00e+00 |
| FZR1 | 0.71 | 0.0016727 | 5.36e-04 |
| G0S2 | 5.41 | 0.0000000 | 0.00e+00 |
| G2E3 | -2.37 | 0.0000000 | 0.00e+00 |
| G3BP2 | -0.63 | 0.0000020 | 4.00e-07 |
| G6PD | -1.46 | 0.0000000 | 0.00e+00 |
| GAA | 1.71 | 0.0000000 | 0.00e+00 |
| GAB2 | 1.93 | 0.0000000 | 0.00e+00 |
| GAB3 | 4.04 | 0.0007592 | 2.27e-04 |
| GABARAPL1 | 3.95 | 0.0000000 | 0.00e+00 |
| GABARAPL3 | 4.53 | 0.0000000 | 0.00e+00 |
| GABPA | -1.50 | 0.0004831 | 1.40e-04 |
| GABPB1 | 0.74 | 0.0012779 | 4.00e-04 |
| GABPB1-AS1 | -1.02 | 0.0763817 | 3.77e-02 |
| GABRA5 | -2.58 | 0.0000000 | 0.00e+00 |
| GABRB3 | -1.91 | 0.0000000 | 0.00e+00 |
| GACAT2 | -3.50 | 0.0000003 | 1.00e-07 |
| GADD45A | 4.10 | 0.0000000 | 0.00e+00 |
| GADD45B | 1.23 | 0.0000000 | 0.00e+00 |
| GADD45G | 2.32 | 0.0908930 | 4.62e-02 |
| GADD45GIP1 | -0.64 | 0.0055902 | 2.02e-03 |
| GAK | 1.36 | 0.0000000 | 0.00e+00 |
| GAL3ST1 | -1.77 | 0.0001398 | 3.72e-05 |
| GALC | -0.85 | 0.0020600 | 6.77e-04 |
| GALK1 | -1.09 | 0.0000046 | 1.00e-06 |
| GALM | -1.43 | 0.0000001 | 0.00e+00 |
| GALNT1 | -1.23 | 0.0000000 | 0.00e+00 |
| GALNT10 | -1.07 | 0.0000000 | 0.00e+00 |
| GALNT11 | -1.98 | 0.0000000 | 0.00e+00 |
| GALNT12 | 2.50 | 0.0000000 | 0.00e+00 |
| GALNT13 | -1.42 | 0.0068028 | 2.50e-03 |
| GALNT2 | -0.51 | 0.0002836 | 7.88e-05 |
| GALNT4 | 1.33 | 0.0000000 | 0.00e+00 |
| GALNT5 | -2.05 | 0.0010225 | 3.15e-04 |
| GALNT7 | -0.72 | 0.0003982 | 1.14e-04 |
| GAMT | -2.12 | 0.0000035 | 7.00e-07 |
| GAPDH | -1.03 | 0.0000000 | 0.00e+00 |
| GAPVD1 | 0.75 | 0.0001220 | 3.21e-05 |
| GAREM | 1.89 | 0.0000003 | 1.00e-07 |
| GAREML | 0.78 | 0.0581361 | 2.75e-02 |
| GARS | 1.00 | 0.0000000 | 0.00e+00 |
| GART | -0.45 | 0.0030121 | 1.02e-03 |
| GAS2L3 | -2.10 | 0.0000158 | 3.70e-06 |
| GAS5 | 1.13 | 0.0000000 | 0.00e+00 |
| GATA3 | 2.86 | 0.0000000 | 0.00e+00 |
| GATA6 | 3.75 | 0.0000000 | 0.00e+00 |
| GATAD1 | 0.50 | 0.0396484 | 1.78e-02 |
| GATAD2A | 0.53 | 0.0041709 | 1.46e-03 |
| GATAD2B | 1.06 | 0.0022780 | 7.57e-04 |
| GATB | -1.20 | 0.0007330 | 2.19e-04 |
| GATC | -0.75 | 0.0003148 | 8.81e-05 |
| GATM | -1.09 | 0.0704250 | 3.42e-02 |
| GATS | -0.50 | 0.0706488 | 3.44e-02 |
| GATSL2 | -2.30 | 0.0000000 | 0.00e+00 |
| GBA2 | 0.46 | 0.0296395 | 1.28e-02 |
| GBAS | -0.94 | 0.0000009 | 2.00e-07 |
| GBE1 | -1.12 | 0.0000023 | 5.00e-07 |
| GBF1 | 1.39 | 0.0000000 | 0.00e+00 |
| GBP1 | 9.71 | 0.0000000 | 0.00e+00 |
| GBP1P1 | 9.65 | 0.0000157 | 3.60e-06 |
| GBP2 | 8.13 | 0.0000000 | 0.00e+00 |
| GBP3 | 5.10 | 0.0000000 | 0.00e+00 |
| GBP4 | 10.38 | 0.0000000 | 0.00e+00 |
| GBP5 | 11.84 | 0.0000000 | 0.00e+00 |
| GBP7 | 7.25 | 0.0038167 | 1.32e-03 |
| GCAT | -2.07 | 0.0775093 | 3.84e-02 |
| GCC1 | 1.22 | 0.0000000 | 0.00e+00 |
| GCFC2 | -0.90 | 0.0109436 | 4.25e-03 |
| GCH1 | 1.54 | 0.0000095 | 2.10e-06 |
| GCKR | 3.84 | 0.0000000 | 0.00e+00 |
| GCLC | -1.10 | 0.0000000 | 0.00e+00 |
| GCLM | -1.20 | 0.0002280 | 6.25e-05 |
| GCN1 | 0.40 | 0.0004943 | 1.44e-04 |
| GCNT3 | 1.16 | 0.0000000 | 0.00e+00 |
| GCNT4 | 3.50 | 0.0089861 | 3.41e-03 |
| GCSH | -2.75 | 0.0000000 | 0.00e+00 |
| GDA | 0.99 | 0.0541680 | 2.53e-02 |
| GDAP1 | -1.58 | 0.0000042 | 9.00e-07 |
| GDF11 | -2.06 | 0.0036834 | 1.27e-03 |
| GDF15 | 1.13 | 0.0000000 | 0.00e+00 |
| GDI1 | 1.00 | 0.0000000 | 0.00e+00 |
| GDI2 | -0.92 | 0.0000000 | 0.00e+00 |
| GEM | 1.83 | 0.0000001 | 0.00e+00 |
| GEMIN2 | -1.15 | 0.0078695 | 2.95e-03 |
| GEMIN4 | -0.90 | 0.0000001 | 0.00e+00 |
| GEMIN5 | -0.77 | 0.0004040 | 1.16e-04 |
| GEMIN6 | -1.30 | 0.0000430 | 1.06e-05 |
| GEN1 | -2.27 | 0.0000000 | 0.00e+00 |
| GFI1B | 4.39 | 0.0349629 | 1.54e-02 |
| GFM1 | -1.09 | 0.0000000 | 0.00e+00 |
| GFM2 | -0.64 | 0.0033926 | 1.17e-03 |
| GFPT1 | 0.72 | 0.0000003 | 1.00e-07 |
| GFPT2 | 3.85 | 0.0000000 | 0.00e+00 |
| GGA1 | 0.50 | 0.0584490 | 2.76e-02 |
| GGA2 | -0.90 | 0.0000066 | 1.50e-06 |
| GGA3 | 1.05 | 0.0000000 | 0.00e+00 |
| GGCT | -2.02 | 0.0000000 | 0.00e+00 |
| GGCX | 0.46 | 0.0602210 | 2.86e-02 |
| GGH | -1.19 | 0.0000012 | 3.00e-07 |
| GGT1 | 1.04 | 0.0000000 | 0.00e+00 |
| GGT3P | 1.31 | 0.0027778 | 9.38e-04 |
| GGT5 | 2.46 | 0.0060031 | 2.18e-03 |
| GGT7 | 1.07 | 0.0040809 | 1.42e-03 |
| GGTLC2 | 0.88 | 0.0156562 | 6.30e-03 |
| GHDC | 0.69 | 0.0175185 | 7.14e-03 |
| GHITM | -0.38 | 0.0009636 | 2.95e-04 |
| GID4 | 0.91 | 0.0006027 | 1.77e-04 |
| GIGYF1 | 0.57 | 0.0037626 | 1.30e-03 |
| GIMAP2 | 4.58 | 0.0232926 | 9.79e-03 |
| GINM1 | 0.77 | 0.0054926 | 1.97e-03 |
| GINS1 | -2.66 | 0.0000000 | 0.00e+00 |
| GINS2 | -3.87 | 0.0000000 | 0.00e+00 |
| GINS3 | -1.33 | 0.0000350 | 8.50e-06 |
| GINS4 | -2.86 | 0.0000000 | 0.00e+00 |
| GIP | -2.75 | 0.0116453 | 4.55e-03 |
| GIPR | 3.01 | 0.0000000 | 0.00e+00 |
| GIT1 | 0.40 | 0.0082517 | 3.10e-03 |
| GJA1 | 1.44 | 0.0000000 | 0.00e+00 |
| GJA9-MYCBP | -1.48 | 0.0002117 | 5.77e-05 |
| GJB2 | 4.62 | 0.0000000 | 0.00e+00 |
| GJB3 | 5.22 | 0.0000000 | 0.00e+00 |
| GJB4 | 4.07 | 0.0000107 | 2.40e-06 |
| GJC1 | 1.19 | 0.0000000 | 0.00e+00 |
| GJD3 | 3.00 | 0.0195312 | 8.04e-03 |
| GK | 3.65 | 0.0000000 | 0.00e+00 |
| GK3P | 4.10 | 0.0005953 | 1.75e-04 |
| GK5 | -1.44 | 0.0009446 | 2.89e-04 |
| GLA | -0.44 | 0.0038977 | 1.36e-03 |
| GLB1 | 0.63 | 0.0000974 | 2.52e-05 |
| GLB1L | -1.21 | 0.0820867 | 4.11e-02 |
| GLB1L2 | -2.72 | 0.0000246 | 5.90e-06 |
| GLDC | 1.90 | 0.0000000 | 0.00e+00 |
| GLE1 | -0.41 | 0.0230652 | 9.68e-03 |
| GLG1 | 0.72 | 0.0000000 | 0.00e+00 |
| GLIPR1 | 3.98 | 0.0000000 | 0.00e+00 |
| GLIS2 | 1.37 | 0.0000118 | 2.70e-06 |
| GLIS3 | 1.21 | 0.0000000 | 0.00e+00 |
| GLMP | 0.45 | 0.0841372 | 4.23e-02 |
| GLO1 | -0.93 | 0.0000000 | 0.00e+00 |
| GLOD4 | -1.37 | 0.0000000 | 0.00e+00 |
| GLP2R | -1.26 | 0.0033427 | 1.15e-03 |
| GLRX | 1.05 | 0.0000000 | 0.00e+00 |
| GLRX3 | -0.70 | 0.0000288 | 6.90e-06 |
| GLRX5 | -1.04 | 0.0000037 | 8.00e-07 |
| GLS | -0.75 | 0.0001945 | 5.27e-05 |
| GLTSCR1 | 1.94 | 0.0000000 | 0.00e+00 |
| GLTSCR2 | 1.17 | 0.0000000 | 0.00e+00 |
| GLUL | 1.44 | 0.0000000 | 0.00e+00 |
| GM2A | 1.73 | 0.0000000 | 0.00e+00 |
| GMCL1 | -1.02 | 0.0190170 | 7.81e-03 |
| GMDS | 0.68 | 0.0761775 | 3.76e-02 |
| GMEB2 | 1.17 | 0.0000078 | 1.70e-06 |
| GMFB | -1.24 | 0.0076021 | 2.83e-03 |
| GMIP | 2.86 | 0.0000000 | 0.00e+00 |
| GMNN | -2.28 | 0.0000000 | 0.00e+00 |
| GMPR2 | -0.42 | 0.0612786 | 2.91e-02 |
| GMPS | -0.41 | 0.0291147 | 1.25e-02 |
| GNA12 | 0.57 | 0.0014553 | 4.61e-04 |
| GNA13 | 0.57 | 0.0003194 | 8.95e-05 |
| GNA15 | 4.11 | 0.0067982 | 2.50e-03 |
| GNAI3 | -0.58 | 0.0017210 | 5.53e-04 |
| GNAL | -1.41 | 0.0022329 | 7.40e-04 |
| GNAQ | -0.49 | 0.0715637 | 3.49e-02 |
| GNAS | 0.19 | 0.0853545 | 4.30e-02 |
| GNAZ | -2.25 | 0.0164216 | 6.64e-03 |
| GNB1 | 0.42 | 0.0000632 | 1.59e-05 |
| GNB2 | 0.27 | 0.0409229 | 1.84e-02 |
| GNB4 | -1.19 | 0.0009347 | 2.86e-04 |
| GNB5 | -0.47 | 0.0532145 | 2.48e-02 |
| GNE | 0.41 | 0.0209624 | 8.69e-03 |
| GNG10 | -1.18 | 0.0001386 | 3.68e-05 |
| GNG12 | -1.06 | 0.0000000 | 0.00e+00 |
| GNG4 | 2.53 | 0.0000000 | 0.00e+00 |
| GNG5 | 1.13 | 0.0000000 | 0.00e+00 |
| GNG8 | 7.12 | 0.0052864 | 1.89e-03 |
| GNL2 | -0.83 | 0.0001174 | 3.08e-05 |
| GNL3 | -0.61 | 0.0003037 | 8.48e-05 |
| GNL3L | -0.65 | 0.0021539 | 7.11e-04 |
| GNPAT | -1.20 | 0.0000000 | 0.00e+00 |
| GNPDA1 | -0.69 | 0.0003894 | 1.11e-04 |
| GNPDA2 | -1.16 | 0.0482718 | 2.22e-02 |
| GNPNAT1 | 1.04 | 0.0000000 | 0.00e+00 |
| GNPTAB | -0.92 | 0.0003083 | 8.61e-05 |
| GNPTG | 1.32 | 0.0000001 | 0.00e+00 |
| GNS | 1.43 | 0.0000000 | 0.00e+00 |
| GOLGA2 | 1.49 | 0.0000000 | 0.00e+00 |
| GOLGA3 | 0.32 | 0.0628952 | 3.00e-02 |
| GOLGA4 | -0.71 | 0.0000399 | 9.80e-06 |
| GOLGA7B | 3.19 | 0.0000000 | 0.00e+00 |
| GOLGA8B | -0.46 | 0.0932513 | 4.77e-02 |
| GOLIM4 | -1.53 | 0.0000010 | 2.00e-07 |
| GOLM1 | 0.89 | 0.0000000 | 0.00e+00 |
| GOLPH3 | 0.65 | 0.0000248 | 5.90e-06 |
| GOLT1B | -0.41 | 0.0935357 | 4.79e-02 |
| GON4L | 0.69 | 0.0003224 | 9.04e-05 |
| GORAB | 0.80 | 0.0861896 | 4.34e-02 |
| GOSR1 | 0.75 | 0.0000396 | 9.70e-06 |
| GOT2 | -0.59 | 0.0000032 | 7.00e-07 |
| GP1BA | 9.31 | 0.0000364 | 8.90e-06 |
| GPAA1 | -0.56 | 0.0017018 | 5.46e-04 |
| GPAM | -2.04 | 0.0000003 | 1.00e-07 |
| GPATCH11 | -1.37 | 0.0004202 | 1.21e-04 |
| GPATCH3 | 1.11 | 0.0027829 | 9.40e-04 |
| GPATCH8 | 0.95 | 0.0000000 | 0.00e+00 |
| GPBP1 | 0.51 | 0.0055585 | 2.00e-03 |
| GPBP1L1 | 0.61 | 0.0018663 | 6.05e-04 |
| GPC1 | -0.52 | 0.0003583 | 1.02e-04 |
| GPC4 | 4.73 | 0.0004576 | 1.32e-04 |
| GPC6 | 1.25 | 0.0000122 | 2.80e-06 |
| GPD1L | -0.98 | 0.0039392 | 1.37e-03 |
| GPI | -0.75 | 0.0000000 | 0.00e+00 |
| GPKOW | 0.78 | 0.0000193 | 4.50e-06 |
| GPLD1 | -2.45 | 0.0513142 | 2.38e-02 |
| GPN1 | -0.76 | 0.0007203 | 2.15e-04 |
| GPN2 | 0.58 | 0.0768946 | 3.80e-02 |
| GPN3 | -1.32 | 0.0000126 | 2.90e-06 |
| GPR107 | 0.87 | 0.0000000 | 0.00e+00 |
| GPR108 | 1.75 | 0.0000000 | 0.00e+00 |
| GPR132 | 4.60 | 0.0008750 | 2.66e-04 |
| GPR137 | 0.81 | 0.0057191 | 2.07e-03 |
| GPR137B | 0.79 | 0.0928324 | 4.74e-02 |
| GPR141 | 4.46 | 0.0301049 | 1.30e-02 |
| GPR156 | 2.79 | 0.0098155 | 3.76e-03 |
| GPR157 | 2.11 | 0.0248219 | 1.05e-02 |
| GPR160 | 1.76 | 0.0003253 | 9.14e-05 |
| GPR162 | -2.87 | 0.0414410 | 1.87e-02 |
| GPR176 | 2.32 | 0.0000000 | 0.00e+00 |
| GPR180 | -0.75 | 0.0974652 | 5.01e-02 |
| GPR37L1 | 4.51 | 0.0000492 | 1.23e-05 |
| GPR84 | 7.89 | 0.0009916 | 3.05e-04 |
| GPRC5A | 1.06 | 0.0000000 | 0.00e+00 |
| GPRC5B | 2.53 | 0.0000000 | 0.00e+00 |
| GPRC5C | 2.33 | 0.0000000 | 0.00e+00 |
| GPRIN2 | -1.45 | 0.0000149 | 3.50e-06 |
| GPS2 | 0.35 | 0.0973765 | 5.01e-02 |
| GPSM1 | 0.72 | 0.0371954 | 1.65e-02 |
| GPSM2 | -2.73 | 0.0000000 | 0.00e+00 |
| GPT2 | 0.93 | 0.0004730 | 1.37e-04 |
| GPX1 | -0.32 | 0.0621421 | 2.96e-02 |
| GPX2 | 0.85 | 0.0000000 | 0.00e+00 |
| GPX3 | -1.08 | 0.0076538 | 2.85e-03 |
| GPX4 | -0.69 | 0.0000004 | 1.00e-07 |
| GPX8 | -0.75 | 0.0032765 | 1.12e-03 |
| GRAMD1A | 3.02 | 0.0000000 | 0.00e+00 |
| GRAMD1B | 1.27 | 0.0000000 | 0.00e+00 |
| GRAMD3 | 1.99 | 0.0199104 | 8.22e-03 |
| GRB10 | 3.14 | 0.0000000 | 0.00e+00 |
| GRB14 | -1.32 | 0.0041617 | 1.46e-03 |
| GRB2 | 0.44 | 0.0001198 | 3.15e-05 |
| GRB7 | 1.64 | 0.0000000 | 0.00e+00 |
| GREB1L | 1.40 | 0.0196016 | 8.08e-03 |
| GREM1 | 4.03 | 0.0000102 | 2.30e-06 |
| GRIK4 | 3.36 | 0.0526647 | 2.45e-02 |
| GRIN2D | 3.54 | 0.0000000 | 0.00e+00 |
| GRIN3B | 1.61 | 0.0734676 | 3.60e-02 |
| GRINA | 2.30 | 0.0000000 | 0.00e+00 |
| GRIP2 | 1.72 | 0.0339007 | 1.49e-02 |
| GRIPAP1 | 1.20 | 0.0000000 | 0.00e+00 |
| GRK6 | -0.55 | 0.0292296 | 1.26e-02 |
| GRN | 2.40 | 0.0000000 | 0.00e+00 |
| GSAP | 1.34 | 0.0012015 | 3.75e-04 |
| GSDMB | 1.32 | 0.0238173 | 1.00e-02 |
| GSDMD | 1.15 | 0.0001225 | 3.23e-05 |
| GSG2 | -1.39 | 0.0004192 | 1.20e-04 |
| GSK3A | 0.63 | 0.0000001 | 0.00e+00 |
| GSKIP | -0.67 | 0.0687207 | 3.33e-02 |
| GSN | 0.79 | 0.0000005 | 1.00e-07 |
| GSPT1 | 0.48 | 0.0000756 | 1.92e-05 |
| GSR | -0.35 | 0.0012579 | 3.94e-04 |
| GSTA4 | -1.87 | 0.0007903 | 2.38e-04 |
| GSTCD | -1.29 | 0.0001782 | 4.81e-05 |
| GSTK1 | 0.45 | 0.0099304 | 3.81e-03 |
| GSTM4 | -1.10 | 0.0876242 | 4.43e-02 |
| GSTP1 | -0.47 | 0.0000000 | 0.00e+00 |
| GTF2A1 | -1.19 | 0.0001076 | 2.81e-05 |
| GTF2A2 | -0.51 | 0.0084324 | 3.18e-03 |
| GTF2B | 0.59 | 0.0592859 | 2.81e-02 |
| GTF2F1 | 0.69 | 0.0000002 | 0.00e+00 |
| GTF2H3 | -1.12 | 0.0002431 | 6.69e-05 |
| GTF2H5 | -1.70 | 0.0001664 | 4.48e-05 |
| GTF2IP1 | -0.37 | 0.0672327 | 3.24e-02 |
| GTF2IP4 | -0.35 | 0.0812630 | 4.06e-02 |
| GTF2IRD1 | 0.90 | 0.0001375 | 3.65e-05 |
| GTF3A | -1.11 | 0.0000000 | 0.00e+00 |
| GTF3C1 | 0.75 | 0.0000000 | 0.00e+00 |
| GTF3C2 | -0.56 | 0.0003439 | 9.69e-05 |
| GTF3C3 | -0.95 | 0.0000081 | 1.80e-06 |
| GTPBP1 | 2.78 | 0.0000000 | 0.00e+00 |
| GTPBP2 | 1.98 | 0.0000000 | 0.00e+00 |
| GTPBP3 | -1.15 | 0.0055873 | 2.01e-03 |
| GTPBP4 | -0.89 | 0.0000000 | 0.00e+00 |
| GTSE1 | -2.78 | 0.0000000 | 0.00e+00 |
| GUCD1 | 0.49 | 0.0054042 | 1.94e-03 |
| GUF1 | -1.87 | 0.0000013 | 3.00e-07 |
| GUK1 | 0.46 | 0.0161878 | 6.54e-03 |
| GULP1 | -0.88 | 0.0026430 | 8.89e-04 |
| GUSB | 0.68 | 0.0023555 | 7.85e-04 |
| GXYLT1 | -0.96 | 0.0017485 | 5.63e-04 |
| GYG2 | -1.88 | 0.0000468 | 1.16e-05 |
| GYS1 | 1.50 | 0.0000000 | 0.00e+00 |
| GZF1 | -0.73 | 0.0313482 | 1.36e-02 |
| H19 | -1.33 | 0.0001997 | 5.42e-05 |
| H1F0 | 2.24 | 0.0000000 | 0.00e+00 |
| H1FX | 0.75 | 0.0000000 | 0.00e+00 |
| H2AFV | -1.24 | 0.0000000 | 0.00e+00 |
| H2AFX | -2.09 | 0.0000000 | 0.00e+00 |
| H2AFY | -0.42 | 0.0006275 | 1.85e-04 |
| H2AFY2 | 1.04 | 0.0000000 | 0.00e+00 |
| H2AFZ | -2.69 | 0.0000000 | 0.00e+00 |
| H3F3A | -1.44 | 0.0000000 | 0.00e+00 |
| H3F3AP4 | -1.44 | 0.0000000 | 0.00e+00 |
| H3F3B | 0.60 | 0.0000000 | 0.00e+00 |
| H3F3C | 0.79 | 0.0000001 | 0.00e+00 |
| H6PD | 1.65 | 0.0000000 | 0.00e+00 |
| HABP2 | -3.81 | 0.0552215 | 2.59e-02 |
| HACD1 | -1.42 | 0.0013089 | 4.11e-04 |
| HACD2 | -1.06 | 0.0013159 | 4.13e-04 |
| HACD3 | -1.40 | 0.0000000 | 0.00e+00 |
| HACL1 | -0.76 | 0.0072629 | 2.69e-03 |
| HADH | -1.94 | 0.0000000 | 0.00e+00 |
| HADHA | -0.41 | 0.0003667 | 1.04e-04 |
| HADHB | -0.63 | 0.0000892 | 2.30e-05 |
| HAGHL | -2.67 | 0.0000767 | 1.96e-05 |
| HAND1 | 3.80 | 0.0000106 | 2.40e-06 |
| HAPLN3 | 4.07 | 0.0000000 | 0.00e+00 |
| HARS | 0.66 | 0.0000012 | 2.00e-07 |
| HAS2 | 1.93 | 0.0000034 | 7.00e-07 |
| HAS3 | 4.10 | 0.0000000 | 0.00e+00 |
| HAT1 | -2.71 | 0.0000000 | 0.00e+00 |
| HAUS1 | -1.65 | 0.0000557 | 1.40e-05 |
| HAUS2 | -0.89 | 0.0031306 | 1.07e-03 |
| HAUS3 | -1.08 | 0.0891268 | 4.52e-02 |
| HAUS4 | -1.26 | 0.0000002 | 0.00e+00 |
| HAUS5 | -1.04 | 0.0009511 | 2.91e-04 |
| HAUS6 | -1.98 | 0.0000000 | 0.00e+00 |
| HAUS7 | -1.45 | 0.0000297 | 7.20e-06 |
| HAUS8 | -1.39 | 0.0017457 | 5.62e-04 |
| HAVCR2 | 2.87 | 0.0651149 | 3.12e-02 |
| HBEGF | 3.29 | 0.0000000 | 0.00e+00 |
| HBP1 | 1.55 | 0.0000000 | 0.00e+00 |
| HBS1L | -1.93 | 0.0000000 | 0.00e+00 |
| HCAR2 | 6.52 | 0.0000000 | 0.00e+00 |
| HCAR3 | 7.80 | 0.0000007 | 1.00e-07 |
| HCFC1 | 0.95 | 0.0000000 | 0.00e+00 |
| HCFC1R1 | -1.77 | 0.0000000 | 0.00e+00 |
| HCK | 7.12 | 0.0000103 | 2.30e-06 |
| HCN2 | 1.36 | 0.0000000 | 0.00e+00 |
| HCP5 | 7.33 | 0.0000000 | 0.00e+00 |
| HDAC1 | -1.04 | 0.0000000 | 0.00e+00 |
| HDAC10 | 1.23 | 0.0245450 | 1.04e-02 |
| HDAC2 | -1.01 | 0.0000000 | 0.00e+00 |
| HDAC3 | -0.53 | 0.0294017 | 1.27e-02 |
| HDAC5 | 2.53 | 0.0000000 | 0.00e+00 |
| HDAC6 | 0.42 | 0.0367529 | 1.63e-02 |
| HDAC7 | 1.49 | 0.0000000 | 0.00e+00 |
| HDAC9 | 3.50 | 0.0000000 | 0.00e+00 |
| HDDC2 | -0.91 | 0.0000867 | 2.23e-05 |
| HDDC3 | -0.57 | 0.0464864 | 2.13e-02 |
| HDGF | 0.62 | 0.0000000 | 0.00e+00 |
| HDGFRP3 | -0.56 | 0.0121693 | 4.77e-03 |
| HDHD2 | -0.91 | 0.0800650 | 3.99e-02 |
| HDHD3 | 0.94 | 0.0001289 | 3.41e-05 |
| HDX | 2.51 | 0.0027761 | 9.37e-04 |
| HEATR1 | -0.94 | 0.0000006 | 1.00e-07 |
| HEATR3 | -0.95 | 0.0042883 | 1.51e-03 |
| HEATR4 | 5.61 | 0.0750936 | 3.69e-02 |
| HEATR5A | -1.22 | 0.0000000 | 0.00e+00 |
| HEATR5B | -0.81 | 0.0085982 | 3.25e-03 |
| HEATR9 | 7.82 | 0.0011695 | 3.64e-04 |
| HECA | 1.82 | 0.0007769 | 2.33e-04 |
| HECTD1 | -0.80 | 0.0000003 | 1.00e-07 |
| HECTD2 | -1.02 | 0.0643207 | 3.08e-02 |
| HECTD3 | 0.53 | 0.0397458 | 1.78e-02 |
| HECTD4 | 0.55 | 0.0042320 | 1.48e-03 |
| HEG1 | 1.27 | 0.0001967 | 5.34e-05 |
| HEIH | 1.19 | 0.0000000 | 0.00e+00 |
| HELB | 0.95 | 0.0494635 | 2.28e-02 |
| HELLS | -1.92 | 0.0000000 | 0.00e+00 |
| HELZ2 | 5.07 | 0.0000000 | 0.00e+00 |
| HENMT1 | -1.68 | 0.0830245 | 4.16e-02 |
| HERC2 | -0.45 | 0.0147532 | 5.90e-03 |
| HERC5 | 5.63 | 0.0000000 | 0.00e+00 |
| HERC6 | 2.95 | 0.0000000 | 0.00e+00 |
| HERPUD1 | 0.40 | 0.0891113 | 4.52e-02 |
| HES4 | 2.22 | 0.0000000 | 0.00e+00 |
| HES7 | 2.47 | 0.0002734 | 7.58e-05 |
| HEXA | 0.45 | 0.0005240 | 1.53e-04 |
| HEXB | 0.45 | 0.0022073 | 7.30e-04 |
| HEXDC | 0.62 | 0.0935135 | 4.78e-02 |
| HEXIM1 | -0.41 | 0.0011089 | 3.43e-04 |
| HEXIM2 | -1.32 | 0.0005837 | 1.71e-04 |
| HFE | -0.81 | 0.0173551 | 7.06e-03 |
| HGF | -5.76 | 0.0886793 | 4.49e-02 |
| HGH1 | -1.25 | 0.0000355 | 8.70e-06 |
| HGS | 1.12 | 0.0000000 | 0.00e+00 |
| HGSNAT | 0.64 | 0.0038106 | 1.32e-03 |
| HHLA2 | 6.76 | 0.0100371 | 3.86e-03 |
| HIAT1 | 1.24 | 0.0000000 | 0.00e+00 |
| HIBCH | -1.25 | 0.0018948 | 6.16e-04 |
| HIC2 | 0.87 | 0.0636589 | 3.04e-02 |
| HID1 | 0.70 | 0.0564995 | 2.66e-02 |
| HIF1A | 0.21 | 0.0818127 | 4.09e-02 |
| HIGD1A | -0.46 | 0.0509720 | 2.36e-02 |
| HINFP | 0.85 | 0.0435893 | 1.98e-02 |
| HINT2 | -0.61 | 0.0279344 | 1.20e-02 |
| HIP1 | -0.46 | 0.0263552 | 1.12e-02 |
| HIP1R | 3.51 | 0.0000000 | 0.00e+00 |
| HIPK1 | 0.46 | 0.0521959 | 2.42e-02 |
| HIRIP3 | -1.53 | 0.0003167 | 8.87e-05 |
| HIST1H1B | -3.12 | 0.0604714 | 2.87e-02 |
| HIST1H1C | 0.59 | 0.0410951 | 1.85e-02 |
| HIST1H1D | -4.43 | 0.0000033 | 7.00e-07 |
| HIST1H1E | -3.12 | 0.0000000 | 0.00e+00 |
| HIST1H2AG | -1.84 | 0.0487739 | 2.24e-02 |
| HIST1H2AH | -1.73 | 0.0774741 | 3.83e-02 |
| HIST1H2AI | -4.74 | 0.0000514 | 1.28e-05 |
| HIST1H2AJ | -4.46 | 0.0312169 | 1.36e-02 |
| HIST1H2AK | -3.20 | 0.0715663 | 3.49e-02 |
| HIST1H2AL | -5.86 | 0.0087318 | 3.30e-03 |
| HIST1H2AM | -4.01 | 0.0044055 | 1.55e-03 |
| HIST1H2BC | -1.53 | 0.0019080 | 6.21e-04 |
| HIST1H2BI | -5.29 | 0.0023425 | 7.80e-04 |
| HIST1H2BK | 0.64 | 0.0036547 | 1.26e-03 |
| HIST1H2BL | -3.96 | 0.0000018 | 4.00e-07 |
| HIST1H2BM | -3.12 | 0.0002356 | 6.47e-05 |
| HIST1H2BN | -1.51 | 0.0403314 | 1.81e-02 |
| HIST1H2BO | -2.58 | 0.0019823 | 6.48e-04 |
| HIST1H3B | -3.61 | 0.0000073 | 1.60e-06 |
| HIST1H3C | -7.76 | 0.0001488 | 3.98e-05 |
| HIST1H3I | -3.17 | 0.0136000 | 5.40e-03 |
| HIST1H3J | -2.58 | 0.0436265 | 1.98e-02 |
| HIST1H4A | -4.76 | 0.0000014 | 3.00e-07 |
| HIST1H4B | -4.31 | 0.0000000 | 0.00e+00 |
| HIST1H4C | -3.30 | 0.0000000 | 0.00e+00 |
| HIST1H4E | -3.15 | 0.0000000 | 0.00e+00 |
| HIST1H4H | -2.73 | 0.0000021 | 4.00e-07 |
| HIST1H4I | -4.43 | 0.0019020 | 6.19e-04 |
| HIST1H4J | -3.00 | 0.0549857 | 2.57e-02 |
| HIST1H4K | -2.37 | 0.0891887 | 4.52e-02 |
| HIST2H2AA3 | 0.72 | 0.0234578 | 9.88e-03 |
| HIST2H2AA4 | 0.72 | 0.0234578 | 9.88e-03 |
| HIST2H2AB | -3.72 | 0.0003876 | 1.11e-04 |
| HIST2H2BA | -3.44 | 0.0121664 | 4.77e-03 |
| HIST2H2BE | -0.87 | 0.0765988 | 3.78e-02 |
| HIST2H2BF | -3.55 | 0.0000000 | 0.00e+00 |
| HIST2H3A | -2.64 | 0.0159852 | 6.45e-03 |
| HIST2H3C | -2.64 | 0.0159852 | 6.45e-03 |
| HIST3H2BB | -3.37 | 0.0285914 | 1.23e-02 |
| HIST4H4 | -3.52 | 0.0000066 | 1.50e-06 |
| HIVEP1 | 0.93 | 0.0011726 | 3.65e-04 |
| HIVEP2 | 3.08 | 0.0000000 | 0.00e+00 |
| HIVEP3 | 3.19 | 0.0000000 | 0.00e+00 |
| HJURP | -3.30 | 0.0000000 | 0.00e+00 |
| HK1 | 1.33 | 0.0000000 | 0.00e+00 |
| HK2 | 4.64 | 0.0000000 | 0.00e+00 |
| HKDC1 | 1.10 | 0.0000000 | 0.00e+00 |
| HLA-A | 4.08 | 0.0000000 | 0.00e+00 |
| HLA-B | 5.72 | 0.0000000 | 0.00e+00 |
| HLA-C | 4.53 | 0.0000000 | 0.00e+00 |
| HLA-E | 3.58 | 0.0000000 | 0.00e+00 |
| HLA-H | 4.63 | 0.0000000 | 0.00e+00 |
| HLCS | -1.00 | 0.0099664 | 3.83e-03 |
| HLF | -3.86 | 0.0199718 | 8.25e-03 |
| HLTF | -2.54 | 0.0000000 | 0.00e+00 |
| HM13 | 1.15 | 0.0000000 | 0.00e+00 |
| HMBS | -1.25 | 0.0000001 | 0.00e+00 |
| HMCES | -0.61 | 0.0144443 | 5.76e-03 |
| HMCN2 | 6.57 | 0.0145416 | 5.81e-03 |
| HMGA2 | 0.58 | 0.0267695 | 1.14e-02 |
| HMGB1 | -1.90 | 0.0000000 | 0.00e+00 |
| HMGB2 | -2.42 | 0.0000000 | 0.00e+00 |
| HMGB3 | -1.40 | 0.0000000 | 0.00e+00 |
| HMGCL | 0.99 | 0.0018397 | 5.96e-04 |
| HMGCR | 1.48 | 0.0000000 | 0.00e+00 |
| HMGCS1 | 1.29 | 0.0000000 | 0.00e+00 |
| HMGN1 | -1.03 | 0.0000001 | 0.00e+00 |
| HMGN2 | -1.02 | 0.0001850 | 5.00e-05 |
| HMGN3 | -1.73 | 0.0000001 | 0.00e+00 |
| HMGXB3 | 0.95 | 0.0000000 | 0.00e+00 |
| HMMR | -3.16 | 0.0000000 | 0.00e+00 |
| HMOX1 | -1.26 | 0.0000000 | 0.00e+00 |
| HNF1A | 0.56 | 0.0816555 | 4.08e-02 |
| HNF1B | 1.14 | 0.0000643 | 1.62e-05 |
| HNRNPA0 | -0.36 | 0.0079217 | 2.97e-03 |
| HNRNPA1 | -0.42 | 0.0031107 | 1.06e-03 |
| HNRNPA1P10 | -0.33 | 0.0151903 | 6.10e-03 |
| HNRNPA2B1 | -1.04 | 0.0000000 | 0.00e+00 |
| HNRNPA3 | -1.17 | 0.0000000 | 0.00e+00 |
| HNRNPA3P1 | -1.31 | 0.0032721 | 1.12e-03 |
| HNRNPAB | -1.20 | 0.0000000 | 0.00e+00 |
| HNRNPC | -0.96 | 0.0000000 | 0.00e+00 |
| HNRNPD | -0.54 | 0.0001672 | 4.50e-05 |
| HNRNPDL | -0.32 | 0.0272083 | 1.16e-02 |
| HNRNPF | -0.37 | 0.0005658 | 1.66e-04 |
| HNRNPH1 | -0.88 | 0.0000000 | 0.00e+00 |
| HNRNPH2 | -0.98 | 0.0000006 | 1.00e-07 |
| HNRNPH3 | -1.09 | 0.0000000 | 0.00e+00 |
| HNRNPK | -0.20 | 0.0579828 | 2.74e-02 |
| HNRNPL | 0.27 | 0.0249592 | 1.06e-02 |
| HNRNPR | -1.18 | 0.0000000 | 0.00e+00 |
| HNRNPU-AS1 | -1.05 | 0.0818267 | 4.09e-02 |
| HNRNPUL2-BSCL2 | -0.36 | 0.0085711 | 3.23e-03 |
| HOMER2 | -0.86 | 0.0569706 | 2.68e-02 |
| HOMER3 | 1.24 | 0.0000675 | 1.71e-05 |
| HOMEZ | 1.90 | 0.0000000 | 0.00e+00 |
| HOOK3 | -0.81 | 0.0060316 | 2.19e-03 |
| HOTAIRM1 | 1.33 | 0.0207153 | 8.58e-03 |
| HOXA1 | 2.34 | 0.0000001 | 0.00e+00 |
| HOXB13 | -1.21 | 0.0188366 | 7.73e-03 |
| HOXB3 | 1.20 | 0.0000062 | 1.40e-06 |
| HOXB4 | 1.76 | 0.0277478 | 1.19e-02 |
| HOXB9 | 1.65 | 0.0000000 | 0.00e+00 |
| HOXD10 | 4.48 | 0.0013415 | 4.22e-04 |
| HOXD11 | 3.04 | 0.0001005 | 2.61e-05 |
| HOXD9 | 6.25 | 0.0258134 | 1.10e-02 |
| HP1BP3 | -1.68 | 0.0000000 | 0.00e+00 |
| HPDL | -6.75 | 0.0004567 | 1.32e-04 |
| HPGD | -0.55 | 0.0037267 | 1.29e-03 |
| HPN | 3.86 | 0.0948273 | 4.86e-02 |
| HPRT1 | -1.26 | 0.0000000 | 0.00e+00 |
| HPS1 | 0.66 | 0.0145598 | 5.82e-03 |
| HPS6 | 0.70 | 0.0022418 | 7.43e-04 |
| HRAS | -0.58 | 0.0767686 | 3.79e-02 |
| HRASLS2 | 4.12 | 0.0000000 | 0.00e+00 |
| HRH1 | 3.46 | 0.0000000 | 0.00e+00 |
| HRH2 | 2.56 | 0.0034923 | 1.20e-03 |
| HRSP12 | -1.35 | 0.0020095 | 6.58e-04 |
| HS1BP3 | 0.71 | 0.0003151 | 8.82e-05 |
| HS2ST1 | -0.54 | 0.0560986 | 2.63e-02 |
| HS3ST1 | 2.78 | 0.0000023 | 5.00e-07 |
| HS3ST3A1 | 4.52 | 0.0011613 | 3.61e-04 |
| HS3ST3B1 | 4.78 | 0.0000000 | 0.00e+00 |
| HSBP1 | -0.65 | 0.0000157 | 3.60e-06 |
| HSCB | 2.17 | 0.0000000 | 0.00e+00 |
| HSD11B1 | 8.69 | 0.0001616 | 4.34e-05 |
| HSD17B10 | -1.05 | 0.0000000 | 0.00e+00 |
| HSD17B14 | -1.01 | 0.0775093 | 3.84e-02 |
| HSD17B4 | -0.78 | 0.0000337 | 8.20e-06 |
| HSD17B7 | 1.08 | 0.0008563 | 2.59e-04 |
| HSD17B7P2 | 2.04 | 0.0000006 | 1.00e-07 |
| HSD3B7 | 1.51 | 0.0000000 | 0.00e+00 |
| HSDL1 | -0.86 | 0.0071421 | 2.64e-03 |
| HSDL2 | -1.50 | 0.0000000 | 0.00e+00 |
| HSF1 | 1.79 | 0.0000000 | 0.00e+00 |
| HSH2D | 4.53 | 0.0000000 | 0.00e+00 |
| HSP90AA1 | -1.95 | 0.0000000 | 0.00e+00 |
| HSP90AB1 | -0.72 | 0.0000000 | 0.00e+00 |
| HSP90B1 | -0.26 | 0.0072037 | 2.67e-03 |
| HSPA14 | -0.74 | 0.0002331 | 6.40e-05 |
| HSPA4 | -0.72 | 0.0000000 | 0.00e+00 |
| HSPA4L | -1.52 | 0.0000234 | 5.60e-06 |
| HSPA5 | 0.75 | 0.0000000 | 0.00e+00 |
| HSPA7 | 3.86 | 0.0926629 | 4.73e-02 |
| HSPA8 | -1.26 | 0.0000000 | 0.00e+00 |
| HSPA9 | -0.44 | 0.0000029 | 6.00e-07 |
| HSPB1 | 0.95 | 0.0000000 | 0.00e+00 |
| HSPB11 | -0.85 | 0.0515073 | 2.39e-02 |
| HSPB8 | 2.90 | 0.0000000 | 0.00e+00 |
| HSPBAP1 | 0.83 | 0.0957874 | 4.91e-02 |
| HSPBP1 | -0.64 | 0.0001273 | 3.36e-05 |
| HSPD1 | -1.69 | 0.0000000 | 0.00e+00 |
| HSPE1 | -1.98 | 0.0000000 | 0.00e+00 |
| HSPE1-MOB4 | -1.29 | 0.0000000 | 0.00e+00 |
| HSPG2 | 1.96 | 0.0000000 | 0.00e+00 |
| HTATSF1 | -1.19 | 0.0000000 | 0.00e+00 |
| HTATSF1P2 | -0.88 | 0.0838955 | 4.21e-02 |
| HTR3E | 6.40 | 0.0199104 | 8.22e-03 |
| HTRA1 | -1.04 | 0.0015743 | 5.02e-04 |
| HUNK | 2.80 | 0.0000000 | 0.00e+00 |
| HUWE1 | 0.37 | 0.0007528 | 2.25e-04 |
| HYAL1 | 1.70 | 0.0001315 | 3.48e-05 |
| HYLS1 | -1.09 | 0.0063130 | 2.31e-03 |
| HYOU1 | 0.72 | 0.0000013 | 3.00e-07 |
| IAH1 | -0.62 | 0.0091685 | 3.49e-03 |
| IARS | -1.17 | 0.0000000 | 0.00e+00 |
| IARS2 | -1.35 | 0.0000000 | 0.00e+00 |
| ICAM1 | 9.70 | 0.0000000 | 0.00e+00 |
| ICAM2 | 6.83 | 0.0087355 | 3.31e-03 |
| ICAM3 | -1.19 | 0.0992567 | 5.12e-02 |
| ICAM4 | 9.14 | 0.0000551 | 1.38e-05 |
| ICAM5 | 2.66 | 0.0001270 | 3.35e-05 |
| ICE1 | -1.08 | 0.0000001 | 0.00e+00 |
| ICE2 | -1.55 | 0.0000000 | 0.00e+00 |
| ICMT | -1.43 | 0.0000000 | 0.00e+00 |
| ICOSLG | 2.47 | 0.0003488 | 9.86e-05 |
| ICT1 | -0.37 | 0.0924371 | 4.72e-02 |
| ID1 | -0.99 | 0.0000000 | 0.00e+00 |
| ID2 | -1.05 | 0.0000047 | 1.00e-06 |
| ID3 | -0.55 | 0.0000426 | 1.05e-05 |
| ID4 | 1.60 | 0.0028291 | 9.56e-04 |
| IDE | -0.80 | 0.0000671 | 1.70e-05 |
| IDH1 | -0.57 | 0.0000007 | 1.00e-07 |
| IDH2 | -0.51 | 0.0228365 | 9.57e-03 |
| IDH3A | -0.89 | 0.0000007 | 1.00e-07 |
| IDH3B | -0.35 | 0.0291636 | 1.26e-02 |
| IDH3G | 0.43 | 0.0462524 | 2.12e-02 |
| IDI1 | 0.56 | 0.0006147 | 1.81e-04 |
| IDO1 | 12.61 | 0.0000000 | 0.00e+00 |
| IDUA | 1.67 | 0.0002309 | 6.34e-05 |
| IER2 | 2.21 | 0.0000000 | 0.00e+00 |
| IER3IP1 | -0.83 | 0.0035528 | 1.22e-03 |
| IER5 | 2.29 | 0.0000000 | 0.00e+00 |
| IER5L | -0.80 | 0.0000039 | 8.00e-07 |
| IFFO1 | 3.39 | 0.0133443 | 5.28e-03 |
| IFI16 | 4.53 | 0.0000000 | 0.00e+00 |
| IFI27 | 9.86 | 0.0000000 | 0.00e+00 |
| IFI30 | 1.96 | 0.0000000 | 0.00e+00 |
| IFI35 | 3.83 | 0.0000000 | 0.00e+00 |
| IFI44 | 9.67 | 0.0000000 | 0.00e+00 |
| IFI44L | 7.80 | 0.0000007 | 1.00e-07 |
| IFI6 | 7.87 | 0.0000000 | 0.00e+00 |
| IFIH1 | 6.63 | 0.0000000 | 0.00e+00 |
| IFIT1 | 7.12 | 0.0000000 | 0.00e+00 |
| IFIT1B | 5.54 | 0.0000000 | 0.00e+00 |
| IFIT2 | 8.61 | 0.0000000 | 0.00e+00 |
| IFIT3 | 7.97 | 0.0000000 | 0.00e+00 |
| IFIT5 | 1.75 | 0.0000000 | 0.00e+00 |
| IFITM1 | 7.47 | 0.0000000 | 0.00e+00 |
| IFITM10 | 2.39 | 0.0016502 | 5.28e-04 |
| IFITM2 | 2.85 | 0.0000000 | 0.00e+00 |
| IFITM3 | 4.91 | 0.0000000 | 0.00e+00 |
| IFNAR1 | 0.48 | 0.0414049 | 1.87e-02 |
| IFNAR2 | 2.41 | 0.0000000 | 0.00e+00 |
| IFNB1 | 10.44 | 0.0000022 | 5.00e-07 |
| IFNGR1 | 2.09 | 0.0000000 | 0.00e+00 |
| IFNGR2 | 2.52 | 0.0000000 | 0.00e+00 |
| IFNL1 | 9.95 | 0.0000000 | 0.00e+00 |
| IFNL2 | 9.33 | 0.0000000 | 0.00e+00 |
| IFNL3 | 12.91 | 0.0000000 | 0.00e+00 |
| IFNL4 | 7.25 | 0.0042118 | 1.48e-03 |
| IFNLR1 | 2.75 | 0.0000000 | 0.00e+00 |
| IFRD1 | 0.92 | 0.0000001 | 0.00e+00 |
| IFRD2 | -0.75 | 0.0000491 | 1.22e-05 |
| IFT122 | -1.34 | 0.0001349 | 3.58e-05 |
| IFT172 | -2.27 | 0.0000013 | 3.00e-07 |
| IFT22 | -1.47 | 0.0000011 | 2.00e-07 |
| IFT27 | -0.91 | 0.0029427 | 9.97e-04 |
| IFT46 | -0.83 | 0.0416964 | 1.88e-02 |
| IFT52 | -0.94 | 0.0062225 | 2.27e-03 |
| IFT74 | -0.81 | 0.0362420 | 1.61e-02 |
| IFT80 | -2.28 | 0.0000000 | 0.00e+00 |
| IFT81 | -1.31 | 0.0000056 | 1.20e-06 |
| IFT88 | -1.60 | 0.0004629 | 1.34e-04 |
| IGBP1 | 0.72 | 0.0000182 | 4.30e-06 |
| IGDCC4 | 3.45 | 0.0000000 | 0.00e+00 |
| IGF2 | 3.77 | 0.0000000 | 0.00e+00 |
| IGF2BP1 | 0.95 | 0.0000000 | 0.00e+00 |
| IGF2BP2 | 1.71 | 0.0000000 | 0.00e+00 |
| IGF2R | 1.27 | 0.0000000 | 0.00e+00 |
| IGFBP1 | 5.91 | 0.0000000 | 0.00e+00 |
| IGFBP3 | 0.91 | 0.0003861 | 1.10e-04 |
| IGFBP4 | 0.22 | 0.0125731 | 4.95e-03 |
| IGFBP5 | -3.81 | 0.0709631 | 3.46e-02 |
| IGFBP6 | 4.34 | 0.0000000 | 0.00e+00 |
| IGFBP7 | 2.13 | 0.0000000 | 0.00e+00 |
| IGFBPL1 | -2.66 | 0.0000800 | 2.04e-05 |
| IGFL2 | 5.76 | 0.0586487 | 2.77e-02 |
| IGFN1 | 4.68 | 0.0000162 | 3.80e-06 |
| IGHMBP2 | 1.33 | 0.0000001 | 0.00e+00 |
| IGSF1 | 1.94 | 0.0863212 | 4.35e-02 |
| IGSF10 | -2.44 | 0.0045004 | 1.59e-03 |
| IGSF11 | -2.11 | 0.0000000 | 0.00e+00 |
| IGSF23 | -5.76 | 0.0886793 | 4.49e-02 |
| IGSF3 | 0.53 | 0.0012260 | 3.83e-04 |
| IGSF8 | 1.60 | 0.0000000 | 0.00e+00 |
| IKBIP | -0.67 | 0.0021460 | 7.08e-04 |
| IKBKAP | -1.15 | 0.0000000 | 0.00e+00 |
| IKBKB | 0.73 | 0.0001408 | 3.75e-05 |
| IKBKE | 3.88 | 0.0000000 | 0.00e+00 |
| IKBKG | 0.88 | 0.0000975 | 2.53e-05 |
| IKZF1 | 6.30 | 0.0241362 | 1.02e-02 |
| IL10RA | 4.46 | 0.0014158 | 4.47e-04 |
| IL10RB | 1.68 | 0.0000000 | 0.00e+00 |
| IL11 | 3.58 | 0.0000000 | 0.00e+00 |
| IL12A | 2.58 | 0.0445722 | 2.03e-02 |
| IL13RA1 | 0.39 | 0.0186118 | 7.63e-03 |
| IL15 | 1.80 | 0.0000016 | 3.00e-07 |
| IL15RA | 4.55 | 0.0000000 | 0.00e+00 |
| IL17RA | 1.32 | 0.0000016 | 3.00e-07 |
| IL18BP | 3.45 | 0.0000000 | 0.00e+00 |
| IL19 | 5.44 | 0.0989393 | 5.10e-02 |
| IL1A | 11.76 | 0.0000001 | 0.00e+00 |
| IL1B | 8.03 | 0.0000000 | 0.00e+00 |
| IL1R1 | -0.82 | 0.0008747 | 2.65e-04 |
| IL1RN | 7.17 | 0.0045032 | 1.59e-03 |
| IL21R | 2.91 | 0.0603928 | 2.87e-02 |
| IL22RA1 | 1.62 | 0.0003852 | 1.10e-04 |
| IL23A | 6.17 | 0.0000000 | 0.00e+00 |
| IL24 | 6.90 | 0.0077875 | 2.91e-03 |
| IL27 | 7.09 | 0.0053339 | 1.91e-03 |
| IL2RG | 6.05 | 0.0000000 | 0.00e+00 |
| IL32 | 6.58 | 0.0000000 | 0.00e+00 |
| IL34 | 2.46 | 0.0062857 | 2.30e-03 |
| IL36G | 8.10 | 0.0006158 | 1.81e-04 |
| IL36RN | 6.09 | 0.0356604 | 1.58e-02 |
| IL3RA | 8.22 | 0.0004738 | 1.37e-04 |
| IL4I1 | 4.15 | 0.0000000 | 0.00e+00 |
| IL4R | 2.75 | 0.0000000 | 0.00e+00 |
| IL6 | 12.74 | 0.0000000 | 0.00e+00 |
| IL7R | 7.75 | 0.0013430 | 4.23e-04 |
| ILDR1 | 6.61 | 0.0134179 | 5.32e-03 |
| ILF2 | -0.68 | 0.0000000 | 0.00e+00 |
| ILF3 | -0.56 | 0.0000007 | 1.00e-07 |
| ILF3-AS1 | -1.96 | 0.0000200 | 4.70e-06 |
| ILK | 0.86 | 0.0000000 | 0.00e+00 |
| IMMP1L | -1.35 | 0.0533384 | 2.49e-02 |
| IMMT | -0.87 | 0.0000000 | 0.00e+00 |
| IMP3 | -0.51 | 0.0079831 | 2.99e-03 |
| IMP4 | -0.92 | 0.0000003 | 0.00e+00 |
| IMPACT | -0.85 | 0.0493331 | 2.27e-02 |
| IMPAD1 | -0.88 | 0.0000004 | 1.00e-07 |
| IMPDH1 | -1.42 | 0.0000000 | 0.00e+00 |
| IMPDH2 | -1.70 | 0.0000000 | 0.00e+00 |
| INA | 0.67 | 0.0009300 | 2.84e-04 |
| INAFM1 | 1.46 | 0.0043469 | 1.53e-03 |
| INCENP | -1.17 | 0.0000001 | 0.00e+00 |
| INF2 | 0.93 | 0.0000000 | 0.00e+00 |
| ING1 | 1.91 | 0.0000000 | 0.00e+00 |
| ING5 | -1.52 | 0.0002249 | 6.16e-05 |
| INHBA | 5.78 | 0.0010037 | 3.09e-04 |
| INHBB | -1.97 | 0.0000000 | 0.00e+00 |
| INHBE | 3.08 | 0.0009216 | 2.81e-04 |
| INO80 | 0.69 | 0.0012093 | 3.77e-04 |
| INO80B | 0.93 | 0.0000226 | 5.40e-06 |
| INO80E | -0.36 | 0.0829337 | 4.15e-02 |
| INPP1 | 1.83 | 0.0000000 | 0.00e+00 |
| INPP5A | -0.96 | 0.0158364 | 6.39e-03 |
| INPP5F | -0.93 | 0.0222122 | 9.27e-03 |
| INPP5K | 0.59 | 0.0048006 | 1.71e-03 |
| INPPL1 | 0.87 | 0.0000000 | 0.00e+00 |
| INS-IGF2 | 3.63 | 0.0000001 | 0.00e+00 |
| INSIG1 | 1.39 | 0.0000000 | 0.00e+00 |
| INSL3 | 5.69 | 0.0644894 | 3.09e-02 |
| INSR | 0.96 | 0.0097700 | 3.74e-03 |
| INTS1 | 0.51 | 0.0002360 | 6.48e-05 |
| INTS10 | -1.76 | 0.0000000 | 0.00e+00 |
| INTS12 | 0.92 | 0.0016799 | 5.39e-04 |
| INTS2 | -1.24 | 0.0000038 | 8.00e-07 |
| INTS3 | 0.50 | 0.0665937 | 3.21e-02 |
| INTS6 | 1.82 | 0.0000000 | 0.00e+00 |
| INTS8 | -1.34 | 0.0000000 | 0.00e+00 |
| IP6K1 | 1.30 | 0.0000000 | 0.00e+00 |
| IP6K2 | 0.47 | 0.0284281 | 1.22e-02 |
| IPO11 | -1.61 | 0.0000000 | 0.00e+00 |
| IPO11-LRRC70 | -1.53 | 0.0008211 | 2.48e-04 |
| IPO4 | -1.53 | 0.0000000 | 0.00e+00 |
| IPO5 | -1.57 | 0.0000000 | 0.00e+00 |
| IPO7 | -0.49 | 0.0000082 | 1.80e-06 |
| IPO8 | -0.63 | 0.0056723 | 2.05e-03 |
| IPO9 | -0.71 | 0.0000026 | 6.00e-07 |
| IPPK | 0.81 | 0.0059636 | 2.17e-03 |
| IPW | -2.12 | 0.0058451 | 2.12e-03 |
| IQCH-AS1 | -2.09 | 0.0709139 | 3.45e-02 |
| IQCJ-SCHIP1 | 1.26 | 0.0000002 | 0.00e+00 |
| IQGAP2 | 1.25 | 0.0014788 | 4.69e-04 |
| IQGAP3 | -1.03 | 0.0000028 | 6.00e-07 |
| IQSEC2 | 0.69 | 0.0029843 | 1.01e-03 |
| IQSEC3 | 6.83 | 0.0088158 | 3.34e-03 |
| IRAK2 | 5.30 | 0.0000000 | 0.00e+00 |
| IRAK3 | 4.61 | 0.0007802 | 2.34e-04 |
| IREB2 | -0.91 | 0.0000000 | 0.00e+00 |
| IRF1 | 5.61 | 0.0000000 | 0.00e+00 |
| IRF2 | 1.70 | 0.0000000 | 0.00e+00 |
| IRF2BP1 | 0.51 | 0.0194445 | 8.00e-03 |
| IRF2BP2 | 1.37 | 0.0000000 | 0.00e+00 |
| IRF2BPL | 2.41 | 0.0000000 | 0.00e+00 |
| IRF3 | 0.94 | 0.0000000 | 0.00e+00 |
| IRF4 | 6.40 | 0.0206040 | 8.53e-03 |
| IRF5 | 1.42 | 0.0000017 | 4.00e-07 |
| IRF7 | 6.56 | 0.0000000 | 0.00e+00 |
| IRF8 | 6.57 | 0.0160575 | 6.48e-03 |
| IRF9 | 3.68 | 0.0000000 | 0.00e+00 |
| IRGQ | 1.13 | 0.0000003 | 1.00e-07 |
| IRS2 | 1.00 | 0.0000000 | 0.00e+00 |
| IRX3 | 1.09 | 0.0548300 | 2.56e-02 |
| ISCA1 | -1.29 | 0.0003883 | 1.11e-04 |
| ISCA2 | 0.51 | 0.0756801 | 3.73e-02 |
| ISG15 | 8.55 | 0.0000000 | 0.00e+00 |
| ISG20 | 5.69 | 0.0000000 | 0.00e+00 |
| ISM1 | 3.68 | 0.0000002 | 0.00e+00 |
| ISOC2 | -0.69 | 0.0007534 | 2.25e-04 |
| IST1 | 1.05 | 0.0000000 | 0.00e+00 |
| ISY1-RAB43 | 1.05 | 0.0000006 | 1.00e-07 |
| ISYNA1 | -1.66 | 0.0000129 | 3.00e-06 |
| ITFG2 | -0.91 | 0.0506298 | 2.34e-02 |
| ITFG3 | 1.15 | 0.0000000 | 0.00e+00 |
| ITGA2 | 2.26 | 0.0000000 | 0.00e+00 |
| ITGA2B | -2.89 | 0.0329262 | 1.44e-02 |
| ITGA3 | 0.76 | 0.0000000 | 0.00e+00 |
| ITGA5 | 5.55 | 0.0000000 | 0.00e+00 |
| ITGA6 | -0.37 | 0.0366540 | 1.63e-02 |
| ITGAE | -1.80 | 0.0000000 | 0.00e+00 |
| ITGAV | 0.38 | 0.0017335 | 5.58e-04 |
| ITGAX | 4.21 | 0.0000000 | 0.00e+00 |
| ITGB1 | -1.16 | 0.0000000 | 0.00e+00 |
| ITGB1BP1 | -1.23 | 0.0000000 | 0.00e+00 |
| ITGB2 | 2.31 | 0.0044931 | 1.58e-03 |
| ITGB3 | 3.88 | 0.0000378 | 9.20e-06 |
| ITGB3BP | -2.51 | 0.0000004 | 1.00e-07 |
| ITGB4 | 0.87 | 0.0000000 | 0.00e+00 |
| ITGB8 | 4.14 | 0.0000000 | 0.00e+00 |
| ITIH2 | 2.70 | 0.0000174 | 4.10e-06 |
| ITIH5 | 4.22 | 0.0000002 | 0.00e+00 |
| ITIH6 | 5.97 | 0.0428851 | 1.94e-02 |
| ITM2B | 1.25 | 0.0000000 | 0.00e+00 |
| ITPKC | 3.25 | 0.0000000 | 0.00e+00 |
| ITPR3 | 1.99 | 0.0000000 | 0.00e+00 |
| ITPRIP | 2.14 | 0.0000000 | 0.00e+00 |
| ITPRIPL2 | 1.93 | 0.0000000 | 0.00e+00 |
| ITSN1 | 0.57 | 0.0240546 | 1.01e-02 |
| ITSN2 | 0.59 | 0.0052025 | 1.86e-03 |
| IVD | -0.87 | 0.0387274 | 1.73e-02 |
| IVNS1ABP | -1.04 | 0.0000007 | 1.00e-07 |
| JADE1 | -1.16 | 0.0000011 | 2.00e-07 |
| JADE2 | 1.67 | 0.0000000 | 0.00e+00 |
| JAG1 | 2.56 | 0.0000000 | 0.00e+00 |
| JAG2 | -2.48 | 0.0000000 | 0.00e+00 |
| JAGN1 | 0.43 | 0.0522702 | 2.43e-02 |
| JAK1 | 0.84 | 0.0000000 | 0.00e+00 |
| JAK2 | 1.68 | 0.0000007 | 1.00e-07 |
| JAKMIP3 | 1.07 | 0.0044099 | 1.55e-03 |
| JARID2 | 0.72 | 0.0024722 | 8.26e-04 |
| JAZF1 | 1.48 | 0.0141013 | 5.62e-03 |
| JDP2 | 0.72 | 0.0064808 | 2.37e-03 |
| JHDM1D-AS1 | 3.25 | 0.0000000 | 0.00e+00 |
| JKAMP | -1.16 | 0.0000002 | 0.00e+00 |
| JMJD1C | 0.41 | 0.0466313 | 2.14e-02 |
| JMJD6 | 0.96 | 0.0000000 | 0.00e+00 |
| JMJD8 | -1.03 | 0.0000006 | 1.00e-07 |
| JOSD1 | 1.52 | 0.0000000 | 0.00e+00 |
| JPH2 | 2.54 | 0.0038517 | 1.34e-03 |
| JPH4 | 5.44 | 0.0911055 | 4.64e-02 |
| JRK | -0.58 | 0.0969762 | 4.98e-02 |
| JTB | -0.47 | 0.0035578 | 1.23e-03 |
| JUN | 4.64 | 0.0000000 | 0.00e+00 |
| JUNB | 3.05 | 0.0000000 | 0.00e+00 |
| JUND | 1.04 | 0.0000000 | 0.00e+00 |
| JUP | 2.08 | 0.0000000 | 0.00e+00 |
| KALRN | 1.63 | 0.0100063 | 3.84e-03 |
| KANK2 | -1.21 | 0.0000011 | 2.00e-07 |
| KANSL1 | 0.99 | 0.0005187 | 1.51e-04 |
| KANSL3 | 0.81 | 0.0000010 | 2.00e-07 |
| KARS | -0.52 | 0.0000191 | 4.50e-06 |
| KAT2A | -0.75 | 0.0000189 | 4.40e-06 |
| KAT2B | -1.22 | 0.0072625 | 2.69e-03 |
| KAT5 | 0.81 | 0.0000140 | 3.20e-06 |
| KAT6B | -0.86 | 0.0353265 | 1.56e-02 |
| KAT7 | 0.84 | 0.0000109 | 2.50e-06 |
| KATNAL1 | -1.80 | 0.0012812 | 4.02e-04 |
| KATNB1 | -0.69 | 0.0310104 | 1.35e-02 |
| KAZN | 1.24 | 0.0052991 | 1.90e-03 |
| KBTBD4 | -0.87 | 0.0047794 | 1.70e-03 |
| KBTBD6 | -1.03 | 0.0680442 | 3.29e-02 |
| KCCAT211 | 3.25 | 0.0228238 | 9.56e-03 |
| KCNAB2 | -1.51 | 0.0000401 | 9.90e-06 |
| KCNC3 | 2.11 | 0.0018870 | 6.13e-04 |
| KCNF1 | 1.12 | 0.0019279 | 6.28e-04 |
| KCNG1 | 1.05 | 0.0090588 | 3.44e-03 |
| KCNIP3 | -4.09 | 0.0738103 | 3.62e-02 |
| KCNJ15 | 3.37 | 0.0143256 | 5.71e-03 |
| KCNK1 | 2.98 | 0.0000000 | 0.00e+00 |
| KCNK5 | 2.58 | 0.0000000 | 0.00e+00 |
| KCNN1 | 4.94 | 0.0000000 | 0.00e+00 |
| KCNN4 | 1.37 | 0.0000000 | 0.00e+00 |
| KCNQ1OT1 | -1.68 | 0.0175214 | 7.14e-03 |
| KCNQ3 | 2.97 | 0.0505797 | 2.34e-02 |
| KCNQ4 | 8.29 | 0.0000000 | 0.00e+00 |
| KCNS3 | -1.80 | 0.0225096 | 9.42e-03 |
| KCNV1 | 3.60 | 0.0062668 | 2.29e-03 |
| KCTD11 | 1.54 | 0.0000000 | 0.00e+00 |
| KCTD13 | 1.84 | 0.0000000 | 0.00e+00 |
| KCTD15 | -1.53 | 0.0000000 | 0.00e+00 |
| KCTD19 | 2.12 | 0.0793708 | 3.94e-02 |
| KCTD21 | 1.00 | 0.0066426 | 2.44e-03 |
| KCTD3 | -1.35 | 0.0000000 | 0.00e+00 |
| KCTD5 | 0.41 | 0.0770095 | 3.81e-02 |
| KCTD6 | -0.93 | 0.0808415 | 4.03e-02 |
| KCTD7 | -1.39 | 0.0015743 | 5.02e-04 |
| KDELC1 | -1.34 | 0.0142431 | 5.68e-03 |
| KDELR1 | 0.84 | 0.0000000 | 0.00e+00 |
| KDM1B | -0.81 | 0.0364626 | 1.62e-02 |
| KDM2A | 1.44 | 0.0000000 | 0.00e+00 |
| KDM2B | 1.18 | 0.0000000 | 0.00e+00 |
| KDM3A | 0.63 | 0.0020635 | 6.78e-04 |
| KDM3B | 0.70 | 0.0000005 | 1.00e-07 |
| KDM4B | 1.85 | 0.0000000 | 0.00e+00 |
| KDM4C | 0.54 | 0.0731941 | 3.59e-02 |
| KDM5A | 0.74 | 0.0000010 | 2.00e-07 |
| KDM5B | 1.85 | 0.0000000 | 0.00e+00 |
| KDM5C | 1.49 | 0.0000000 | 0.00e+00 |
| KDM6A | 2.14 | 0.0000000 | 0.00e+00 |
| KDM6B | 4.46 | 0.0000000 | 0.00e+00 |
| KDM7A | 3.58 | 0.0000000 | 0.00e+00 |
| KDSR | 1.31 | 0.0000000 | 0.00e+00 |
| KHK | -5.25 | 0.0000000 | 0.00e+00 |
| KHNYN | 1.54 | 0.0000000 | 0.00e+00 |
| KIAA0020 | -1.09 | 0.0000004 | 1.00e-07 |
| KIAA0040 | 3.33 | 0.0000000 | 0.00e+00 |
| KIAA0100 | -0.44 | 0.0001873 | 5.07e-05 |
| KIAA0101 | -3.30 | 0.0000000 | 0.00e+00 |
| KIAA0195 | 0.78 | 0.0000037 | 8.00e-07 |
| KIAA0196 | -1.22 | 0.0000000 | 0.00e+00 |
| KIAA0226 | 1.49 | 0.0000077 | 1.70e-06 |
| KIAA0319L | 0.74 | 0.0003280 | 9.21e-05 |
| KIAA0355 | 1.29 | 0.0000000 | 0.00e+00 |
| KIAA0556 | 0.57 | 0.0158352 | 6.38e-03 |
| KIAA0586 | -1.69 | 0.0001517 | 4.06e-05 |
| KIAA0895L | -1.06 | 0.0771063 | 3.81e-02 |
| KIAA1109 | -1.29 | 0.0000062 | 1.40e-06 |
| KIAA1143 | -0.97 | 0.0106705 | 4.13e-03 |
| KIAA1147 | -1.12 | 0.0004472 | 1.29e-04 |
| KIAA1191 | 0.71 | 0.0000000 | 0.00e+00 |
| KIAA1211 | 1.58 | 0.0010238 | 3.15e-04 |
| KIAA1217 | 3.41 | 0.0000000 | 0.00e+00 |
| KIAA1429 | -0.56 | 0.0056163 | 2.02e-03 |
| KIAA1462 | 1.60 | 0.0000081 | 1.80e-06 |
| KIAA1468 | -0.54 | 0.0635652 | 3.04e-02 |
| KIAA1522 | 1.21 | 0.0000000 | 0.00e+00 |
| KIAA1524 | -2.70 | 0.0000000 | 0.00e+00 |
| KIAA1614 | 2.32 | 0.0625768 | 2.99e-02 |
| KIAA1644 | 3.05 | 0.0000298 | 7.20e-06 |
| KIAA1671 | 1.19 | 0.0000000 | 0.00e+00 |
| KIAA1715 | -1.65 | 0.0000822 | 2.10e-05 |
| KIAA1804 | -0.91 | 0.0025581 | 8.58e-04 |
| KIAA1841 | -0.86 | 0.0259521 | 1.11e-02 |
| KIDINS220 | -0.42 | 0.0533974 | 2.49e-02 |
| KIF11 | -3.25 | 0.0000000 | 0.00e+00 |
| KIF13A | -0.92 | 0.0000541 | 1.35e-05 |
| KIF13B | 1.18 | 0.0000000 | 0.00e+00 |
| KIF14 | -3.55 | 0.0000000 | 0.00e+00 |
| KIF15 | -4.02 | 0.0000000 | 0.00e+00 |
| KIF18A | -3.25 | 0.0000000 | 0.00e+00 |
| KIF18B | -2.25 | 0.0000000 | 0.00e+00 |
| KIF1B | 0.52 | 0.0130205 | 5.14e-03 |
| KIF1C | -0.25 | 0.0770668 | 3.81e-02 |
| KIF20A | -3.98 | 0.0000000 | 0.00e+00 |
| KIF20B | -2.34 | 0.0000000 | 0.00e+00 |
| KIF21A | -0.49 | 0.0246647 | 1.04e-02 |
| KIF21B | 1.69 | 0.0000000 | 0.00e+00 |
| KIF22 | -2.09 | 0.0000000 | 0.00e+00 |
| KIF23 | -2.47 | 0.0000000 | 0.00e+00 |
| KIF24 | -1.44 | 0.0034974 | 1.20e-03 |
| KIF26A | -1.20 | 0.0814750 | 4.07e-02 |
| KIF26B | 5.44 | 0.0956866 | 4.91e-02 |
| KIF2A | 0.34 | 0.0991671 | 5.12e-02 |
| KIF2C | -1.93 | 0.0000000 | 0.00e+00 |
| KIF3A | -1.56 | 0.0000219 | 5.20e-06 |
| KIF3C | 1.17 | 0.0000000 | 0.00e+00 |
| KIF4A | -2.75 | 0.0000000 | 0.00e+00 |
| KIF4B | -3.38 | 0.0000019 | 4.00e-07 |
| KIF5B | -1.18 | 0.0000000 | 0.00e+00 |
| KIFAP3 | 0.95 | 0.0013462 | 4.24e-04 |
| KIFC1 | -2.47 | 0.0000000 | 0.00e+00 |
| KIFC3 | 0.36 | 0.0015215 | 4.84e-04 |
| KIRREL | 0.65 | 0.0002036 | 5.53e-05 |
| KIRREL2 | 4.45 | 0.0000000 | 0.00e+00 |
| KITLG | -1.84 | 0.0000361 | 8.80e-06 |
| KLC1 | 0.34 | 0.0452574 | 2.07e-02 |
| KLC2 | 1.11 | 0.0000000 | 0.00e+00 |
| KLF10 | 0.43 | 0.0245768 | 1.04e-02 |
| KLF11 | 0.98 | 0.0146709 | 5.86e-03 |
| KLF12 | -1.50 | 0.0573582 | 2.71e-02 |
| KLF15 | 6.65 | 0.0124880 | 4.91e-03 |
| KLF16 | 0.81 | 0.0011784 | 3.67e-04 |
| KLF2 | 3.83 | 0.0000000 | 0.00e+00 |
| KLF3 | 2.19 | 0.0000000 | 0.00e+00 |
| KLF4 | 5.17 | 0.0000000 | 0.00e+00 |
| KLF5 | 1.18 | 0.0000000 | 0.00e+00 |
| KLF6 | 3.06 | 0.0000000 | 0.00e+00 |
| KLF7 | 1.67 | 0.0000000 | 0.00e+00 |
| KLF9 | 3.30 | 0.0000000 | 0.00e+00 |
| KLHDC2 | -0.47 | 0.0758294 | 3.74e-02 |
| KLHDC3 | -0.47 | 0.0834777 | 4.19e-02 |
| KLHDC7B | 7.09 | 0.0057398 | 2.08e-03 |
| KLHL11 | -1.20 | 0.0029312 | 9.92e-04 |
| KLHL12 | -0.88 | 0.0012194 | 3.81e-04 |
| KLHL13 | -2.34 | 0.0713836 | 3.48e-02 |
| KLHL21 | 1.98 | 0.0000000 | 0.00e+00 |
| KLHL23 | -1.92 | 0.0000103 | 2.30e-06 |
| KLHL24 | 0.83 | 0.0642854 | 3.08e-02 |
| KLHL29 | 1.59 | 0.0001097 | 2.86e-05 |
| KLHL38 | 6.09 | 0.0350409 | 1.55e-02 |
| KLHL5 | 0.59 | 0.0001177 | 3.09e-05 |
| KLHL7 | -0.88 | 0.0156967 | 6.32e-03 |
| KLHL9 | -0.47 | 0.0493331 | 2.27e-02 |
| KLK10 | 10.63 | 0.0000000 | 0.00e+00 |
| KLRC1 | 6.09 | 0.0346150 | 1.53e-02 |
| KLRC2 | 2.79 | 0.0203411 | 8.41e-03 |
| KMT2B | 2.37 | 0.0000000 | 0.00e+00 |
| KMT2C | 0.48 | 0.0359286 | 1.59e-02 |
| KMT2D | 0.72 | 0.0000000 | 0.00e+00 |
| KMT2E | 0.77 | 0.0001868 | 5.05e-05 |
| KNOP1 | -0.56 | 0.0266545 | 1.14e-02 |
| KNSTRN | -1.43 | 0.0000003 | 1.00e-07 |
| KNTC1 | -2.94 | 0.0000000 | 0.00e+00 |
| KPNA1 | 0.37 | 0.0263687 | 1.13e-02 |
| KPNA2 | -1.65 | 0.0000000 | 0.00e+00 |
| KPNA4 | 0.34 | 0.0412408 | 1.86e-02 |
| KPNB1 | -0.28 | 0.0069086 | 2.54e-03 |
| KRAS | 0.42 | 0.0470443 | 2.16e-02 |
| KRBA1 | 1.48 | 0.0012409 | 3.88e-04 |
| KRCC1 | 1.01 | 0.0029576 | 1.00e-03 |
| KRIT1 | -1.20 | 0.0001432 | 3.82e-05 |
| KRR1 | -1.05 | 0.0000356 | 8.70e-06 |
| KRT10 | -0.79 | 0.0020558 | 6.75e-04 |
| KRT14 | 9.84 | 0.0000100 | 2.20e-06 |
| KRT15 | 1.89 | 0.0000175 | 4.10e-06 |
| KRT16 | 2.71 | 0.0060775 | 2.21e-03 |
| KRT16P1 | 5.61 | 0.0717731 | 3.50e-02 |
| KRT16P2 | 5.76 | 0.0578985 | 2.73e-02 |
| KRT17 | 8.00 | 0.0000000 | 0.00e+00 |
| KRT18 | -1.00 | 0.0000000 | 0.00e+00 |
| KRT18P55 | -0.72 | 0.0122479 | 4.81e-03 |
| KRT20 | -6.97 | 0.0038251 | 1.33e-03 |
| KRT4 | -5.16 | 0.0000000 | 0.00e+00 |
| KRT6A | 3.08 | 0.0000000 | 0.00e+00 |
| KRT6B | 2.52 | 0.0000076 | 1.70e-06 |
| KRT6C | 1.62 | 0.0198874 | 8.21e-03 |
| KRT8 | -1.03 | 0.0000000 | 0.00e+00 |
| KRT80 | -0.61 | 0.0003751 | 1.07e-04 |
| KRT81 | -0.97 | 0.0000000 | 0.00e+00 |
| KRT83 | -0.60 | 0.0002245 | 6.15e-05 |
| KRT85 | -0.78 | 0.0000193 | 4.50e-06 |
| KRT86 | -0.76 | 0.0000000 | 0.00e+00 |
| KRT8P41 | -1.25 | 0.0370594 | 1.65e-02 |
| KRTCAP2 | 1.24 | 0.0000000 | 0.00e+00 |
| KSR1 | 0.76 | 0.0127906 | 5.04e-03 |
| KTN1 | -0.46 | 0.0002775 | 7.70e-05 |
| KY | 3.25 | 0.0677694 | 3.27e-02 |
| KYNU | 0.86 | 0.0000000 | 0.00e+00 |
| L1CAM | 1.85 | 0.0000000 | 0.00e+00 |
| L2HGDH | -0.84 | 0.0251369 | 1.07e-02 |
| L3HYPDH | -0.93 | 0.0034985 | 1.21e-03 |
| L3MBTL4 | 7.30 | 0.0034407 | 1.18e-03 |
| LACC1 | 1.27 | 0.0121747 | 4.78e-03 |
| LACTB | 0.54 | 0.0073074 | 2.71e-03 |
| LACTB2 | -0.76 | 0.0019097 | 6.22e-04 |
| LAD1 | 3.30 | 0.0000000 | 0.00e+00 |
| LAGE3 | -1.03 | 0.0008116 | 2.45e-04 |
| LAMA5 | 0.80 | 0.0000000 | 0.00e+00 |
| LAMB1 | -0.85 | 0.0000000 | 0.00e+00 |
| LAMB2 | 1.44 | 0.0000000 | 0.00e+00 |
| LAMB3 | 6.20 | 0.0000000 | 0.00e+00 |
| LAMC1 | 0.62 | 0.0000000 | 0.00e+00 |
| LAMC2 | 6.57 | 0.0000000 | 0.00e+00 |
| LAMP1 | 0.75 | 0.0000001 | 0.00e+00 |
| LAMP3 | 9.06 | 0.0000000 | 0.00e+00 |
| LAMTOR1 | 0.37 | 0.0337500 | 1.48e-02 |
| LAMTOR4 | -0.73 | 0.0012865 | 4.03e-04 |
| LANCL1 | -1.66 | 0.0000000 | 0.00e+00 |
| LANCL2 | -0.78 | 0.0684229 | 3.31e-02 |
| LAP3 | 2.53 | 0.0000000 | 0.00e+00 |
| LAPTM4A | -0.30 | 0.0500737 | 2.31e-02 |
| LAPTM4B | -0.94 | 0.0000000 | 0.00e+00 |
| LAPTM5 | 7.61 | 0.0018018 | 5.82e-04 |
| LARGE | 1.18 | 0.0002445 | 6.73e-05 |
| LARP1 | 0.39 | 0.0007063 | 2.10e-04 |
| LARP6 | 2.10 | 0.0000000 | 0.00e+00 |
| LARP7 | -0.63 | 0.0337398 | 1.48e-02 |
| LARS | -1.11 | 0.0000000 | 0.00e+00 |
| LAS1L | -1.49 | 0.0000000 | 0.00e+00 |
| LASP1 | -0.21 | 0.0270407 | 1.16e-02 |
| LATS2 | 1.30 | 0.0000000 | 0.00e+00 |
| LAYN | 1.89 | 0.0202583 | 8.37e-03 |
| LBH | -1.46 | 0.0000000 | 0.00e+00 |
| LBP | 6.35 | 0.0000000 | 0.00e+00 |
| LBR | -2.04 | 0.0000000 | 0.00e+00 |
| LBX2-AS1 | 1.32 | 0.0452460 | 2.06e-02 |
| LCE3D | 8.26 | 0.0004255 | 1.22e-04 |
| LCE3E | 6.69 | 0.0125482 | 4.94e-03 |
| LCE5A | 5.90 | 0.0466313 | 2.14e-02 |
| LCLAT1 | -1.34 | 0.0000028 | 6.00e-07 |
| LCN2 | 8.55 | 0.0000000 | 0.00e+00 |
| LCP1 | 5.94 | 0.0006173 | 1.82e-04 |
| LCP2 | 6.20 | 0.0283388 | 1.22e-02 |
| LDAH | -1.21 | 0.0000027 | 6.00e-07 |
| LDHA | -1.12 | 0.0000000 | 0.00e+00 |
| LDHB | -1.54 | 0.0000000 | 0.00e+00 |
| LDLR | 3.50 | 0.0000000 | 0.00e+00 |
| LDLRAD2 | 5.76 | 0.0612786 | 2.91e-02 |
| LDLRAP1 | -0.72 | 0.0751642 | 3.70e-02 |
| LDOC1 | -0.55 | 0.0692871 | 3.36e-02 |
| LDOC1L | 0.53 | 0.0189028 | 7.76e-03 |
| LEMD2 | 0.98 | 0.0000000 | 0.00e+00 |
| LEMD3 | -0.57 | 0.0835554 | 4.19e-02 |
| LENG8 | 0.76 | 0.0044609 | 1.57e-03 |
| LENG9 | 2.21 | 0.0003870 | 1.10e-04 |
| LEPR | -1.04 | 0.0976391 | 5.02e-02 |
| LEPROT | -0.58 | 0.0146853 | 5.87e-03 |
| LEPROTL1 | -0.84 | 0.0002603 | 7.18e-05 |
| LETM1 | -0.41 | 0.0622248 | 2.97e-02 |
| LGALS1 | -0.39 | 0.0005945 | 1.75e-04 |
| LGALS3BP | 5.02 | 0.0000000 | 0.00e+00 |
| LGALS8 | 0.82 | 0.0000078 | 1.70e-06 |
| LGALS9 | 6.78 | 0.0000000 | 0.00e+00 |
| LGALS9B | 4.91 | 0.0108522 | 4.21e-03 |
| LGALS9C | 8.04 | 0.0007056 | 2.10e-04 |
| LGALSL | 1.14 | 0.0004015 | 1.15e-04 |
| LGI2 | 2.91 | 0.0598138 | 2.83e-02 |
| LGMN | 1.80 | 0.0000000 | 0.00e+00 |
| LGR4 | -1.83 | 0.0000000 | 0.00e+00 |
| LHFP | 2.06 | 0.0303943 | 1.32e-02 |
| LHFPL2 | 1.55 | 0.0000000 | 0.00e+00 |
| LHX8 | 1.41 | 0.0384316 | 1.72e-02 |
| LIAS | -1.92 | 0.0018289 | 5.92e-04 |
| LIF | 0.69 | 0.0000011 | 2.00e-07 |
| LIFR | 1.42 | 0.0000001 | 0.00e+00 |
| LIG1 | -1.85 | 0.0000000 | 0.00e+00 |
| LIG3 | -0.60 | 0.0052924 | 1.90e-03 |
| LIMA1 | 1.70 | 0.0000000 | 0.00e+00 |
| LIMCH1 | -0.58 | 0.0000150 | 3.50e-06 |
| LIMD2 | 0.91 | 0.0270641 | 1.16e-02 |
| LIMK1 | 1.02 | 0.0000001 | 0.00e+00 |
| LIMK2 | 1.36 | 0.0000011 | 2.00e-07 |
| LIN37 | 2.34 | 0.0000000 | 0.00e+00 |
| LIN54 | -1.26 | 0.0046498 | 1.65e-03 |
| LIN7C | -1.08 | 0.0006976 | 2.07e-04 |
| LIN9 | -1.97 | 0.0000231 | 5.50e-06 |
| LINC-PINT | 0.77 | 0.0376030 | 1.67e-02 |
| LINC00094 | -1.44 | 0.0015022 | 4.77e-04 |
| LINC00116 | -0.59 | 0.0283590 | 1.22e-02 |
| LINC00173 | -2.30 | 0.0582061 | 2.75e-02 |
| LINC00265 | 1.38 | 0.0784064 | 3.88e-02 |
| LINC00284 | -5.90 | 0.0687493 | 3.33e-02 |
| LINC00341 | 7.61 | 0.0018018 | 5.82e-04 |
| LINC00346 | 2.32 | 0.0984054 | 5.07e-02 |
| LINC00471 | -3.32 | 0.0528472 | 2.46e-02 |
| LINC00473 | -0.82 | 0.0011372 | 3.53e-04 |
| LINC00493 | 0.53 | 0.0064520 | 2.36e-03 |
| LINC00520 | 6.32 | 0.0001764 | 4.76e-05 |
| LINC00540 | 2.32 | 0.0124428 | 4.89e-03 |
| LINC00598 | 5.53 | 0.0810935 | 4.05e-02 |
| LINC00641 | -1.73 | 0.0004357 | 1.26e-04 |
| LINC00657 | -0.26 | 0.0903986 | 4.59e-02 |
| LINC00673 | 2.01 | 0.0000000 | 0.00e+00 |
| LINC00707 | 1.83 | 0.0000000 | 0.00e+00 |
| LINC00842 | -1.46 | 0.0013115 | 4.12e-04 |
| LINC00941 | 2.01 | 0.0007185 | 2.14e-04 |
| LINC00963 | 1.05 | 0.0233595 | 9.83e-03 |
| LINC00998 | -1.28 | 0.0090443 | 3.43e-03 |
| LINC01011 | -2.49 | 0.0973344 | 5.00e-02 |
| LINC01106 | -2.72 | 0.0041066 | 1.44e-03 |
| LINC01123 | -2.38 | 0.0011425 | 3.55e-04 |
| LINC01138 | 1.47 | 0.0948273 | 4.86e-02 |
| LINC01207 | 2.52 | 0.0321874 | 1.40e-02 |
| LINC01239 | 6.65 | 0.0131266 | 5.19e-03 |
| LINC01270 | -2.67 | 0.0002158 | 5.89e-05 |
| LINC01296 | -0.97 | 0.0025448 | 8.53e-04 |
| LINC01420 | 0.69 | 0.0058415 | 2.11e-03 |
| LINC01468 | -1.69 | 0.0044939 | 1.59e-03 |
| LINC01521 | -1.57 | 0.0741308 | 3.64e-02 |
| LINC01539 | 6.61 | 0.0133613 | 5.29e-03 |
| LINC01559 | 2.37 | 0.0147069 | 5.88e-03 |
| LINC01578 | 1.54 | 0.0000000 | 0.00e+00 |
| LINC01589 | 1.79 | 0.0181505 | 7.42e-03 |
| LINC01604 | -1.73 | 0.0039039 | 1.36e-03 |
| LINGO2 | 5.76 | 0.0578985 | 2.73e-02 |
| LIPA | 0.36 | 0.0246769 | 1.04e-02 |
| LIPG | 6.62 | 0.0000000 | 0.00e+00 |
| LIPH | 2.21 | 0.0000000 | 0.00e+00 |
| LLGL1 | -0.55 | 0.0442196 | 2.01e-02 |
| LLGL2 | -0.53 | 0.0037063 | 1.28e-03 |
| LMAN1 | -0.80 | 0.0000002 | 0.00e+00 |
| LMAN2 | 0.53 | 0.0000147 | 3.40e-06 |
| LMAN2L | -0.46 | 0.0603406 | 2.86e-02 |
| LMBR1 | -0.57 | 0.0168581 | 6.83e-03 |
| LMBR1L | 1.19 | 0.0000299 | 7.20e-06 |
| LMBRD2 | -1.07 | 0.0910442 | 4.63e-02 |
| LMCD1 | 2.04 | 0.0000000 | 0.00e+00 |
| LMF1 | 1.90 | 0.0000000 | 0.00e+00 |
| LMF2 | 1.19 | 0.0000000 | 0.00e+00 |
| LMNB1 | -2.43 | 0.0000000 | 0.00e+00 |
| LMNB2 | -1.05 | 0.0000000 | 0.00e+00 |
| LMO2 | 3.62 | 0.0002386 | 6.56e-05 |
| LMO7 | 1.42 | 0.0000000 | 0.00e+00 |
| LMTK2 | 0.62 | 0.0005121 | 1.49e-04 |
| LMX1B | 1.19 | 0.0502065 | 2.32e-02 |
| LOC100126784 | 4.77 | 0.0003615 | 1.03e-04 |
| LOC100130417 | -2.58 | 0.0600549 | 2.85e-02 |
| LOC100130476 | 5.53 | 0.0806408 | 4.02e-02 |
| LOC100287042 | -2.19 | 0.0250969 | 1.07e-02 |
| LOC100288637 | -1.75 | 0.0396348 | 1.78e-02 |
| LOC100288778 | 0.93 | 0.0416964 | 1.88e-02 |
| LOC100419583 | 2.66 | 0.0000000 | 0.00e+00 |
| LOC100505666 | -2.30 | 0.0691330 | 3.35e-02 |
| LOC100505817 | 2.03 | 0.0000000 | 0.00e+00 |
| LOC100506100 | -2.78 | 0.0007436 | 2.22e-04 |
| LOC100506127 | -2.73 | 0.0052810 | 1.89e-03 |
| LOC100506548 | -0.81 | 0.0336974 | 1.48e-02 |
| LOC100507002 | 1.34 | 0.0040132 | 1.40e-03 |
| LOC100507144 | 2.91 | 0.0598138 | 2.83e-02 |
| LOC100507412 | 1.89 | 0.0497074 | 2.29e-02 |
| LOC100507487 | -1.67 | 0.0538859 | 2.52e-02 |
| LOC100630923 | 0.53 | 0.0499624 | 2.31e-02 |
| LOC101060091 | 6.80 | 0.0093623 | 3.57e-03 |
| LOC101243545 | 2.58 | 0.0835900 | 4.20e-02 |
| LOC101927021 | -1.28 | 0.0333788 | 1.46e-02 |
| LOC101927204 | 1.71 | 0.0515700 | 2.39e-02 |
| LOC101927550 | 2.87 | 0.0066430 | 2.44e-03 |
| LOC101927746 | -2.36 | 0.0024170 | 8.06e-04 |
| LOC101927811 | 5.44 | 0.0918945 | 4.69e-02 |
| LOC101927865 | 5.97 | 0.0417361 | 1.88e-02 |
| LOC101928841 | 3.30 | 0.0185170 | 7.58e-03 |
| LOC101929427 | 4.04 | 0.0672812 | 3.25e-02 |
| LOC101929709 | 2.50 | 0.0001076 | 2.80e-05 |
| LOC102723354 | -2.47 | 0.0286905 | 1.23e-02 |
| LOC103021296 | 3.86 | 0.0917429 | 4.68e-02 |
| LOC105274304 | -6.61 | 0.0119251 | 4.67e-03 |
| LOC146880 | -1.06 | 0.0000000 | 0.00e+00 |
| LOC171391 | -1.88 | 0.0865905 | 4.37e-02 |
| LOC283335 | -1.85 | 0.0448012 | 2.04e-02 |
| LOC339803 | -0.90 | 0.0127646 | 5.03e-03 |
| LOC341056 | -1.08 | 0.0531035 | 2.47e-02 |
| LOC344967 | 1.29 | 0.0156967 | 6.32e-03 |
| LOC388282 | -1.69 | 0.0046321 | 1.64e-03 |
| LOC389602 | 6.53 | 0.0157810 | 6.36e-03 |
| LOC400043 | 2.19 | 0.0632569 | 3.02e-02 |
| LOC400684 | -2.12 | 0.0855536 | 4.31e-02 |
| LOC400927-CSNK1E | 0.84 | 0.0000010 | 2.00e-07 |
| LOC407835 | 2.56 | 0.0000000 | 0.00e+00 |
| LOC440311 | 1.44 | 0.0000000 | 0.00e+00 |
| LOC440434 | 0.86 | 0.0680787 | 3.29e-02 |
| LOC440896 | 11.40 | 0.0000002 | 0.00e+00 |
| LOC441454 | -1.04 | 0.0265718 | 1.13e-02 |
| LOC642846 | -1.18 | 0.0030523 | 1.04e-03 |
| LOC646626 | 7.17 | 0.0044928 | 1.58e-03 |
| LOC652276 | 1.14 | 0.0450600 | 2.06e-02 |
| LOC727896 | -1.06 | 0.0063746 | 2.33e-03 |
| LOC728554 | -2.10 | 0.0000000 | 0.00e+00 |
| LOC728743 | 1.22 | 0.0305539 | 1.32e-02 |
| LOC729080 | -3.81 | 0.0020199 | 6.61e-04 |
| LOC730102 | -1.74 | 0.0024747 | 8.27e-04 |
| LOC731424 | 7.00 | 0.0064268 | 2.35e-03 |
| LOC81691 | -1.20 | 0.0682915 | 3.30e-02 |
| LOC93622 | -1.98 | 0.0000643 | 1.62e-05 |
| LOH12CR1 | 0.90 | 0.0522057 | 2.42e-02 |
| LOH12CR2 | -5.76 | 0.0886793 | 4.49e-02 |
| LONP1 | 0.33 | 0.0174722 | 7.12e-03 |
| LONRF1 | 1.17 | 0.0003593 | 1.02e-04 |
| LONRF2 | -0.86 | 0.0044348 | 1.56e-03 |
| LONRF3 | 0.98 | 0.0489314 | 2.25e-02 |
| LOX | 5.27 | 0.0047038 | 1.67e-03 |
| LOXL2 | 0.54 | 0.0001900 | 5.15e-05 |
| LPAR5 | 1.96 | 0.0765571 | 3.78e-02 |
| LPCAT1 | 0.65 | 0.0085211 | 3.21e-03 |
| LPCAT2 | -1.26 | 0.0137849 | 5.48e-03 |
| LPIN1 | 0.37 | 0.0619141 | 2.95e-02 |
| LPIN2 | 0.59 | 0.0107573 | 4.16e-03 |
| LPXN | 2.59 | 0.0000045 | 1.00e-06 |
| LRCH1 | 1.00 | 0.0001350 | 3.58e-05 |
| LRCH3 | 0.80 | 0.0246175 | 1.04e-02 |
| LRCH4 | 0.87 | 0.0064742 | 2.37e-03 |
| LRFN1 | 1.37 | 0.0000079 | 1.80e-06 |
| LRFN3 | 1.90 | 0.0000000 | 0.00e+00 |
| LRFN4 | -0.55 | 0.0710938 | 3.46e-02 |
| LRG1 | 3.57 | 0.0000194 | 4.60e-06 |
| LRIF1 | 0.91 | 0.0016477 | 5.27e-04 |
| LRIG1 | 1.91 | 0.0000000 | 0.00e+00 |
| LRIG3 | 1.60 | 0.0000000 | 0.00e+00 |
| LRP1 | 1.76 | 0.0000000 | 0.00e+00 |
| LRP10 | 2.87 | 0.0000000 | 0.00e+00 |
| LRP12 | 0.57 | 0.0330904 | 1.45e-02 |
| LRP3 | -0.60 | 0.0049225 | 1.75e-03 |
| LRP4 | -0.87 | 0.0146601 | 5.86e-03 |
| LRP5 | 0.53 | 0.0003742 | 1.06e-04 |
| LRP6 | -1.33 | 0.0000000 | 0.00e+00 |
| LRPAP1 | 0.29 | 0.0947081 | 4.85e-02 |
| LRPPRC | -1.35 | 0.0000000 | 0.00e+00 |
| LRR1 | -1.27 | 0.0001218 | 3.20e-05 |
| LRRC1 | 0.83 | 0.0539790 | 2.52e-02 |
| LRRC14 | -0.60 | 0.0285100 | 1.23e-02 |
| LRRC15 | 5.90 | 0.0462524 | 2.12e-02 |
| LRRC20 | -1.52 | 0.0000021 | 4.00e-07 |
| LRRC23 | -1.07 | 0.0089415 | 3.39e-03 |
| LRRC31 | 5.44 | 0.0933961 | 4.78e-02 |
| LRRC32 | 6.20 | 0.0000000 | 0.00e+00 |
| LRRC38 | 10.40 | 0.0000023 | 5.00e-07 |
| LRRC40 | -1.70 | 0.0000868 | 2.23e-05 |
| LRRC41 | 1.13 | 0.0000000 | 0.00e+00 |
| LRRC45 | -1.88 | 0.0000001 | 0.00e+00 |
| LRRC47 | 0.47 | 0.0347966 | 1.53e-02 |
| LRRC49 | -1.54 | 0.0009446 | 2.89e-04 |
| LRRC55 | 6.53 | 0.0000000 | 0.00e+00 |
| LRRC58 | -1.32 | 0.0000001 | 0.00e+00 |
| LRRC59 | 0.46 | 0.0000040 | 9.00e-07 |
| LRRC75A | 1.80 | 0.0236212 | 9.95e-03 |
| LRRC8A | 1.45 | 0.0000000 | 0.00e+00 |
| LRRC8B | -1.02 | 0.0526560 | 2.45e-02 |
| LRRC8E | 1.03 | 0.0401372 | 1.80e-02 |
| LRRCC1 | -1.76 | 0.0425357 | 1.92e-02 |
| LRRFIP2 | 0.95 | 0.0000007 | 1.00e-07 |
| LRRK1 | 0.82 | 0.0000381 | 9.30e-06 |
| LRRK2 | -0.63 | 0.0412532 | 1.86e-02 |
| LRRN3 | 5.98 | 0.0005209 | 1.52e-04 |
| LRSAM1 | 0.77 | 0.0150412 | 6.03e-03 |
| LSAMP | 5.61 | 0.0771151 | 3.81e-02 |
| LSG1 | -0.67 | 0.0026145 | 8.78e-04 |
| LSM10 | -0.59 | 0.0818732 | 4.09e-02 |
| LSM3 | -1.56 | 0.0000000 | 0.00e+00 |
| LSM4 | -1.76 | 0.0000000 | 0.00e+00 |
| LSM5 | -1.78 | 0.0000000 | 0.00e+00 |
| LSM7 | -0.64 | 0.0393742 | 1.76e-02 |
| LSM8 | -0.67 | 0.0351625 | 1.55e-02 |
| LSR | 3.68 | 0.0000000 | 0.00e+00 |
| LSS | 0.81 | 0.0005759 | 1.69e-04 |
| LTA4H | -1.40 | 0.0000000 | 0.00e+00 |
| LTB4R | -0.74 | 0.0948345 | 4.86e-02 |
| LTBP2 | 3.20 | 0.0000000 | 0.00e+00 |
| LTBP3 | 1.92 | 0.0000000 | 0.00e+00 |
| LTBP4 | 0.77 | 0.0001660 | 4.47e-05 |
| LTBR | 1.26 | 0.0000000 | 0.00e+00 |
| LTN1 | -1.26 | 0.0000034 | 7.00e-07 |
| LTV1 | -0.88 | 0.0009042 | 2.75e-04 |
| LUC7L3 | -1.55 | 0.0000000 | 0.00e+00 |
| LURAP1L | 0.93 | 0.0076816 | 2.87e-03 |
| LUZP1 | 1.08 | 0.0000000 | 0.00e+00 |
| LUZP6 | -0.49 | 0.0003449 | 9.73e-05 |
| LXN | -0.98 | 0.0003770 | 1.07e-04 |
| LY6E | 1.71 | 0.0000000 | 0.00e+00 |
| LYAR | -0.68 | 0.0501437 | 2.32e-02 |
| LYG1 | 2.37 | 0.0526501 | 2.45e-02 |
| LYN | 2.81 | 0.0000000 | 0.00e+00 |
| LYPD1 | 4.64 | 0.0000000 | 0.00e+00 |
| LYPD3 | 3.97 | 0.0010963 | 3.39e-04 |
| LYPLA1 | -0.96 | 0.0000207 | 4.90e-06 |
| LYRM2 | -0.68 | 0.0343344 | 1.51e-02 |
| LYRM5 | -1.22 | 0.0767624 | 3.79e-02 |
| LYRM7 | -1.61 | 0.0055843 | 2.01e-03 |
| LYSMD2 | 1.52 | 0.0000000 | 0.00e+00 |
| LYZ | 7.35 | 0.0031811 | 1.09e-03 |
| LZIC | -0.84 | 0.0791471 | 3.92e-02 |
| LZTFL1 | -1.11 | 0.0261719 | 1.12e-02 |
| LZTS3 | 1.58 | 0.0000022 | 4.00e-07 |
| MACF1 | 0.57 | 0.0000109 | 2.50e-06 |
| MAD1L1 | 0.93 | 0.0020439 | 6.70e-04 |
| MAD2L1 | -3.44 | 0.0000000 | 0.00e+00 |
| MAD2L1BP | 0.70 | 0.0221739 | 9.25e-03 |
| MAD2L2 | 0.47 | 0.0837918 | 4.21e-02 |
| MADD | 0.56 | 0.0033243 | 1.14e-03 |
| MAEA | -1.00 | 0.0000207 | 4.90e-06 |
| MAF | 4.70 | 0.0173878 | 7.08e-03 |
| MAFA | 2.95 | 0.0246218 | 1.04e-02 |
| MAFB | 4.21 | 0.0494635 | 2.28e-02 |
| MAFF | 5.19 | 0.0000000 | 0.00e+00 |
| MAFG | 1.01 | 0.0000000 | 0.00e+00 |
| MAFK | 1.08 | 0.0000000 | 0.00e+00 |
| MAGEA6 | 1.09 | 0.0231618 | 9.73e-03 |
| MAGED2 | 0.81 | 0.0000001 | 0.00e+00 |
| MAGEH1 | -1.42 | 0.0494635 | 2.28e-02 |
| MAGI1 | 0.79 | 0.0563893 | 2.65e-02 |
| MAGOH | -0.68 | 0.0973445 | 5.00e-02 |
| MAGOHB | -1.09 | 0.0034942 | 1.20e-03 |
| MAK16 | -1.11 | 0.0000066 | 1.50e-06 |
| MAL2 | 3.02 | 0.0180406 | 7.37e-03 |
| MALAT1 | -1.67 | 0.0000000 | 0.00e+00 |
| MALL | -1.89 | 0.0000000 | 0.00e+00 |
| MAML1 | 1.41 | 0.0000000 | 0.00e+00 |
| MAML2 | 1.57 | 0.0002669 | 7.39e-05 |
| MAML3 | 1.10 | 0.0000001 | 0.00e+00 |
| MAMLD1 | 3.76 | 0.0000000 | 0.00e+00 |
| MAN1A1 | 4.04 | 0.0672812 | 3.25e-02 |
| MAN1A2 | -0.74 | 0.0299273 | 1.29e-02 |
| MAN2A1 | -0.56 | 0.0183044 | 7.49e-03 |
| MAN2B1 | 2.10 | 0.0000000 | 0.00e+00 |
| MAN2B2 | 0.99 | 0.0003366 | 9.47e-05 |
| MAN2C1 | 0.45 | 0.0245280 | 1.04e-02 |
| MANBAL | -0.34 | 0.0569434 | 2.68e-02 |
| MANEA | -1.57 | 0.0195082 | 8.03e-03 |
| MANEAL | -0.83 | 0.0156181 | 6.28e-03 |
| MANF | 0.37 | 0.0825074 | 4.13e-02 |
| MAP1A | -2.94 | 0.0142197 | 5.67e-03 |
| MAP1LC3A | 3.25 | 0.0000000 | 0.00e+00 |
| MAP1LC3B | 1.52 | 0.0000000 | 0.00e+00 |
| MAP1LC3B2 | 1.26 | 0.0000750 | 1.91e-05 |
| MAP1S | 2.79 | 0.0000000 | 0.00e+00 |
| MAP2 | 3.00 | 0.0000000 | 0.00e+00 |
| MAP2K1 | 1.00 | 0.0000000 | 0.00e+00 |
| MAP2K2 | 2.52 | 0.0000000 | 0.00e+00 |
| MAP2K3 | 2.02 | 0.0000000 | 0.00e+00 |
| MAP2K4 | -0.52 | 0.0232810 | 9.79e-03 |
| MAP2K6 | -2.92 | 0.0000000 | 0.00e+00 |
| MAP2K7 | 0.64 | 0.0116219 | 4.54e-03 |
| MAP3K1 | 0.82 | 0.0121167 | 4.75e-03 |
| MAP3K10 | 2.39 | 0.0000000 | 0.00e+00 |
| MAP3K11 | 1.46 | 0.0000000 | 0.00e+00 |
| MAP3K12 | -1.03 | 0.0039090 | 1.36e-03 |
| MAP3K13 | 0.85 | 0.0076021 | 2.83e-03 |
| MAP3K14 | 0.99 | 0.0000000 | 0.00e+00 |
| MAP3K15 | -1.25 | 0.0580068 | 2.74e-02 |
| MAP3K5 | 1.11 | 0.0030868 | 1.05e-03 |
| MAP3K7 | -0.78 | 0.0032518 | 1.11e-03 |
| MAP3K8 | 2.75 | 0.0000000 | 0.00e+00 |
| MAP4 | 0.82 | 0.0000000 | 0.00e+00 |
| MAP4K2 | -1.01 | 0.0153121 | 6.15e-03 |
| MAP4K4 | 2.79 | 0.0000000 | 0.00e+00 |
| MAP6D1 | -2.29 | 0.0450474 | 2.05e-02 |
| MAP7 | -0.49 | 0.0042589 | 1.49e-03 |
| MAP7D1 | 1.16 | 0.0000000 | 0.00e+00 |
| MAP7D2 | -1.35 | 0.0025238 | 8.45e-04 |
| MAP7D3 | -1.33 | 0.0004733 | 1.37e-04 |
| MAP9 | -2.21 | 0.0000088 | 2.00e-06 |
| MAPK1 | -0.33 | 0.0726720 | 3.56e-02 |
| MAPK12 | -0.79 | 0.0410755 | 1.85e-02 |
| MAPK3 | 0.99 | 0.0000001 | 0.00e+00 |
| MAPK6 | 0.37 | 0.0197231 | 8.13e-03 |
| MAPK7 | 1.09 | 0.0002524 | 6.96e-05 |
| MAPK8IP2 | 1.73 | 0.0003401 | 9.58e-05 |
| MAPK8IP3 | 1.26 | 0.0000000 | 0.00e+00 |
| MAPK9 | -0.67 | 0.0040163 | 1.40e-03 |
| MAPKAP1 | -1.49 | 0.0000000 | 0.00e+00 |
| MAPKAPK2 | 0.62 | 0.0000938 | 2.42e-05 |
| MAPKAPK3 | -1.39 | 0.0000011 | 2.00e-07 |
| MAPKAPK5-AS1 | -1.05 | 0.0017871 | 5.76e-04 |
| MAPKBP1 | 2.39 | 0.0000000 | 0.00e+00 |
| MAPRE1 | -0.48 | 0.0002837 | 7.88e-05 |
| MAPRE2 | -1.09 | 0.0000057 | 1.20e-06 |
| MAPRE3 | 1.82 | 0.0000000 | 0.00e+00 |
| MARC1 | -3.56 | 0.0000148 | 3.40e-06 |
| MARCH2 | 1.30 | 0.0000204 | 4.80e-06 |
| MARCH3 | 4.75 | 0.0000109 | 2.50e-06 |
| MARCH4 | 2.37 | 0.0000000 | 0.00e+00 |
| MARCH6 | -0.56 | 0.0027036 | 9.11e-04 |
| MARCH9 | 1.21 | 0.0023274 | 7.75e-04 |
| MARCKS | 1.84 | 0.0000000 | 0.00e+00 |
| MARCKSL1 | 0.90 | 0.0000000 | 0.00e+00 |
| MARK1 | 7.20 | 0.0042580 | 1.49e-03 |
| MARK2 | 2.32 | 0.0000000 | 0.00e+00 |
| MARK3 | 0.32 | 0.0928878 | 4.75e-02 |
| MARK4 | 1.57 | 0.0000001 | 0.00e+00 |
| MARS | 0.64 | 0.0000000 | 0.00e+00 |
| MARS2 | -1.36 | 0.0046821 | 1.66e-03 |
| MAST2 | 0.95 | 0.0000000 | 0.00e+00 |
| MAST4 | 3.03 | 0.0000000 | 0.00e+00 |
| MASTL | -0.73 | 0.0026177 | 8.79e-04 |
| MAT2A | -0.51 | 0.0000206 | 4.90e-06 |
| MATK | 7.51 | 0.0022179 | 7.34e-04 |
| MATN2 | -1.51 | 0.0000003 | 1.00e-07 |
| MATN3 | -1.04 | 0.0722667 | 3.53e-02 |
| MATR3 | -1.11 | 0.0000000 | 0.00e+00 |
| MAU2 | 1.10 | 0.0000006 | 1.00e-07 |
| MAVS | 0.35 | 0.0095713 | 3.65e-03 |
| MAX | 0.72 | 0.0004689 | 1.36e-04 |
| MB | -2.01 | 0.0040895 | 1.43e-03 |
| MB21D1 | 2.01 | 0.0000001 | 0.00e+00 |
| MB21D2 | 2.26 | 0.0000000 | 0.00e+00 |
| MBD1 | 1.10 | 0.0000000 | 0.00e+00 |
| MBD6 | 1.90 | 0.0000000 | 0.00e+00 |
| MBLAC2 | -1.55 | 0.0766122 | 3.78e-02 |
| MBNL1 | -0.46 | 0.0140241 | 5.58e-03 |
| MBP | 0.88 | 0.0000018 | 4.00e-07 |
| MBTD1 | -1.01 | 0.0208218 | 8.63e-03 |
| MBTPS1 | -0.33 | 0.0350935 | 1.55e-02 |
| MBTPS2 | -0.48 | 0.0281628 | 1.21e-02 |
| MCAM | 2.05 | 0.0000000 | 0.00e+00 |
| MCCC2 | -0.62 | 0.0014152 | 4.47e-04 |
| MCEE | -1.25 | 0.0878625 | 4.44e-02 |
| MCL1 | 1.84 | 0.0000000 | 0.00e+00 |
| MCM10 | -3.15 | 0.0000000 | 0.00e+00 |
| MCM2 | -2.47 | 0.0000000 | 0.00e+00 |
| MCM3 | -1.90 | 0.0000000 | 0.00e+00 |
| MCM3AP | 0.49 | 0.0039285 | 1.37e-03 |
| MCM4 | -2.84 | 0.0000000 | 0.00e+00 |
| MCM5 | -2.24 | 0.0000000 | 0.00e+00 |
| MCM6 | -3.56 | 0.0000000 | 0.00e+00 |
| MCM7 | -1.92 | 0.0000000 | 0.00e+00 |
| MCM8 | -2.07 | 0.0000000 | 0.00e+00 |
| MCMBP | -0.93 | 0.0000000 | 0.00e+00 |
| MCOLN1 | 0.65 | 0.0964040 | 4.95e-02 |
| MCTP1 | 1.56 | 0.0000146 | 3.40e-06 |
| MCTP2 | 5.90 | 0.0480835 | 2.21e-02 |
| MCTS1 | -0.91 | 0.0000233 | 5.60e-06 |
| MCUR1 | -0.74 | 0.0234111 | 9.86e-03 |
| MDFI | 3.03 | 0.0435621 | 1.98e-02 |
| MDFIC | -0.74 | 0.0269221 | 1.15e-02 |
| MDGA1 | 2.09 | 0.0000000 | 0.00e+00 |
| MDH1 | -1.23 | 0.0000000 | 0.00e+00 |
| MDK | 3.52 | 0.0000000 | 0.00e+00 |
| MDM1 | -1.20 | 0.0058749 | 2.13e-03 |
| MDM2 | 1.11 | 0.0000000 | 0.00e+00 |
| MDN1 | -1.11 | 0.0000080 | 1.80e-06 |
| MDP1 | -1.03 | 0.0335461 | 1.47e-02 |
| ME1 | -1.09 | 0.0000000 | 0.00e+00 |
| ME2 | -1.35 | 0.0000000 | 0.00e+00 |
| MEA1 | -0.48 | 0.0243347 | 1.03e-02 |
| MECP2 | 0.34 | 0.0906092 | 4.61e-02 |
| MECR | -0.91 | 0.0436659 | 1.98e-02 |
| MED11 | -0.64 | 0.0357910 | 1.58e-02 |
| MED12 | 1.38 | 0.0000000 | 0.00e+00 |
| MED13 | 0.51 | 0.0009274 | 2.83e-04 |
| MED15 | 2.63 | 0.0000000 | 0.00e+00 |
| MED16 | 1.31 | 0.0000277 | 6.70e-06 |
| MED19 | 0.74 | 0.0122013 | 4.79e-03 |
| MED20 | -1.29 | 0.0001500 | 4.01e-05 |
| MED21 | -0.74 | 0.0145494 | 5.81e-03 |
| MED23 | -0.83 | 0.0383604 | 1.71e-02 |
| MED24 | 0.80 | 0.0000016 | 3.00e-07 |
| MED25 | 1.08 | 0.0000000 | 0.00e+00 |
| MED26 | 0.75 | 0.0799928 | 3.98e-02 |
| MED29 | 0.54 | 0.0000824 | 2.11e-05 |
| MED31 | -0.77 | 0.0755804 | 3.72e-02 |
| MED4 | -0.60 | 0.0959602 | 4.92e-02 |
| MEF2A | 0.62 | 0.0187834 | 7.71e-03 |
| MEF2BNB-MEF2B | 0.82 | 0.0664974 | 3.20e-02 |
| MEF2D | 1.71 | 0.0000000 | 0.00e+00 |
| MEFV | 8.68 | 0.0000000 | 0.00e+00 |
| MEG3 | 0.93 | 0.0563581 | 2.65e-02 |
| MEGF11 | 2.44 | 0.0424725 | 1.92e-02 |
| MEGF8 | 0.77 | 0.0000002 | 0.00e+00 |
| MEGF9 | -0.95 | 0.0000026 | 6.00e-07 |
| MEIS1 | 1.53 | 0.0375189 | 1.67e-02 |
| MEIS3 | 1.23 | 0.0000000 | 0.00e+00 |
| MEIS3P1 | 1.09 | 0.0037875 | 1.31e-03 |
| MELK | -1.92 | 0.0000000 | 0.00e+00 |
| MEMO1 | -1.29 | 0.0000005 | 1.00e-07 |
| MEN1 | -0.42 | 0.0984634 | 5.07e-02 |
| MESDC1 | 2.51 | 0.0000000 | 0.00e+00 |
| MEST | -0.95 | 0.0001243 | 3.27e-05 |
| METAP1 | -0.99 | 0.0000028 | 6.00e-07 |
| METAP2 | -0.74 | 0.0000025 | 5.00e-07 |
| METRN | -2.24 | 0.0000000 | 0.00e+00 |
| METRNL | 3.56 | 0.0000000 | 0.00e+00 |
| METTL10 | -1.08 | 0.0122885 | 4.82e-03 |
| METTL13 | -0.40 | 0.0735269 | 3.61e-02 |
| METTL15 | -0.92 | 0.0329885 | 1.44e-02 |
| METTL16 | -0.35 | 0.0573031 | 2.70e-02 |
| METTL17 | -0.62 | 0.0083308 | 3.13e-03 |
| METTL3 | -0.91 | 0.0000020 | 4.00e-07 |
| METTL5 | -1.11 | 0.0000765 | 1.95e-05 |
| METTL7B | -1.91 | 0.0000401 | 9.90e-06 |
| METTL8 | -1.61 | 0.0001099 | 2.87e-05 |
| MEX3D | 0.97 | 0.0161708 | 6.53e-03 |
| MFAP1 | -0.47 | 0.0849193 | 4.27e-02 |
| MFF | -1.14 | 0.0000000 | 0.00e+00 |
| MFGE8 | 1.02 | 0.0000000 | 0.00e+00 |
| MFHAS1 | 1.51 | 0.0000000 | 0.00e+00 |
| MFI2 | 1.17 | 0.0000000 | 0.00e+00 |
| MFN1 | -0.87 | 0.0022260 | 7.37e-04 |
| MFSD10 | 0.54 | 0.0126911 | 5.00e-03 |
| MFSD11 | 0.64 | 0.0431345 | 1.95e-02 |
| MFSD12 | 2.43 | 0.0000000 | 0.00e+00 |
| MFSD2A | 1.99 | 0.0000002 | 0.00e+00 |
| MFSD5 | 0.44 | 0.0502082 | 2.32e-02 |
| MGA | -0.52 | 0.0649273 | 3.11e-02 |
| MGAT1 | 0.44 | 0.0082026 | 3.08e-03 |
| MGAT2 | 0.77 | 0.0000168 | 3.90e-06 |
| MGAT4B | 0.59 | 0.0000033 | 7.00e-07 |
| MGAT5 | 1.35 | 0.0000000 | 0.00e+00 |
| MGC27345 | -1.72 | 0.0105268 | 4.07e-03 |
| MGLL | 0.83 | 0.0000000 | 0.00e+00 |
| MGME1 | -0.96 | 0.0000532 | 1.33e-05 |
| MGRN1 | 1.99 | 0.0000000 | 0.00e+00 |
| MGST1 | -0.45 | 0.0002056 | 5.59e-05 |
| MGST3 | -1.07 | 0.0000000 | 0.00e+00 |
| MIA-RAB4B | 1.84 | 0.0000000 | 0.00e+00 |
| MIA3 | 0.77 | 0.0006523 | 1.93e-04 |
| MIAT | -1.95 | 0.0407850 | 1.83e-02 |
| MIB1 | -0.81 | 0.0059143 | 2.14e-03 |
| MICAL1 | 1.29 | 0.0000251 | 6.00e-06 |
| MICAL2 | 1.30 | 0.0000000 | 0.00e+00 |
| MICAL3 | 1.20 | 0.0000000 | 0.00e+00 |
| MICALCL | 4.09 | 0.0621421 | 2.96e-02 |
| MICALL1 | 1.54 | 0.0000000 | 0.00e+00 |
| MICALL2 | 1.08 | 0.0006216 | 1.83e-04 |
| MID1 | 0.56 | 0.0000034 | 7.00e-07 |
| MID1IP1 | 0.48 | 0.0034111 | 1.17e-03 |
| MID2 | 1.56 | 0.0000016 | 3.00e-07 |
| MIDN | 0.96 | 0.0000002 | 0.00e+00 |
| MIER2 | 1.29 | 0.0000204 | 4.80e-06 |
| MIF | -1.16 | 0.0000000 | 0.00e+00 |
| MIIP | 2.30 | 0.0000000 | 0.00e+00 |
| MILR1 | 1.60 | 0.0734316 | 3.60e-02 |
| MINA | -0.66 | 0.0057948 | 2.10e-03 |
| MINK1 | 0.98 | 0.0000000 | 0.00e+00 |
| MINOS1 | -0.66 | 0.0253639 | 1.08e-02 |
| MINOS1-NBL1 | 1.61 | 0.0000000 | 0.00e+00 |
| MIPEP | -1.43 | 0.0000595 | 1.50e-05 |
| MIR100HG | 2.08 | 0.0023296 | 7.76e-04 |
| MIR17HG | -2.24 | 0.0103318 | 3.98e-03 |
| MIR22HG | 1.16 | 0.0007177 | 2.14e-04 |
| MIRLET7BHG | -1.58 | 0.0277845 | 1.19e-02 |
| MIS18A | -0.81 | 0.0948345 | 4.86e-02 |
| MIS18BP1 | -2.03 | 0.0000000 | 0.00e+00 |
| MISP | 1.15 | 0.0000000 | 0.00e+00 |
| MITF | -1.17 | 0.0160934 | 6.50e-03 |
| MKI67 | -2.70 | 0.0000000 | 0.00e+00 |
| MKL1 | 2.14 | 0.0000000 | 0.00e+00 |
| MKL2 | -0.56 | 0.0308859 | 1.34e-02 |
| MKLN1 | 0.46 | 0.0055054 | 1.98e-03 |
| MKLN1-AS | 1.66 | 0.0012382 | 3.87e-04 |
| MKNK1 | 0.81 | 0.0097773 | 3.74e-03 |
| MKS1 | -1.26 | 0.0050162 | 1.79e-03 |
| MLEC | -0.87 | 0.0000000 | 0.00e+00 |
| MLF2 | 0.24 | 0.0457285 | 2.09e-02 |
| MLH1 | -0.98 | 0.0000005 | 1.00e-07 |
| MLH3 | -1.11 | 0.0002381 | 6.54e-05 |
| MLKL | 2.16 | 0.0000000 | 0.00e+00 |
| MLLT1 | 0.82 | 0.0000422 | 1.04e-05 |
| MLLT4 | 0.79 | 0.0000260 | 6.20e-06 |
| MLLT6 | 1.57 | 0.0000000 | 0.00e+00 |
| MLPH | -0.46 | 0.0007385 | 2.21e-04 |
| MLST8 | -0.91 | 0.0003374 | 9.49e-05 |
| MLX | 0.61 | 0.0001153 | 3.02e-05 |
| MLXIP | 0.79 | 0.0000077 | 1.70e-06 |
| MMAB | -0.47 | 0.0794412 | 3.95e-02 |
| MMADHC | -0.77 | 0.0000067 | 1.50e-06 |
| MMGT1 | -0.88 | 0.0038996 | 1.36e-03 |
| MMP1 | 12.06 | 0.0000000 | 0.00e+00 |
| MMP10 | 11.22 | 0.0000003 | 0.00e+00 |
| MMP12 | 7.35 | 0.0031187 | 1.06e-03 |
| MMP13 | 5.83 | 0.0515073 | 2.39e-02 |
| MMP19 | 8.03 | 0.0007340 | 2.19e-04 |
| MMP24 | -1.25 | 0.0000000 | 0.00e+00 |
| MMP25-AS1 | 2.49 | 0.0004975 | 1.45e-04 |
| MMP3 | 5.90 | 0.0480835 | 2.21e-02 |
| MMP7 | 2.65 | 0.0000000 | 0.00e+00 |
| MMP9 | 3.32 | 0.0169611 | 6.88e-03 |
| MMS19 | -0.74 | 0.0002926 | 8.15e-05 |
| MMS22L | -2.74 | 0.0000000 | 0.00e+00 |
| MN1 | 1.69 | 0.0000865 | 2.22e-05 |
| MND1 | -2.33 | 0.0021264 | 7.00e-04 |
| MNS1 | -3.57 | 0.0000901 | 2.32e-05 |
| MNT | 1.88 | 0.0000000 | 0.00e+00 |
| MNX1-AS1 | -2.51 | 0.0026511 | 8.92e-04 |
| MOB2 | 0.71 | 0.0021111 | 6.95e-04 |
| MOB3A | 1.43 | 0.0000028 | 6.00e-07 |
| MOB3B | 1.94 | 0.0002206 | 6.04e-05 |
| MOB3C | 2.74 | 0.0000000 | 0.00e+00 |
| MOB4 | -0.54 | 0.0423108 | 1.91e-02 |
| MOCS2 | -1.53 | 0.0000159 | 3.70e-06 |
| MOGS | 0.82 | 0.0000011 | 2.00e-07 |
| MON1B | 0.99 | 0.0000001 | 0.00e+00 |
| MORC2 | 0.58 | 0.0145362 | 5.80e-03 |
| MORC3 | 1.91 | 0.0000000 | 0.00e+00 |
| MORC4 | -1.57 | 0.0000000 | 0.00e+00 |
| MORF4L1 | -1.14 | 0.0000000 | 0.00e+00 |
| MORF4L2 | -0.24 | 0.0296149 | 1.28e-02 |
| MORN2 | -1.01 | 0.0705188 | 3.43e-02 |
| MOSPD2 | -0.69 | 0.0886793 | 4.49e-02 |
| MOV10 | 1.80 | 0.0000000 | 0.00e+00 |
| MPC2 | -0.47 | 0.0135375 | 5.37e-03 |
| MPDU1 | -0.41 | 0.0163764 | 6.62e-03 |
| MPDZ | -0.53 | 0.0544580 | 2.54e-02 |
| MPHOSPH10 | -0.92 | 0.0002923 | 8.14e-05 |
| MPHOSPH6 | -1.11 | 0.0139878 | 5.57e-03 |
| MPHOSPH9 | -2.21 | 0.0000000 | 0.00e+00 |
| MPP1 | 0.88 | 0.0269341 | 1.15e-02 |
| MPP2 | -2.94 | 0.0000000 | 0.00e+00 |
| MPP3 | -1.36 | 0.0017045 | 5.47e-04 |
| MPP6 | -1.21 | 0.0017431 | 5.61e-04 |
| MPPE1 | 1.43 | 0.0000018 | 4.00e-07 |
| MPRIP | -0.57 | 0.0000000 | 0.00e+00 |
| MPZL1 | 0.73 | 0.0000000 | 0.00e+00 |
| MR1 | 1.85 | 0.0000000 | 0.00e+00 |
| MRC2 | 0.57 | 0.0042471 | 1.49e-03 |
| MRE11A | -1.99 | 0.0000000 | 0.00e+00 |
| MREG | 2.07 | 0.0000000 | 0.00e+00 |
| MRFAP1 | -0.92 | 0.0000000 | 0.00e+00 |
| MRFAP1L1 | -0.97 | 0.0006797 | 2.02e-04 |
| MRGPRX3 | 9.08 | 0.0000632 | 1.60e-05 |
| MRI1 | -0.74 | 0.0719698 | 3.52e-02 |
| MROH1 | 1.42 | 0.0000129 | 3.00e-06 |
| MRPL1 | -1.39 | 0.0008110 | 2.45e-04 |
| MRPL10 | 0.42 | 0.0182959 | 7.48e-03 |
| MRPL11 | -1.44 | 0.0000000 | 0.00e+00 |
| MRPL12 | -1.42 | 0.0000000 | 0.00e+00 |
| MRPL13 | -1.20 | 0.0000006 | 1.00e-07 |
| MRPL15 | -0.98 | 0.0000005 | 1.00e-07 |
| MRPL17 | -0.49 | 0.0036896 | 1.28e-03 |
| MRPL18 | -0.95 | 0.0000001 | 0.00e+00 |
| MRPL19 | -1.49 | 0.0000000 | 0.00e+00 |
| MRPL20 | -0.85 | 0.0001184 | 3.11e-05 |
| MRPL21 | -0.92 | 0.0000107 | 2.40e-06 |
| MRPL23 | -0.69 | 0.0236026 | 9.94e-03 |
| MRPL24 | -1.83 | 0.0000000 | 0.00e+00 |
| MRPL27 | -0.68 | 0.0006475 | 1.92e-04 |
| MRPL3 | -1.72 | 0.0000000 | 0.00e+00 |
| MRPL30 | -1.57 | 0.0000000 | 0.00e+00 |
| MRPL33 | -0.50 | 0.0030575 | 1.04e-03 |
| MRPL34 | -0.63 | 0.0615557 | 2.93e-02 |
| MRPL35 | -1.35 | 0.0000000 | 0.00e+00 |
| MRPL36 | -0.86 | 0.0020322 | 6.66e-04 |
| MRPL37 | -0.90 | 0.0000014 | 3.00e-07 |
| MRPL39 | -0.95 | 0.0102129 | 3.93e-03 |
| MRPL40 | -0.95 | 0.0015636 | 4.98e-04 |
| MRPL41 | -0.51 | 0.0106838 | 4.13e-03 |
| MRPL42 | -1.11 | 0.0000291 | 7.00e-06 |
| MRPL43 | -0.68 | 0.0012118 | 3.78e-04 |
| MRPL45 | -0.62 | 0.0276113 | 1.18e-02 |
| MRPL46 | 0.53 | 0.0077328 | 2.89e-03 |
| MRPL47 | -0.82 | 0.0109174 | 4.24e-03 |
| MRPL48 | -1.15 | 0.0006616 | 1.96e-04 |
| MRPL49 | 0.31 | 0.0678502 | 3.28e-02 |
| MRPL50 | -0.75 | 0.0120818 | 4.73e-03 |
| MRPL51 | -1.40 | 0.0000000 | 0.00e+00 |
| MRPL52 | -0.53 | 0.0241234 | 1.02e-02 |
| MRPL53 | -0.38 | 0.0798648 | 3.97e-02 |
| MRPL55 | -0.71 | 0.0281541 | 1.21e-02 |
| MRPL57 | -0.89 | 0.0006681 | 1.98e-04 |
| MRPS12 | -0.99 | 0.0000000 | 0.00e+00 |
| MRPS15 | -0.49 | 0.0220640 | 9.20e-03 |
| MRPS16 | -1.21 | 0.0000000 | 0.00e+00 |
| MRPS17 | -1.10 | 0.0015158 | 4.82e-04 |
| MRPS18A | 0.70 | 0.0011001 | 3.41e-04 |
| MRPS21 | -0.87 | 0.0000213 | 5.10e-06 |
| MRPS22 | -0.71 | 0.0109297 | 4.24e-03 |
| MRPS23 | -0.90 | 0.0000002 | 0.00e+00 |
| MRPS24 | -0.41 | 0.0199718 | 8.25e-03 |
| MRPS25 | -1.49 | 0.0000000 | 0.00e+00 |
| MRPS26 | -0.76 | 0.0000730 | 1.86e-05 |
| MRPS27 | -0.51 | 0.0018289 | 5.92e-04 |
| MRPS28 | -0.89 | 0.0051488 | 1.84e-03 |
| MRPS33 | -0.71 | 0.0066408 | 2.44e-03 |
| MRPS34 | -1.97 | 0.0000000 | 0.00e+00 |
| MRPS35 | -0.77 | 0.0000047 | 1.00e-06 |
| MRPS5 | -0.99 | 0.0000001 | 0.00e+00 |
| MRPS6 | -0.62 | 0.0917237 | 4.68e-02 |
| MRPS7 | -0.75 | 0.0000010 | 2.00e-07 |
| MRPS9 | -0.87 | 0.0010906 | 3.37e-04 |
| MRRF | -0.65 | 0.0068515 | 2.52e-03 |
| MRS2 | -0.69 | 0.0304007 | 1.32e-02 |
| MRTO4 | -1.58 | 0.0000000 | 0.00e+00 |
| MSANTD3 | 0.81 | 0.0000378 | 9.30e-06 |
| MSANTD3-TMEFF1 | 1.06 | 0.0003963 | 1.13e-04 |
| MSC | 3.53 | 0.0000000 | 0.00e+00 |
| MSC-AS1 | 1.53 | 0.0089037 | 3.38e-03 |
| MSH2 | -2.21 | 0.0000000 | 0.00e+00 |
| MSH3 | -0.68 | 0.0621167 | 2.96e-02 |
| MSH6 | -1.87 | 0.0000000 | 0.00e+00 |
| MSI2 | -0.89 | 0.0039598 | 1.38e-03 |
| MSL3 | -1.18 | 0.0002286 | 6.27e-05 |
| MSMB | 7.20 | 0.0042760 | 1.50e-03 |
| MSMO1 | 1.93 | 0.0000000 | 0.00e+00 |
| MSN | 1.55 | 0.0000000 | 0.00e+00 |
| MSRB1 | 0.40 | 0.0799209 | 3.98e-02 |
| MSRB3 | -1.33 | 0.0000000 | 0.00e+00 |
| MST1R | 2.07 | 0.0000000 | 0.00e+00 |
| MSTO1 | -0.59 | 0.0450459 | 2.05e-02 |
| MSTO2P | -0.48 | 0.0523357 | 2.43e-02 |
| MSX1 | 1.36 | 0.0015136 | 4.81e-04 |
| MSX2 | 1.56 | 0.0001494 | 3.99e-05 |
| MT1F | 2.84 | 0.0002050 | 5.57e-05 |
| MT1G | 4.46 | 0.0000721 | 1.83e-05 |
| MT1X | 1.72 | 0.0001016 | 2.64e-05 |
| MT2A | 3.21 | 0.0000000 | 0.00e+00 |
| MTA2 | 0.30 | 0.0398669 | 1.79e-02 |
| MTBP | -2.27 | 0.0006501 | 1.92e-04 |
| MTCH2 | -1.46 | 0.0000000 | 0.00e+00 |
| MTDH | -0.74 | 0.0000017 | 3.00e-07 |
| MTERF1 | -1.15 | 0.0223434 | 9.34e-03 |
| MTERF2 | -1.56 | 0.0207652 | 8.61e-03 |
| MTERF3 | -1.16 | 0.0019935 | 6.52e-04 |
| MTERF4 | -0.75 | 0.0246890 | 1.05e-02 |
| MTF1 | 0.95 | 0.0014309 | 4.53e-04 |
| MTFMT | -0.90 | 0.0059484 | 2.16e-03 |
| MTFR2 | -2.32 | 0.0000835 | 2.14e-05 |
| MTHFD1 | -1.29 | 0.0000000 | 0.00e+00 |
| MTHFD2 | 0.52 | 0.0000058 | 1.30e-06 |
| MTHFR | 1.64 | 0.0000000 | 0.00e+00 |
| MTHFS | 0.54 | 0.0074973 | 2.79e-03 |
| MTIF2 | -1.24 | 0.0000000 | 0.00e+00 |
| MTMR11 | 1.29 | 0.0000000 | 0.00e+00 |
| MTMR14 | 0.97 | 0.0000001 | 0.00e+00 |
| MTMR2 | -0.91 | 0.0021921 | 7.24e-04 |
| MTMR3 | 1.31 | 0.0000000 | 0.00e+00 |
| MTMR4 | 1.09 | 0.0000000 | 0.00e+00 |
| MTMR6 | -0.60 | 0.0496483 | 2.29e-02 |
| MTO1 | -1.28 | 0.0036706 | 1.27e-03 |
| MTPN | -0.49 | 0.0003449 | 9.73e-05 |
| MTR | -0.81 | 0.0026287 | 8.84e-04 |
| MTRNR2L2 | -0.47 | 0.0917237 | 4.68e-02 |
| MTRNR2L8 | -0.70 | 0.0000000 | 0.00e+00 |
| MTRNR2L9 | -0.69 | 0.0001843 | 4.98e-05 |
| MTRR | -0.63 | 0.0079303 | 2.97e-03 |
| MTSS1 | 4.20 | 0.0000000 | 0.00e+00 |
| MTSS1L | -0.89 | 0.0000756 | 1.92e-05 |
| MTUS1 | -1.40 | 0.0000000 | 0.00e+00 |
| MTX2 | -0.61 | 0.0116992 | 4.57e-03 |
| MTX3 | -1.37 | 0.0005396 | 1.57e-04 |
| MUC1 | 7.39 | 0.0000000 | 0.00e+00 |
| MUC13 | 2.42 | 0.0000000 | 0.00e+00 |
| MUC16 | 4.01 | 0.0000000 | 0.00e+00 |
| MUC17 | 6.25 | 0.0258620 | 1.10e-02 |
| MUC2 | 7.55 | 0.0000020 | 4.00e-07 |
| MUC3A | 2.78 | 0.0000000 | 0.00e+00 |
| MUC5AC | 0.79 | 0.0000000 | 0.00e+00 |
| MUC5B | 3.21 | 0.0000000 | 0.00e+00 |
| MUL1 | 0.66 | 0.0022050 | 7.29e-04 |
| MUM1 | -0.67 | 0.0247667 | 1.05e-02 |
| MUS81 | 0.94 | 0.0000308 | 7.40e-06 |
| MVD | 1.79 | 0.0000000 | 0.00e+00 |
| MVP | 1.29 | 0.0000000 | 0.00e+00 |
| MX1 | 9.24 | 0.0000000 | 0.00e+00 |
| MX2 | 10.91 | 0.0000006 | 1.00e-07 |
| MXD1 | 5.41 | 0.0000000 | 0.00e+00 |
| MXD3 | -2.68 | 0.0000001 | 0.00e+00 |
| MXD4 | 0.96 | 0.0001770 | 4.78e-05 |
| MXI1 | 0.76 | 0.0367529 | 1.63e-02 |
| MYADM | 2.18 | 0.0000000 | 0.00e+00 |
| MYB | 2.43 | 0.0666908 | 3.21e-02 |
| MYBBP1A | 0.42 | 0.0077255 | 2.88e-03 |
| MYBL1 | -2.68 | 0.0000086 | 1.90e-06 |
| MYBL2 | -2.16 | 0.0000000 | 0.00e+00 |
| MYC | 0.32 | 0.0305605 | 1.33e-02 |
| MYCBP | -1.54 | 0.0001191 | 3.13e-05 |
| MYCN | 5.97 | 0.0423108 | 1.91e-02 |
| MYD88 | 2.37 | 0.0000000 | 0.00e+00 |
| MYDGF | 1.17 | 0.0000000 | 0.00e+00 |
| MYEF2 | -0.70 | 0.0201889 | 8.34e-03 |
| MYEOV | 1.08 | 0.0000000 | 0.00e+00 |
| MYEOV2 | -0.50 | 0.0795802 | 3.95e-02 |
| MYH10 | -1.59 | 0.0000000 | 0.00e+00 |
| MYH11 | 4.29 | 0.0000000 | 0.00e+00 |
| MYH14 | 1.14 | 0.0000005 | 1.00e-07 |
| MYH16 | 4.32 | 0.0397683 | 1.78e-02 |
| MYH9 | 2.02 | 0.0000000 | 0.00e+00 |
| MYL12A | 0.90 | 0.0000000 | 0.00e+00 |
| MYL12B | 0.25 | 0.0643462 | 3.08e-02 |
| MYL5 | 1.62 | 0.0018143 | 5.86e-04 |
| MYL9 | 2.25 | 0.0097700 | 3.74e-03 |
| MYLK | -1.92 | 0.0000000 | 0.00e+00 |
| MYO10 | 0.61 | 0.0000000 | 0.00e+00 |
| MYO15B | 0.40 | 0.0152402 | 6.12e-03 |
| MYO19 | -0.50 | 0.0161043 | 6.50e-03 |
| MYO1B | -0.41 | 0.0002084 | 5.67e-05 |
| MYO1C | 0.55 | 0.0000000 | 0.00e+00 |
| MYO1D | 1.10 | 0.0071392 | 2.64e-03 |
| MYO1E | 1.11 | 0.0000000 | 0.00e+00 |
| MYO1G | 5.17 | 0.0055880 | 2.01e-03 |
| MYO5A | -0.84 | 0.0000398 | 9.80e-06 |
| MYO5B | 0.96 | 0.0018090 | 5.85e-04 |
| MYO5C | -1.83 | 0.0000000 | 0.00e+00 |
| MYO6 | 1.03 | 0.0000050 | 1.10e-06 |
| MYO9A | -1.42 | 0.0000305 | 7.40e-06 |
| MYO9B | 1.43 | 0.0000000 | 0.00e+00 |
| MYOCD | 3.39 | 0.0000000 | 0.00e+00 |
| MYOF | -0.47 | 0.0000007 | 1.00e-07 |
| MYPOP | 1.12 | 0.0084056 | 3.17e-03 |
| MYRIP | 6.03 | 0.0377916 | 1.68e-02 |
| MZF1 | 0.61 | 0.0616471 | 2.94e-02 |
| MZT1 | -1.65 | 0.0006173 | 1.82e-04 |
| MZT2A | -0.69 | 0.0022380 | 7.42e-04 |
| MZT2B | -0.59 | 0.0004357 | 1.26e-04 |
| N4BP1 | 2.77 | 0.0000000 | 0.00e+00 |
| N4BP2 | -1.25 | 0.0038300 | 1.33e-03 |
| N4BP2L1 | 4.52 | 0.0010847 | 3.35e-04 |
| N4BP3 | 4.43 | 0.0000000 | 0.00e+00 |
| NAA15 | -1.03 | 0.0000002 | 0.00e+00 |
| NAA20 | -0.66 | 0.0001007 | 2.61e-05 |
| NAA25 | -1.03 | 0.0000013 | 3.00e-07 |
| NAA38 | -1.65 | 0.0000000 | 0.00e+00 |
| NAA40 | -0.56 | 0.0520406 | 2.42e-02 |
| NAA50 | -0.70 | 0.0000069 | 1.50e-06 |
| NAA60 | 1.26 | 0.0000000 | 0.00e+00 |
| NAAA | -0.93 | 0.0304320 | 1.32e-02 |
| NAB1 | 0.66 | 0.0010216 | 3.15e-04 |
| NAB2 | 2.57 | 0.0000000 | 0.00e+00 |
| NABP1 | 0.82 | 0.0226999 | 9.51e-03 |
| NABP2 | -1.20 | 0.0000000 | 0.00e+00 |
| NACA | -0.26 | 0.0147069 | 5.88e-03 |
| NACC1 | 0.48 | 0.0013233 | 4.16e-04 |
| NADK2 | -0.93 | 0.0195082 | 8.03e-03 |
| NAE1 | -1.85 | 0.0000000 | 0.00e+00 |
| NAGLU | 0.71 | 0.0002603 | 7.19e-05 |
| NALCN | -2.55 | 0.0012287 | 3.84e-04 |
| NAMPT | 0.43 | 0.0000202 | 4.80e-06 |
| NANP | -0.67 | 0.0691518 | 3.35e-02 |
| NANS | -0.82 | 0.0002151 | 5.87e-05 |
| NAP1L1 | -0.25 | 0.0257980 | 1.10e-02 |
| NAP1L2 | -3.29 | 0.0000371 | 9.10e-06 |
| NAP1L4 | -0.85 | 0.0000132 | 3.00e-06 |
| NAPA | 0.96 | 0.0000000 | 0.00e+00 |
| NARF | -0.47 | 0.0682915 | 3.30e-02 |
| NARR | 0.49 | 0.0414681 | 1.87e-02 |
| NARS | -0.95 | 0.0000000 | 0.00e+00 |
| NARS2 | -1.49 | 0.0000019 | 4.00e-07 |
| NASP | -1.41 | 0.0000000 | 0.00e+00 |
| NAT9 | 0.42 | 0.0820379 | 4.10e-02 |
| NATD1 | 4.34 | 0.0000000 | 0.00e+00 |
| NAV1 | 1.85 | 0.0000000 | 0.00e+00 |
| NAV2 | 3.54 | 0.0000000 | 0.00e+00 |
| NAV3 | 3.58 | 0.0000000 | 0.00e+00 |
| NBEA | -1.03 | 0.0164077 | 6.64e-03 |
| NBL1 | 1.64 | 0.0000000 | 0.00e+00 |
| NBN | 0.36 | 0.0278431 | 1.19e-02 |
| NBR1 | 1.37 | 0.0000000 | 0.00e+00 |
| NCAPD2 | -1.96 | 0.0000000 | 0.00e+00 |
| NCAPD3 | -2.63 | 0.0000000 | 0.00e+00 |
| NCAPG | -3.46 | 0.0000000 | 0.00e+00 |
| NCAPG2 | -2.22 | 0.0000000 | 0.00e+00 |
| NCAPH | -2.67 | 0.0000000 | 0.00e+00 |
| NCBP1 | -1.86 | 0.0000000 | 0.00e+00 |
| NCBP2 | -0.48 | 0.0455565 | 2.08e-02 |
| NCCRP1 | 5.68 | 0.0000000 | 0.00e+00 |
| NCEH1 | 1.21 | 0.0000000 | 0.00e+00 |
| NCF2 | 7.83 | 0.0000000 | 0.00e+00 |
| NCF4 | 6.61 | 0.0133613 | 5.29e-03 |
| NCK1 | 1.35 | 0.0000002 | 0.00e+00 |
| NCK2 | 0.55 | 0.0540534 | 2.52e-02 |
| NCKAP1 | -1.03 | 0.0000000 | 0.00e+00 |
| NCKAP5L | 1.44 | 0.0000003 | 1.00e-07 |
| NCKIPSD | 0.79 | 0.0303300 | 1.31e-02 |
| NCL | -1.34 | 0.0000000 | 0.00e+00 |
| NCOA1 | 0.75 | 0.0003756 | 1.07e-04 |
| NCOA4 | 0.88 | 0.0000000 | 0.00e+00 |
| NCOA5 | -0.52 | 0.0651149 | 3.12e-02 |
| NCOA6 | 1.00 | 0.0000010 | 2.00e-07 |
| NCOA7 | 1.45 | 0.0000000 | 0.00e+00 |
| NCOR1 | 0.37 | 0.0029787 | 1.01e-03 |
| NCOR2 | 0.80 | 0.0000000 | 0.00e+00 |
| NCR3LG1 | 1.71 | 0.0000001 | 0.00e+00 |
| NCS1 | -1.21 | 0.0000009 | 2.00e-07 |
| NCSTN | 1.45 | 0.0000000 | 0.00e+00 |
| NDC1 | -2.21 | 0.0000000 | 0.00e+00 |
| NDC80 | -2.76 | 0.0000000 | 0.00e+00 |
| NDE1 | 1.66 | 0.0000000 | 0.00e+00 |
| NDEL1 | 0.92 | 0.0000072 | 1.60e-06 |
| NDNF | 5.53 | 0.0835040 | 4.19e-02 |
| NDOR1 | 0.54 | 0.0355473 | 1.57e-02 |
| NDRG1 | 1.39 | 0.0000001 | 0.00e+00 |
| NDRG2 | 1.83 | 0.0000029 | 6.00e-07 |
| NDRG3 | -0.84 | 0.0021962 | 7.26e-04 |
| NDRG4 | 0.58 | 0.0714886 | 3.49e-02 |
| NDST1 | 1.20 | 0.0000000 | 0.00e+00 |
| NDUFA1 | -0.62 | 0.0000836 | 2.14e-05 |
| NDUFA10 | -0.84 | 0.0000020 | 4.00e-07 |
| NDUFA11 | -0.37 | 0.0878814 | 4.44e-02 |
| NDUFA12 | -0.55 | 0.0271651 | 1.16e-02 |
| NDUFA2 | -1.00 | 0.0000031 | 7.00e-07 |
| NDUFA4 | -0.95 | 0.0185731 | 7.61e-03 |
| NDUFA5 | -1.08 | 0.0029416 | 9.96e-04 |
| NDUFA8 | -0.53 | 0.0106758 | 4.13e-03 |
| NDUFA9 | -1.17 | 0.0000000 | 0.00e+00 |
| NDUFAB1 | -1.02 | 0.0000011 | 2.00e-07 |
| NDUFAF2 | -0.70 | 0.0261198 | 1.11e-02 |
| NDUFAF3 | -0.55 | 0.0512041 | 2.37e-02 |
| NDUFAF7 | -0.67 | 0.0552215 | 2.59e-02 |
| NDUFB1 | -0.62 | 0.0169080 | 6.85e-03 |
| NDUFB10 | -0.80 | 0.0000078 | 1.70e-06 |
| NDUFB2 | -1.25 | 0.0000000 | 0.00e+00 |
| NDUFB3 | -1.46 | 0.0000000 | 0.00e+00 |
| NDUFB4 | -0.41 | 0.0158364 | 6.39e-03 |
| NDUFB5 | -0.69 | 0.0150219 | 6.02e-03 |
| NDUFB6 | -1.23 | 0.0000005 | 1.00e-07 |
| NDUFB7 | -0.66 | 0.0058836 | 2.13e-03 |
| NDUFB8 | -0.81 | 0.0000000 | 0.00e+00 |
| NDUFB9 | -0.53 | 0.0003141 | 8.79e-05 |
| NDUFC2 | -0.96 | 0.0000018 | 4.00e-07 |
| NDUFC2-KCTD14 | -0.82 | 0.0001859 | 5.03e-05 |
| NDUFS1 | -0.52 | 0.0013375 | 4.20e-04 |
| NDUFS3 | -0.94 | 0.0000190 | 4.50e-06 |
| NDUFS5 | -0.34 | 0.0977381 | 5.03e-02 |
| NDUFS6 | -0.90 | 0.0000135 | 3.10e-06 |
| NDUFS8 | -0.54 | 0.0020662 | 6.79e-04 |
| NDUFV1 | -1.08 | 0.0000000 | 0.00e+00 |
| NDUFV2 | 1.11 | 0.0000000 | 0.00e+00 |
| NEAT1 | -0.66 | 0.0000000 | 0.00e+00 |
| NEB | -2.34 | 0.0437211 | 1.99e-02 |
| NEBL | -0.91 | 0.0131692 | 5.21e-03 |
| NECAP1 | 0.48 | 0.0910166 | 4.63e-02 |
| NECAP2 | 1.46 | 0.0000000 | 0.00e+00 |
| NEDD4L | 1.13 | 0.0000000 | 0.00e+00 |
| NEDD8 | -0.50 | 0.0019778 | 6.46e-04 |
| NEDD9 | 0.81 | 0.0000000 | 0.00e+00 |
| NEFL | -1.58 | 0.0199718 | 8.25e-03 |
| NEIL2 | -0.79 | 0.0302679 | 1.31e-02 |
| NEIL3 | -4.54 | 0.0000000 | 0.00e+00 |
| NEK1 | -1.23 | 0.0858619 | 4.33e-02 |
| NEK2 | -3.56 | 0.0000000 | 0.00e+00 |
| NELFA | 0.87 | 0.0005104 | 1.49e-04 |
| NELFCD | -0.59 | 0.0048836 | 1.74e-03 |
| NEMF | -1.32 | 0.0000002 | 0.00e+00 |
| NEMP1 | -0.37 | 0.0931764 | 4.76e-02 |
| NEMP2 | -1.07 | 0.0566746 | 2.67e-02 |
| NEO1 | 0.98 | 0.0000000 | 0.00e+00 |
| NET1 | 0.30 | 0.0387702 | 1.73e-02 |
| NETO2 | -1.32 | 0.0000000 | 0.00e+00 |
| NEURL1B | -2.84 | 0.0000492 | 1.22e-05 |
| NEURL3 | 7.07 | 0.0000000 | 0.00e+00 |
| NF2 | -1.17 | 0.0000242 | 5.80e-06 |
| NFAM1 | 5.61 | 0.0717731 | 3.50e-02 |
| NFAT5 | 0.99 | 0.0000000 | 0.00e+00 |
| NFATC1 | 2.34 | 0.0000000 | 0.00e+00 |
| NFATC2 | 1.49 | 0.0090112 | 3.42e-03 |
| NFATC2IP | 0.44 | 0.0284614 | 1.22e-02 |
| NFATC4 | 3.75 | 0.0000164 | 3.80e-06 |
| NFE2L1 | 1.30 | 0.0000000 | 0.00e+00 |
| NFE2L2 | 0.63 | 0.0000025 | 5.00e-07 |
| NFE2L3 | 3.32 | 0.0000000 | 0.00e+00 |
| NFIA | -1.30 | 0.0009165 | 2.79e-04 |
| NFIC | 0.75 | 0.0000067 | 1.50e-06 |
| NFIL3 | 2.04 | 0.0000000 | 0.00e+00 |
| NFIX | 1.42 | 0.0025953 | 8.71e-04 |
| NFKB1 | 3.01 | 0.0000000 | 0.00e+00 |
| NFKB2 | 3.90 | 0.0000000 | 0.00e+00 |
| NFKBIA | 4.23 | 0.0000000 | 0.00e+00 |
| NFKBIB | 1.69 | 0.0000000 | 0.00e+00 |
| NFKBID | 1.34 | 0.0207127 | 8.58e-03 |
| NFKBIE | 3.57 | 0.0000000 | 0.00e+00 |
| NFKBIZ | 5.68 | 0.0000000 | 0.00e+00 |
| NFS1 | -0.79 | 0.0026344 | 8.86e-04 |
| NFU1 | -0.51 | 0.0567366 | 2.67e-02 |
| NFX1 | 0.51 | 0.0063607 | 2.33e-03 |
| NFYA | -0.57 | 0.0814192 | 4.07e-02 |
| NFYB | -1.06 | 0.0013699 | 4.32e-04 |
| NGEF | 0.88 | 0.0104188 | 4.02e-03 |
| NGFR | 6.16 | 0.0000000 | 0.00e+00 |
| NGFRAP1 | -0.59 | 0.0008955 | 2.72e-04 |
| NGRN | -0.69 | 0.0000000 | 0.00e+00 |
| NHEJ1 | -0.89 | 0.0326700 | 1.43e-02 |
| NHLRC2 | -0.59 | 0.0551168 | 2.58e-02 |
| NHP2 | -1.49 | 0.0000000 | 0.00e+00 |
| NHS | 1.09 | 0.0044971 | 1.59e-03 |
| NHSL1 | 1.19 | 0.0211366 | 8.78e-03 |
| NID1 | 1.47 | 0.0003598 | 1.02e-04 |
| NID2 | 2.31 | 0.0000000 | 0.00e+00 |
| NIF3L1 | -1.80 | 0.0000000 | 0.00e+00 |
| NIFK | -1.04 | 0.0000160 | 3.70e-06 |
| NIM1K | 6.90 | 0.0076582 | 2.86e-03 |
| NINJ1 | 1.94 | 0.0000000 | 0.00e+00 |
| NIP7 | -0.87 | 0.0000620 | 1.56e-05 |
| NIPA1 | 0.95 | 0.0000000 | 0.00e+00 |
| NIPAL4 | 3.49 | 0.0000000 | 0.00e+00 |
| NIPSNAP1 | -0.97 | 0.0000000 | 0.00e+00 |
| NIPSNAP3A | -1.16 | 0.0137612 | 5.47e-03 |
| NIT1 | 0.85 | 0.0000994 | 2.58e-05 |
| NIT2 | -0.92 | 0.0000157 | 3.70e-06 |
| NKIRAS2 | 1.18 | 0.0000000 | 0.00e+00 |
| NKTR | -0.89 | 0.0027144 | 9.15e-04 |
| NKX2-8 | 2.82 | 0.0002636 | 7.29e-05 |
| NKX3-1 | 4.39 | 0.0000000 | 0.00e+00 |
| NLE1 | -1.12 | 0.0000124 | 2.90e-06 |
| NLGN3 | 2.35 | 0.0031165 | 1.06e-03 |
| NLGN4Y | 1.83 | 0.0229942 | 9.65e-03 |
| NLN | -1.78 | 0.0000000 | 0.00e+00 |
| NLRC5 | 5.02 | 0.0000000 | 0.00e+00 |
| NLRP3 | 6.25 | 0.0259984 | 1.11e-02 |
| NMD3 | -0.71 | 0.0012328 | 3.85e-04 |
| NME1 | -2.27 | 0.0000000 | 0.00e+00 |
| NME1-NME2 | -1.01 | 0.0000000 | 0.00e+00 |
| NME2 | -0.80 | 0.0000000 | 0.00e+00 |
| NME4 | -2.09 | 0.0000000 | 0.00e+00 |
| NME7 | -1.22 | 0.0059361 | 2.15e-03 |
| NMI | 2.56 | 0.0000000 | 0.00e+00 |
| NMNAT2 | 1.77 | 0.0208705 | 8.65e-03 |
| NMT2 | -0.96 | 0.0245481 | 1.04e-02 |
| NNMT | 0.98 | 0.0000000 | 0.00e+00 |
| NOC2L | -0.74 | 0.0000010 | 2.00e-07 |
| NOCT | 4.58 | 0.0000000 | 0.00e+00 |
| NOD2 | 9.80 | 0.0000107 | 2.40e-06 |
| NOL10 | -1.19 | 0.0000000 | 0.00e+00 |
| NOL11 | -1.17 | 0.0000000 | 0.00e+00 |
| NOL12 | -1.18 | 0.0005240 | 1.53e-04 |
| NOL3 | -1.15 | 0.0001963 | 5.32e-05 |
| NOL4L | 1.85 | 0.0000000 | 0.00e+00 |
| NOL6 | 0.30 | 0.0837531 | 4.20e-02 |
| NOL8 | -1.56 | 0.0000000 | 0.00e+00 |
| NOL9 | -1.21 | 0.0002545 | 7.02e-05 |
| NOLC1 | -0.64 | 0.0000000 | 0.00e+00 |
| NOMO1 | 1.34 | 0.0000000 | 0.00e+00 |
| NOMO2 | 1.49 | 0.0000000 | 0.00e+00 |
| NOMO3 | 1.48 | 0.0000000 | 0.00e+00 |
| NOP14-AS1 | 1.64 | 0.0003086 | 8.63e-05 |
| NOP16 | -0.93 | 0.0000016 | 3.00e-07 |
| NOP2 | -0.30 | 0.0394779 | 1.77e-02 |
| NOP56 | -1.24 | 0.0000000 | 0.00e+00 |
| NOP58 | -1.16 | 0.0000000 | 0.00e+00 |
| NOP9 | -0.39 | 0.0831997 | 4.17e-02 |
| NOS1 | 4.39 | 0.0000000 | 0.00e+00 |
| NOS2 | 7.86 | 0.0000000 | 0.00e+00 |
| NOSIP | 0.58 | 0.0005868 | 1.72e-04 |
| NOTCH2 | 1.22 | 0.0000000 | 0.00e+00 |
| NOTCH2NL | 0.89 | 0.0117231 | 4.58e-03 |
| NOTCH3 | -0.35 | 0.0553377 | 2.59e-02 |
| NOX1 | 4.53 | 0.0000000 | 0.00e+00 |
| NPAS1 | 1.60 | 0.0051388 | 1.84e-03 |
| NPAT | -1.10 | 0.0086461 | 3.27e-03 |
| NPC1 | 1.82 | 0.0000000 | 0.00e+00 |
| NPC1L1 | 5.61 | 0.0735043 | 3.61e-02 |
| NPC2 | 0.60 | 0.0006256 | 1.85e-04 |
| NPDC1 | 0.74 | 0.0014195 | 4.49e-04 |
| NPEPPS | 1.09 | 0.0000000 | 0.00e+00 |
| NPFFR1 | 5.53 | 0.0806408 | 4.02e-02 |
| NPHP1 | -2.33 | 0.0177626 | 7.25e-03 |
| NPHP3-ACAD11 | -0.79 | 0.0369744 | 1.64e-02 |
| NPHS1 | 5.74 | 0.0000000 | 0.00e+00 |
| NPLOC4 | 0.56 | 0.0000025 | 5.00e-07 |
| NPM1 | -1.69 | 0.0000000 | 0.00e+00 |
| NPM3 | -1.61 | 0.0000000 | 0.00e+00 |
| NPPA-AS1 | 1.57 | 0.0129344 | 5.10e-03 |
| NPPC | 5.90 | 0.0480835 | 2.21e-02 |
| NPR1 | 4.19 | 0.0000000 | 0.00e+00 |
| NPR3 | -2.00 | 0.0000000 | 0.00e+00 |
| NPTN | 0.50 | 0.0001020 | 2.65e-05 |
| NPTX1 | 0.92 | 0.0000006 | 1.00e-07 |
| NPY4R | -2.34 | 0.0000000 | 0.00e+00 |
| NQO1 | -1.80 | 0.0000000 | 0.00e+00 |
| NQO2 | -0.62 | 0.0089123 | 3.38e-03 |
| NR0B1 | -0.75 | 0.0011454 | 3.56e-04 |
| NR1D1 | 4.60 | 0.0000000 | 0.00e+00 |
| NR1D2 | 0.76 | 0.0077612 | 2.90e-03 |
| NR1H2 | 1.13 | 0.0000000 | 0.00e+00 |
| NR2C1 | -0.69 | 0.0411690 | 1.85e-02 |
| NR2C2AP | -1.12 | 0.0028927 | 9.78e-04 |
| NR2F1 | 2.09 | 0.0000001 | 0.00e+00 |
| NR2F1-AS1 | 1.78 | 0.0036469 | 1.26e-03 |
| NR2F6 | -0.40 | 0.0431304 | 1.95e-02 |
| NR4A1 | 3.23 | 0.0000000 | 0.00e+00 |
| NR4A2 | 2.12 | 0.0000000 | 0.00e+00 |
| NR4A3 | 1.88 | 0.0001592 | 4.28e-05 |
| NR5A1 | 6.44 | 0.0183337 | 7.50e-03 |
| NR5A2 | 1.32 | 0.0000044 | 9.00e-07 |
| NRAS | -0.44 | 0.0393565 | 1.76e-02 |
| NRAV | -0.70 | 0.0250748 | 1.06e-02 |
| NRBF2 | 1.04 | 0.0002400 | 6.60e-05 |
| NRCAM | -0.57 | 0.0000530 | 1.32e-05 |
| NRD1 | -0.61 | 0.0000072 | 1.60e-06 |
| NRG1 | 1.11 | 0.0000000 | 0.00e+00 |
| NRG2 | 3.18 | 0.0000000 | 0.00e+00 |
| NRG4 | -2.85 | 0.0134232 | 5.32e-03 |
| NRGN | -2.00 | 0.0884698 | 4.47e-02 |
| NRIP1 | -0.50 | 0.0063376 | 2.32e-03 |
| NRIR | 6.25 | 0.0262457 | 1.12e-02 |
| NRP1 | -0.46 | 0.0003502 | 9.90e-05 |
| NRP2 | 2.38 | 0.0000000 | 0.00e+00 |
| NRROS | 6.90 | 0.0077309 | 2.89e-03 |
| NSA2 | -0.78 | 0.0000341 | 8.30e-06 |
| NSDHL | 0.98 | 0.0000000 | 0.00e+00 |
| NSFL1C | 0.66 | 0.0000001 | 0.00e+00 |
| NSL1 | -0.81 | 0.0012241 | 3.82e-04 |
| NSMCE1 | -1.07 | 0.0000243 | 5.80e-06 |
| NSMCE4A | -0.81 | 0.0000192 | 4.50e-06 |
| NSUN4 | 0.61 | 0.0499147 | 2.30e-02 |
| NSUN5 | 0.87 | 0.0000279 | 6.70e-06 |
| NT5C2 | 0.95 | 0.0000000 | 0.00e+00 |
| NT5C3A | 1.69 | 0.0000000 | 0.00e+00 |
| NT5C3B | -0.65 | 0.0002006 | 5.45e-05 |
| NT5DC1 | -0.81 | 0.0061949 | 2.26e-03 |
| NT5DC2 | -3.62 | 0.0000000 | 0.00e+00 |
| NT5DC3 | -1.78 | 0.0006925 | 2.06e-04 |
| NT5E | 2.26 | 0.0000000 | 0.00e+00 |
| NT5M | -1.62 | 0.0075242 | 2.80e-03 |
| NTHL1 | -1.52 | 0.0000001 | 0.00e+00 |
| NTMT1 | -0.70 | 0.0007239 | 2.16e-04 |
| NTN1 | 6.73 | 0.0000000 | 0.00e+00 |
| NTN4 | -0.43 | 0.0267110 | 1.14e-02 |
| NTNG2 | 1.86 | 0.0043469 | 1.53e-03 |
| NTPCR | -0.71 | 0.0027761 | 9.37e-04 |
| NTRK3 | -1.45 | 0.0000001 | 0.00e+00 |
| NUAK2 | 1.90 | 0.0000000 | 0.00e+00 |
| NUB1 | 1.51 | 0.0000000 | 0.00e+00 |
| NUBP2 | -1.10 | 0.0000095 | 2.20e-06 |
| NUBPL | -2.28 | 0.0000056 | 1.20e-06 |
| NUCB1 | 1.61 | 0.0000000 | 0.00e+00 |
| NUCKS1 | -1.39 | 0.0000000 | 0.00e+00 |
| NUDC | -0.38 | 0.0098776 | 3.79e-03 |
| NUDCD1 | -0.85 | 0.0001802 | 4.87e-05 |
| NUDCD2 | -1.09 | 0.0011663 | 3.63e-04 |
| NUDT1 | -2.22 | 0.0000000 | 0.00e+00 |
| NUDT11 | -2.22 | 0.0102478 | 3.95e-03 |
| NUDT15 | -1.78 | 0.0000000 | 0.00e+00 |
| NUDT16 | -0.73 | 0.0340107 | 1.49e-02 |
| NUDT16L1 | -0.68 | 0.0960173 | 4.93e-02 |
| NUDT19 | -0.48 | 0.0798676 | 3.97e-02 |
| NUDT2 | -1.09 | 0.0141676 | 5.64e-03 |
| NUDT21 | -0.92 | 0.0000000 | 0.00e+00 |
| NUDT3 | -0.69 | 0.0475959 | 2.18e-02 |
| NUDT5 | -0.91 | 0.0000040 | 9.00e-07 |
| NUDT8 | -1.37 | 0.0799928 | 3.98e-02 |
| NUF2 | -3.23 | 0.0000000 | 0.00e+00 |
| NUMA1 | 0.59 | 0.0000045 | 1.00e-06 |
| NUMB | 1.03 | 0.0000000 | 0.00e+00 |
| NUP107 | -1.35 | 0.0000000 | 0.00e+00 |
| NUP133 | -1.18 | 0.0000001 | 0.00e+00 |
| NUP153 | -0.41 | 0.0299107 | 1.29e-02 |
| NUP155 | -1.54 | 0.0000000 | 0.00e+00 |
| NUP160 | -0.60 | 0.0006376 | 1.88e-04 |
| NUP205 | -1.26 | 0.0000000 | 0.00e+00 |
| NUP214 | 0.70 | 0.0001066 | 2.78e-05 |
| NUP35 | -1.45 | 0.0000044 | 9.00e-07 |
| NUP37 | -1.64 | 0.0000000 | 0.00e+00 |
| NUP43 | -1.05 | 0.0002835 | 7.87e-05 |
| NUP50 | -0.80 | 0.0020270 | 6.64e-04 |
| NUP62 | 0.56 | 0.0000083 | 1.80e-06 |
| NUP62CL | -1.00 | 0.0666741 | 3.21e-02 |
| NUP85 | -1.33 | 0.0000000 | 0.00e+00 |
| NUP88 | -1.07 | 0.0000000 | 0.00e+00 |
| NUP93 | -0.47 | 0.0103318 | 3.98e-03 |
| NUPL1 | -0.72 | 0.0014535 | 4.60e-04 |
| NUPR1 | 4.03 | 0.0000000 | 0.00e+00 |
| NUSAP1 | -1.99 | 0.0000000 | 0.00e+00 |
| NXPE2 | 6.20 | 0.0294725 | 1.27e-02 |
| NXPE3 | -1.09 | 0.0000016 | 3.00e-07 |
| NXPH3 | 5.96 | 0.0000005 | 1.00e-07 |
| NXT2 | -0.82 | 0.0698902 | 3.39e-02 |
| NYAP1 | 4.90 | 0.0000000 | 0.00e+00 |
| NYNRIN | 1.81 | 0.0555710 | 2.61e-02 |
| OAF | 1.53 | 0.0000044 | 9.00e-07 |
| OARD1 | -0.92 | 0.0508467 | 2.35e-02 |
| OAS1 | 3.55 | 0.0000000 | 0.00e+00 |
| OAS2 | 10.64 | 0.0000000 | 0.00e+00 |
| OAS3 | 3.81 | 0.0000000 | 0.00e+00 |
| OASL | 9.37 | 0.0000000 | 0.00e+00 |
| OAT | -1.96 | 0.0000000 | 0.00e+00 |
| OAZ1 | -0.40 | 0.0007635 | 2.29e-04 |
| OCEL1 | 0.83 | 0.0852511 | 4.29e-02 |
| OCIAD2 | 0.58 | 0.0134127 | 5.31e-03 |
| OCLN | 1.59 | 0.0005124 | 1.49e-04 |
| ODC1 | -1.06 | 0.0000000 | 0.00e+00 |
| ODF2 | -0.83 | 0.0004188 | 1.20e-04 |
| ODF3B | 4.10 | 0.0000000 | 0.00e+00 |
| OGDH | 1.42 | 0.0000000 | 0.00e+00 |
| OGFR | 1.99 | 0.0000000 | 0.00e+00 |
| OGFRL1 | 1.63 | 0.0000000 | 0.00e+00 |
| OGG1 | -1.27 | 0.0018529 | 6.00e-04 |
| OIP5 | -3.12 | 0.0000002 | 0.00e+00 |
| OIP5-AS1 | -0.58 | 0.0562727 | 2.64e-02 |
| OLA1 | -0.70 | 0.0000125 | 2.90e-06 |
| OLFM1 | 1.56 | 0.0000001 | 0.00e+00 |
| OLFML2A | -1.93 | 0.0000015 | 3.00e-07 |
| OLFML3 | -1.98 | 0.0123410 | 4.85e-03 |
| OLR1 | 7.52 | 0.0000000 | 0.00e+00 |
| OMA1 | -1.76 | 0.0007352 | 2.20e-04 |
| OPA1 | -2.02 | 0.0000000 | 0.00e+00 |
| OPA3 | 0.86 | 0.0000016 | 3.00e-07 |
| OPLAH | 0.98 | 0.0102804 | 3.96e-03 |
| OPN3 | -0.33 | 0.0741198 | 3.64e-02 |
| OPTN | 3.10 | 0.0000000 | 0.00e+00 |
| OR51E1 | -2.46 | 0.0713595 | 3.48e-02 |
| OR7E14P | 2.06 | 0.0569570 | 2.68e-02 |
| ORAI1 | 1.57 | 0.0000000 | 0.00e+00 |
| ORAI3 | 1.10 | 0.0025325 | 8.48e-04 |
| ORC1 | -2.72 | 0.0000000 | 0.00e+00 |
| ORC2 | -1.00 | 0.0002127 | 5.80e-05 |
| ORC3 | -1.48 | 0.0000013 | 3.00e-07 |
| ORC4 | -0.92 | 0.0013669 | 4.31e-04 |
| ORC5 | -1.57 | 0.0000005 | 1.00e-07 |
| ORC6 | -1.38 | 0.0000291 | 7.00e-06 |
| ORM1 | 9.90 | 0.0000086 | 1.90e-06 |
| ORM2 | 5.81 | 0.0008831 | 2.68e-04 |
| ORMDL3 | 0.52 | 0.0277980 | 1.19e-02 |
| OS9 | 1.06 | 0.0000000 | 0.00e+00 |
| OSBP | 0.94 | 0.0000000 | 0.00e+00 |
| OSBP2 | 2.20 | 0.0000000 | 0.00e+00 |
| OSBPL10 | 0.84 | 0.0002874 | 7.99e-05 |
| OSBPL1A | -1.14 | 0.0079517 | 2.98e-03 |
| OSBPL2 | 0.97 | 0.0000434 | 1.07e-05 |
| OSBPL5 | 1.45 | 0.0000015 | 3.00e-07 |
| OSBPL8 | -1.40 | 0.0000002 | 0.00e+00 |
| OSBPL9 | -0.39 | 0.0381272 | 1.70e-02 |
| OSER1 | 0.90 | 0.0000009 | 2.00e-07 |
| OSGEP | -0.96 | 0.0069818 | 2.58e-03 |
| OSGEPL1 | -2.01 | 0.0005948 | 1.75e-04 |
| OSGIN1 | -1.36 | 0.0000000 | 0.00e+00 |
| OSMR | 3.11 | 0.0000000 | 0.00e+00 |
| OST4 | -0.41 | 0.0066579 | 2.44e-03 |
| OSTC | -0.71 | 0.0106660 | 4.12e-03 |
| OTOA | 4.13 | 0.0579864 | 2.74e-02 |
| OTUD1 | 4.05 | 0.0000000 | 0.00e+00 |
| OTUD3 | -1.04 | 0.0874485 | 4.42e-02 |
| OTUD5 | 0.71 | 0.0000031 | 7.00e-07 |
| OTUD6B | -1.63 | 0.0000287 | 6.90e-06 |
| OTUD6B-AS1 | -2.08 | 0.0000080 | 1.80e-06 |
| OTUD7B | 1.00 | 0.0000337 | 8.20e-06 |
| OVCA2 | 0.80 | 0.0000008 | 2.00e-07 |
| OXCT1 | -3.17 | 0.0609468 | 2.90e-02 |
| OXNAD1 | -1.04 | 0.0137849 | 5.48e-03 |
| OXR1 | -0.73 | 0.0635784 | 3.04e-02 |
| OXSR1 | 0.83 | 0.0000002 | 0.00e+00 |
| OXTR | 2.67 | 0.0000000 | 0.00e+00 |
| P2RX4 | 2.61 | 0.0000000 | 0.00e+00 |
| P2RX5 | -1.30 | 0.0130316 | 5.15e-03 |
| P2RX5-TAX1BP3 | 0.51 | 0.0001487 | 3.97e-05 |
| P2RY11 | 1.70 | 0.0036896 | 1.28e-03 |
| P2RY2 | 1.58 | 0.0000007 | 1.00e-07 |
| P2RY6 | 2.09 | 0.0000000 | 0.00e+00 |
| P3H1 | 0.64 | 0.0082536 | 3.10e-03 |
| P3H2 | 1.44 | 0.0000000 | 0.00e+00 |
| P4HA1 | 0.58 | 0.0043152 | 1.52e-03 |
| P4HA2 | 1.75 | 0.0000000 | 0.00e+00 |
| P4HA3 | 1.15 | 0.0001775 | 4.79e-05 |
| P4HB | 0.68 | 0.0000000 | 0.00e+00 |
| PA2G4 | -1.16 | 0.0000000 | 0.00e+00 |
| PA2G4P4 | -1.27 | 0.0000107 | 2.40e-06 |
| PAAF1 | -0.68 | 0.0796420 | 3.96e-02 |
| PABPC1 | 0.70 | 0.0000000 | 0.00e+00 |
| PABPC1L | -1.14 | 0.0475087 | 2.18e-02 |
| PABPC3 | 0.61 | 0.0567555 | 2.67e-02 |
| PABPC4 | -0.38 | 0.0332735 | 1.46e-02 |
| PACRGL | -1.18 | 0.0906092 | 4.61e-02 |
| PACS1 | 2.11 | 0.0000000 | 0.00e+00 |
| PADI1 | -3.85 | 0.0109101 | 4.23e-03 |
| PADI3 | -3.35 | 0.0025704 | 8.62e-04 |
| PAEP | 6.44 | 0.0210405 | 8.73e-03 |
| PAF1 | 0.65 | 0.0000054 | 1.20e-06 |
| PAFAH1B3 | -1.64 | 0.0000000 | 0.00e+00 |
| PAGR1 | -1.59 | 0.0000000 | 0.00e+00 |
| PAICS | -2.23 | 0.0000000 | 0.00e+00 |
| PAIP1 | 0.30 | 0.0826754 | 4.14e-02 |
| PAIP2 | 0.71 | 0.0000059 | 1.30e-06 |
| PAK1IP1 | -1.36 | 0.0000247 | 5.90e-06 |
| PAK4 | 1.68 | 0.0000000 | 0.00e+00 |
| PAK6 | -2.69 | 0.0989897 | 5.11e-02 |
| PALB2 | -0.72 | 0.0374984 | 1.67e-02 |
| PALD1 | -2.30 | 0.0000048 | 1.00e-06 |
| PALLD | 0.56 | 0.0000233 | 5.50e-06 |
| PALM | 0.75 | 0.0066185 | 2.43e-03 |
| PALM2-AKAP2 | 3.42 | 0.0000000 | 0.00e+00 |
| PALM3 | -3.49 | 0.0330648 | 1.45e-02 |
| PAM | 0.99 | 0.0004799 | 1.39e-04 |
| PAN2 | -0.75 | 0.0097518 | 3.73e-03 |
| PANK3 | -1.07 | 0.0000106 | 2.40e-06 |
| PANX1 | 1.48 | 0.0000000 | 0.00e+00 |
| PANX2 | 1.43 | 0.0029862 | 1.01e-03 |
| PAPD5 | -0.70 | 0.0704447 | 3.42e-02 |
| PAPLN | 1.71 | 0.0000000 | 0.00e+00 |
| PAPOLA | -0.96 | 0.0000000 | 0.00e+00 |
| PAPPA | 1.20 | 0.0000000 | 0.00e+00 |
| PAPSS1 | -1.31 | 0.0000082 | 1.80e-06 |
| PAQR4 | -2.28 | 0.0000000 | 0.00e+00 |
| PAQR5 | 0.51 | 0.0007707 | 2.31e-04 |
| PAQR8 | 3.26 | 0.0000000 | 0.00e+00 |
| PARD3 | -0.63 | 0.0000006 | 1.00e-07 |
| PARD6A | -2.82 | 0.0154039 | 6.19e-03 |
| PARK7 | -1.09 | 0.0000000 | 0.00e+00 |
| PARL | -0.77 | 0.0022340 | 7.40e-04 |
| PARM1 | 2.66 | 0.0000000 | 0.00e+00 |
| PARN | -0.90 | 0.0007239 | 2.16e-04 |
| PARP1 | -1.16 | 0.0000000 | 0.00e+00 |
| PARP10 | 6.80 | 0.0000000 | 0.00e+00 |
| PARP12 | 3.80 | 0.0000000 | 0.00e+00 |
| PARP14 | 3.45 | 0.0000000 | 0.00e+00 |
| PARP2 | -0.82 | 0.0031109 | 1.06e-03 |
| PARP3 | 0.91 | 0.0097994 | 3.75e-03 |
| PARP4 | 0.47 | 0.0016861 | 5.41e-04 |
| PARP6 | 0.59 | 0.0008585 | 2.60e-04 |
| PARP8 | 4.03 | 0.0009757 | 2.99e-04 |
| PARP9 | 3.73 | 0.0000000 | 0.00e+00 |
| PARPBP | -3.13 | 0.0000000 | 0.00e+00 |
| PARVB | -0.73 | 0.0189096 | 7.77e-03 |
| PASK | -2.21 | 0.0000000 | 0.00e+00 |
| PATL1 | 1.37 | 0.0000000 | 0.00e+00 |
| PATL2 | 4.95 | 0.0001584 | 4.26e-05 |
| PATZ1 | 0.69 | 0.0048076 | 1.71e-03 |
| PAWR | 0.63 | 0.0039115 | 1.36e-03 |
| PAX7 | 1.09 | 0.0452460 | 2.06e-02 |
| PAX8 | 2.13 | 0.0071490 | 2.64e-03 |
| PAX8-AS1 | 2.58 | 0.0009711 | 2.98e-04 |
| PAXBP1 | -1.12 | 0.0008485 | 2.57e-04 |
| PAXIP1 | -0.90 | 0.0038879 | 1.35e-03 |
| PBK | -4.11 | 0.0000000 | 0.00e+00 |
| PBRM1 | -0.72 | 0.0210288 | 8.72e-03 |
| PBX1 | -1.03 | 0.0000000 | 0.00e+00 |
| PBX3 | -1.23 | 0.0054022 | 1.94e-03 |
| PBX4 | 2.40 | 0.0005637 | 1.65e-04 |
| PBXIP1 | 1.19 | 0.0000000 | 0.00e+00 |
| PCBD1 | -1.07 | 0.0000000 | 0.00e+00 |
| PCBD2 | -0.98 | 0.0812688 | 4.06e-02 |
| PCBP1 | 0.37 | 0.0002265 | 6.21e-05 |
| PCBP2 | 0.51 | 0.0000000 | 0.00e+00 |
| PCBP2-OT1 | 0.80 | 0.0005081 | 1.48e-04 |
| PCCA | -0.81 | 0.0793708 | 3.94e-02 |
| PCCB | -0.66 | 0.0067816 | 2.49e-03 |
| PCDH1 | 4.74 | 0.0000000 | 0.00e+00 |
| PCDH12 | 8.00 | 0.0007764 | 2.33e-04 |
| PCDH7 | 2.79 | 0.0104735 | 4.04e-03 |
| PCDH9 | -2.03 | 0.0000000 | 0.00e+00 |
| PCED1A | 1.32 | 0.0000000 | 0.00e+00 |
| PCED1B | 0.38 | 0.0828196 | 4.15e-02 |
| PCF11 | -1.12 | 0.0000115 | 2.60e-06 |
| PCGF1 | 0.71 | 0.0341430 | 1.50e-02 |
| PCGF2 | 1.35 | 0.0000000 | 0.00e+00 |
| PCIF1 | 1.00 | 0.0000021 | 4.00e-07 |
| PCK1 | 5.83 | 0.0555609 | 2.61e-02 |
| PCK2 | 1.23 | 0.0000000 | 0.00e+00 |
| PCM1 | -1.34 | 0.0000000 | 0.00e+00 |
| PCMT1 | -0.63 | 0.0064944 | 2.38e-03 |
| PCMTD2 | -1.17 | 0.0464864 | 2.13e-02 |
| PCNA | -2.11 | 0.0000000 | 0.00e+00 |
| PCNP | -0.57 | 0.0036296 | 1.25e-03 |
| PCNT | -0.92 | 0.0021108 | 6.95e-04 |
| PCNXL3 | 0.86 | 0.0000009 | 2.00e-07 |
| PCNXL4 | -1.02 | 0.0008820 | 2.68e-04 |
| PCOLCE2 | -1.32 | 0.0012491 | 3.91e-04 |
| PCSK5 | 0.75 | 0.0779122 | 3.86e-02 |
| PCSK9 | 1.49 | 0.0000000 | 0.00e+00 |
| PCYOX1 | -2.32 | 0.0000000 | 0.00e+00 |
| PCYOX1L | -1.98 | 0.0031986 | 1.09e-03 |
| PCYT2 | 0.68 | 0.0044193 | 1.56e-03 |
| PDAP1 | -0.47 | 0.0018549 | 6.01e-04 |
| PDCD10 | -0.55 | 0.0712500 | 3.47e-02 |
| PDCD1LG2 | 5.83 | 0.0530340 | 2.47e-02 |
| PDCD2 | -0.60 | 0.0296395 | 1.28e-02 |
| PDCD2L | -0.91 | 0.0026620 | 8.97e-04 |
| PDCD4 | -1.07 | 0.0002896 | 8.06e-05 |
| PDCD6 | -0.58 | 0.0004785 | 1.39e-04 |
| PDCD6IP | 0.76 | 0.0000000 | 0.00e+00 |
| PDCD7 | 0.85 | 0.0004193 | 1.20e-04 |
| PDCL3 | -0.77 | 0.0531246 | 2.47e-02 |
| PDDC1 | -1.28 | 0.0000009 | 2.00e-07 |
| PDE1A | -3.23 | 0.0024054 | 8.02e-04 |
| PDE1C | -2.39 | 0.0000001 | 0.00e+00 |
| PDE3A | -0.70 | 0.0014452 | 4.58e-04 |
| PDE3B | -2.08 | 0.0135464 | 5.38e-03 |
| PDE4A | 1.17 | 0.0278021 | 1.19e-02 |
| PDE4B | 1.38 | 0.0000000 | 0.00e+00 |
| PDE4C | 3.09 | 0.0020811 | 6.84e-04 |
| PDE4D | -0.68 | 0.0000001 | 0.00e+00 |
| PDE5A | 1.84 | 0.0405639 | 1.82e-02 |
| PDE7A | -0.90 | 0.0273840 | 1.17e-02 |
| PDE7B | -1.17 | 0.0802852 | 4.00e-02 |
| PDE9A | 2.42 | 0.0000006 | 1.00e-07 |
| -0.65 | 0.0486649 | 2.24e-02 | |
| PDGFA | 1.07 | 0.0001561 | 4.19e-05 |
| PDGFB | 2.48 | 0.0000000 | 0.00e+00 |
| PDGFD | -0.88 | 0.0151571 | 6.08e-03 |
| PDGFRA | 2.76 | 0.0062507 | 2.28e-03 |
| PDGFRL | 1.88 | 0.0000000 | 0.00e+00 |
| PDHA1 | -1.05 | 0.0000000 | 0.00e+00 |
| PDHB | -0.86 | 0.0000313 | 7.60e-06 |
| PDIA3 | 0.58 | 0.0000002 | 0.00e+00 |
| PDIA3P1 | 0.58 | 0.0013122 | 4.12e-04 |
| PDIA4 | 0.62 | 0.0000000 | 0.00e+00 |
| PDIA5 | 1.07 | 0.0000351 | 8.60e-06 |
| PDK2 | -1.18 | 0.0039174 | 1.36e-03 |
| PDK3 | -2.32 | 0.0000000 | 0.00e+00 |
| PDK4 | -1.58 | 0.0000000 | 0.00e+00 |
| PDLIM1 | 1.43 | 0.0000000 | 0.00e+00 |
| PDLIM2 | -0.90 | 0.0101758 | 3.92e-03 |
| PDLIM5 | -0.31 | 0.0038255 | 1.33e-03 |
| PDLIM7 | 4.11 | 0.0000000 | 0.00e+00 |
| PDP1 | 2.61 | 0.0000000 | 0.00e+00 |
| PDPR | -0.51 | 0.0649947 | 3.12e-02 |
| PDS5A | -1.28 | 0.0000000 | 0.00e+00 |
| PDSS1 | -2.42 | 0.0000028 | 6.00e-07 |
| PDXK | -0.49 | 0.0000193 | 4.60e-06 |
| PDXP | -1.04 | 0.0062675 | 2.29e-03 |
| PDZD11 | -0.54 | 0.0357502 | 1.58e-02 |
| PDZD2 | 9.04 | 0.0000704 | 1.79e-05 |
| PDZD8 | -0.96 | 0.0000001 | 0.00e+00 |
| PDZK1 | -3.46 | 0.0628559 | 3.00e-02 |
| PDZK1IP1 | 6.03 | 0.0000000 | 0.00e+00 |
| PDZK1P1 | -4.09 | 0.0874338 | 4.42e-02 |
| PEA15 | 1.28 | 0.0000000 | 0.00e+00 |
| PEAK1 | 1.26 | 0.0000000 | 0.00e+00 |
| PEAR1 | 1.75 | 0.0020590 | 6.76e-04 |
| PEBP1 | -1.53 | 0.0000000 | 0.00e+00 |
| PECAM1 | 3.42 | 0.0127605 | 5.03e-03 |
| PECR | -1.08 | 0.0008568 | 2.59e-04 |
| PEF1 | 0.40 | 0.0429556 | 1.95e-02 |
| PEG10 | -1.97 | 0.0000000 | 0.00e+00 |
| PELI1 | 2.55 | 0.0000000 | 0.00e+00 |
| PELO | 0.83 | 0.0000121 | 2.80e-06 |
| PELP1 | 0.75 | 0.0000004 | 1.00e-07 |
| PEPD | -0.76 | 0.0000010 | 2.00e-07 |
| PER1 | 2.27 | 0.0000000 | 0.00e+00 |
| PER2 | 0.55 | 0.0752443 | 3.70e-02 |
| PERP | 0.78 | 0.0000000 | 0.00e+00 |
| PES1 | -0.93 | 0.0000000 | 0.00e+00 |
| PET100 | -0.70 | 0.0752097 | 3.70e-02 |
| PEX1 | -1.15 | 0.0003388 | 9.54e-05 |
| PEX10 | -0.63 | 0.0842902 | 4.24e-02 |
| PEX14 | 0.61 | 0.0660333 | 3.18e-02 |
| PEX16 | 1.03 | 0.0003596 | 1.02e-04 |
| PFAS | -1.04 | 0.0000001 | 0.00e+00 |
| PFDN5 | -0.23 | 0.0878814 | 4.44e-02 |
| PFKFB2 | -0.85 | 0.0329919 | 1.44e-02 |
| PFKFB3 | -0.35 | 0.0014219 | 4.50e-04 |
| PFKFB4 | 1.92 | 0.0000311 | 7.50e-06 |
| PFKM | -0.90 | 0.0000009 | 2.00e-07 |
| PFN1 | -0.24 | 0.0069416 | 2.56e-03 |
| PFN2 | -0.81 | 0.0000000 | 0.00e+00 |
| PGAM1 | -0.32 | 0.0112882 | 4.40e-03 |
| PGAM4 | -0.39 | 0.0587317 | 2.78e-02 |
| PGAM5 | -0.76 | 0.0000059 | 1.30e-06 |
| PGBD5 | 3.75 | 0.0005613 | 1.64e-04 |
| PGD | -0.23 | 0.0061084 | 2.22e-03 |
| PGF | 2.55 | 0.0000063 | 1.40e-06 |
| PGK1 | -0.37 | 0.0000244 | 5.80e-06 |
| PGLS | 0.58 | 0.0102286 | 3.94e-03 |
| PGLYRP2 | 6.35 | 0.0217322 | 9.05e-03 |
| PGM2 | -0.83 | 0.0156169 | 6.28e-03 |
| PGM3 | 0.61 | 0.0045866 | 1.62e-03 |
| PGP | -1.12 | 0.0000007 | 1.00e-07 |
| PGRMC1 | -0.36 | 0.0173551 | 7.06e-03 |
| PGRMC2 | 0.36 | 0.0313929 | 1.37e-02 |
| PHACTR4 | 0.85 | 0.0000852 | 2.19e-05 |
| PHB | -1.16 | 0.0000000 | 0.00e+00 |
| PHB2 | -0.67 | 0.0000000 | 0.00e+00 |
| PHC1 | 0.57 | 0.0875616 | 4.42e-02 |
| PHC2 | 1.84 | 0.0000000 | 0.00e+00 |
| PHF1 | 1.25 | 0.0040530 | 1.41e-03 |
| PHF11 | 2.37 | 0.0000000 | 0.00e+00 |
| PHF12 | 0.88 | 0.0000037 | 8.00e-07 |
| PHF14 | -0.72 | 0.0030179 | 1.03e-03 |
| PHF19 | -1.65 | 0.0000000 | 0.00e+00 |
| PHF2 | 0.61 | 0.0084964 | 3.20e-03 |
| PHF20L1 | -0.63 | 0.0058462 | 2.12e-03 |
| PHF21A | 1.74 | 0.0000000 | 0.00e+00 |
| PHF23 | 0.42 | 0.0088158 | 3.34e-03 |
| PHF24 | 8.79 | 0.0001273 | 3.36e-05 |
| PHF3 | -0.69 | 0.0043556 | 1.53e-03 |
| PHF5A | -0.68 | 0.0093793 | 3.58e-03 |
| PHF6 | -1.42 | 0.0000000 | 0.00e+00 |
| PHF8 | 0.58 | 0.0228695 | 9.59e-03 |
| PHGDH | -0.54 | 0.0127109 | 5.01e-03 |
| PHIP | -1.95 | 0.0000000 | 0.00e+00 |
| PHKA1 | -0.47 | 0.0333843 | 1.46e-02 |
| PHKA2 | -0.56 | 0.0576916 | 2.72e-02 |
| PHKB | -1.40 | 0.0000000 | 0.00e+00 |
| PHLDA1 | 3.13 | 0.0000000 | 0.00e+00 |
| PHLDA2 | 1.32 | 0.0000200 | 4.70e-06 |
| PHLDA3 | 1.15 | 0.0000000 | 0.00e+00 |
| PHLDB1 | 0.90 | 0.0024790 | 8.29e-04 |
| PHLDB2 | 1.59 | 0.0000000 | 0.00e+00 |
| PHLDB3 | 0.85 | 0.0378050 | 1.69e-02 |
| PHLPP1 | 0.99 | 0.0005636 | 1.65e-04 |
| PHOSPHO2-KLHL23 | -1.88 | 0.0000135 | 3.10e-06 |
| PHPT1 | -1.05 | 0.0000000 | 0.00e+00 |
| PHRF1 | 0.82 | 0.0000600 | 1.51e-05 |
| PHTF2 | -1.52 | 0.0000017 | 3.00e-07 |
| PHYH | -0.61 | 0.0458054 | 2.09e-02 |
| PHYKPL | 1.44 | 0.0000307 | 7.40e-06 |
| PI16 | 5.83 | 0.0515073 | 2.39e-02 |
| PI3 | 8.37 | 0.0000000 | 0.00e+00 |
| PI4K2A | 1.20 | 0.0000000 | 0.00e+00 |
| PI4KB | 1.14 | 0.0000000 | 0.00e+00 |
| PIAS2 | -1.70 | 0.0015500 | 4.94e-04 |
| PIAS3 | 0.67 | 0.0182439 | 7.46e-03 |
| PIAS4 | 0.91 | 0.0003388 | 9.54e-05 |
| PIBF1 | -1.29 | 0.0401683 | 1.81e-02 |
| PICALM | 1.15 | 0.0000000 | 0.00e+00 |
| PIEZO1 | 1.70 | 0.0000000 | 0.00e+00 |
| PIF1 | -4.12 | 0.0000000 | 0.00e+00 |
| PIGA | 0.82 | 0.0022837 | 7.59e-04 |
| PIGF | -1.12 | 0.0081370 | 3.06e-03 |
| PIGG | 0.51 | 0.0567746 | 2.67e-02 |
| PIGK | -1.90 | 0.0000000 | 0.00e+00 |
| PIGM | -1.39 | 0.0017457 | 5.62e-04 |
| PIGN | -0.97 | 0.0056171 | 2.03e-03 |
| PIGR | 9.29 | 0.0000393 | 9.60e-06 |
| PIGS | 0.63 | 0.0001020 | 2.65e-05 |
| PIGT | 0.94 | 0.0000000 | 0.00e+00 |
| PIGW | -1.98 | 0.0007851 | 2.36e-04 |
| PIGX | -1.29 | 0.0006726 | 2.00e-04 |
| PIGY | -0.57 | 0.0213989 | 8.89e-03 |
| PIK3AP1 | 4.93 | 0.0000000 | 0.00e+00 |
| PIK3C2A | -0.73 | 0.0003764 | 1.07e-04 |
| PIK3CD | 1.09 | 0.0007935 | 2.39e-04 |
| PIK3IP1 | 3.43 | 0.0000000 | 0.00e+00 |
| PIK3R3 | 1.46 | 0.0003744 | 1.07e-04 |
| PIK3R4 | -0.74 | 0.0129805 | 5.12e-03 |
| PIK3R5 | 11.85 | 0.0000000 | 0.00e+00 |
| PIKFYVE | -0.80 | 0.0007055 | 2.10e-04 |
| PIM1 | 2.35 | 0.0000000 | 0.00e+00 |
| PIM2 | 0.94 | 0.0114846 | 4.48e-03 |
| PIM3 | 1.98 | 0.0000000 | 0.00e+00 |
| PIN1 | -0.75 | 0.0006334 | 1.87e-04 |
| PIN4 | -0.63 | 0.0637961 | 3.05e-02 |
| PINK1 | 1.09 | 0.0000000 | 0.00e+00 |
| PINLYP | 4.75 | 0.0166937 | 6.76e-03 |
| PINX1 | -1.01 | 0.0115907 | 4.52e-03 |
| PIP | 4.82 | 0.0002845 | 7.91e-05 |
| PIP4K2A | 0.51 | 0.0447641 | 2.04e-02 |
| PIP4K2B | -0.69 | 0.0029199 | 9.88e-04 |
| PIP4K2C | 0.65 | 0.0003075 | 8.59e-05 |
| PIP5K1A | 0.85 | 0.0000005 | 1.00e-07 |
| PIP5K1C | 1.40 | 0.0000002 | 0.00e+00 |
| PIR | -1.07 | 0.0000000 | 0.00e+00 |
| PIR-FIGF | -1.10 | 0.0000013 | 3.00e-07 |
| PISD | 1.29 | 0.0000001 | 0.00e+00 |
| PITPNA-AS1 | -0.68 | 0.0784921 | 3.89e-02 |
| PITPNC1 | 3.17 | 0.0000000 | 0.00e+00 |
| PITPNM1 | 0.99 | 0.0000000 | 0.00e+00 |
| PITPNM2 | 1.58 | 0.0000000 | 0.00e+00 |
| PITX1 | 1.24 | 0.0000001 | 0.00e+00 |
| PITX3 | 3.17 | 0.0800797 | 3.99e-02 |
| PJA1 | 0.93 | 0.0000571 | 1.43e-05 |
| PKD1 | 0.65 | 0.0000676 | 1.71e-05 |
| PKD1L2 | -2.07 | 0.0194956 | 8.03e-03 |
| PKD1P6 | 0.86 | 0.0029109 | 9.85e-04 |
| PKD2 | -1.10 | 0.0001553 | 4.16e-05 |
| PKDCC | -1.21 | 0.0000002 | 0.00e+00 |
| PKI55 | -2.60 | 0.0004359 | 1.26e-04 |
| PKM | -0.74 | 0.0000000 | 0.00e+00 |
| PKMYT1 | -1.76 | 0.0000001 | 0.00e+00 |
| PKN1 | 0.64 | 0.0000079 | 1.80e-06 |
| PKN2 | -0.53 | 0.0398088 | 1.79e-02 |
| PKP1 | 5.97 | 0.0428851 | 1.94e-02 |
| PKP3 | -0.48 | 0.0592219 | 2.80e-02 |
| PKP4 | -0.50 | 0.0105487 | 4.08e-03 |
| PLA1A | 6.04 | 0.0000000 | 0.00e+00 |
| PLA2G12A | -1.95 | 0.0033174 | 1.14e-03 |
| PLA2G15 | 1.08 | 0.0100583 | 3.87e-03 |
| PLA2G4A | 0.48 | 0.0046890 | 1.66e-03 |
| PLA2G4C | 4.42 | 0.0000000 | 0.00e+00 |
| PLA2G4E | 7.73 | 0.0014195 | 4.49e-04 |
| PLAGL2 | 1.26 | 0.0000001 | 0.00e+00 |
| PLAT | 4.57 | 0.0000000 | 0.00e+00 |
| PLAU | 3.97 | 0.0000000 | 0.00e+00 |
| PLAUR | 4.67 | 0.0000000 | 0.00e+00 |
| PLBD2 | 1.05 | 0.0000008 | 2.00e-07 |
| PLCD3 | 0.39 | 0.0029681 | 1.01e-03 |
| PLCE1 | -2.04 | 0.0000010 | 2.00e-07 |
| PLCG1 | -0.60 | 0.0185780 | 7.61e-03 |
| PLCG2 | 2.61 | 0.0000000 | 0.00e+00 |
| PLCL2 | 1.38 | 0.0016870 | 5.41e-04 |
| PLCXD1 | -1.28 | 0.0000002 | 0.00e+00 |
| PLCXD3 | -4.83 | 0.0002159 | 5.90e-05 |
| PLD1 | 0.66 | 0.0118791 | 4.65e-03 |
| PLD2 | 0.83 | 0.0000915 | 2.36e-05 |
| PLD3 | 0.94 | 0.0000000 | 0.00e+00 |
| PLEC | 1.45 | 0.0000000 | 0.00e+00 |
| PLEK | 5.53 | 0.0820025 | 4.10e-02 |
| PLEK2 | 2.90 | 0.0000000 | 0.00e+00 |
| PLEKHA1 | -0.58 | 0.0455565 | 2.08e-02 |
| PLEKHA3 | 0.71 | 0.0285880 | 1.23e-02 |
| PLEKHA4 | 8.28 | 0.0000000 | 0.00e+00 |
| PLEKHA6 | 1.38 | 0.0000000 | 0.00e+00 |
| PLEKHA7 | 1.88 | 0.0000000 | 0.00e+00 |
| PLEKHB2 | 0.26 | 0.0926629 | 4.73e-02 |
| PLEKHF1 | 1.57 | 0.0001272 | 3.36e-05 |
| PLEKHF2 | 0.76 | 0.0703673 | 3.42e-02 |
| PLEKHG1 | 2.60 | 0.0128163 | 5.05e-03 |
| PLEKHG2 | 2.29 | 0.0000000 | 0.00e+00 |
| PLEKHG3 | 1.04 | 0.0000000 | 0.00e+00 |
| PLEKHG4 | 0.88 | 0.0017831 | 5.75e-04 |
| PLEKHG5 | 2.07 | 0.0000316 | 7.70e-06 |
| PLEKHG6 | 4.71 | 0.0000134 | 3.10e-06 |
| PLEKHH2 | -1.53 | 0.0005980 | 1.76e-04 |
| PLEKHM1 | 2.21 | 0.0000000 | 0.00e+00 |
| PLEKHM1P | 1.45 | 0.0002160 | 5.90e-05 |
| PLEKHM2 | 1.92 | 0.0000000 | 0.00e+00 |
| PLEKHN1 | 1.85 | 0.0031159 | 1.06e-03 |
| PLEKHO2 | 1.44 | 0.0000403 | 9.90e-06 |
| PLIN2 | 0.64 | 0.0030919 | 1.05e-03 |
| PLIN3 | 1.53 | 0.0000000 | 0.00e+00 |
| PLK1 | -2.50 | 0.0000000 | 0.00e+00 |
| PLK2 | 1.13 | 0.0000000 | 0.00e+00 |
| PLK3 | 2.38 | 0.0000000 | 0.00e+00 |
| PLK4 | -2.82 | 0.0000000 | 0.00e+00 |
| PLOD1 | 0.60 | 0.0000632 | 1.59e-05 |
| PLOD2 | 1.00 | 0.0000000 | 0.00e+00 |
| PLOD3 | 0.88 | 0.0000275 | 6.60e-06 |
| PLP2 | -0.47 | 0.0009725 | 2.98e-04 |
| PLRG1 | -1.04 | 0.0000006 | 1.00e-07 |
| PLS1 | -0.41 | 0.0526859 | 2.45e-02 |
| PLSCR1 | 2.98 | 0.0000000 | 0.00e+00 |
| PLXNA1 | 1.67 | 0.0000000 | 0.00e+00 |
| PLXNA2 | 4.06 | 0.0000000 | 0.00e+00 |
| PLXNA3 | 2.12 | 0.0000001 | 0.00e+00 |
| PLXNA4 | 4.23 | 0.0000002 | 0.00e+00 |
| PLXNB2 | 1.96 | 0.0000000 | 0.00e+00 |
| PLXND1 | 0.41 | 0.0076816 | 2.87e-03 |
| PM20D2 | -0.90 | 0.0009350 | 2.86e-04 |
| PMAIP1 | 4.77 | 0.0000000 | 0.00e+00 |
| PMEPA1 | 1.13 | 0.0000000 | 0.00e+00 |
| PMF1 | -0.57 | 0.0314908 | 1.37e-02 |
| PML | 4.91 | 0.0000000 | 0.00e+00 |
| PMM2 | -0.76 | 0.0000343 | 8.40e-06 |
| PMPCA | -0.79 | 0.0000928 | 2.39e-05 |
| PMS1 | -1.85 | 0.0000711 | 1.80e-05 |
| PMS2 | -1.23 | 0.0009439 | 2.89e-04 |
| PMVK | -1.60 | 0.0000000 | 0.00e+00 |
| PNISR | -1.11 | 0.0000028 | 6.00e-07 |
| PNKD | 0.97 | 0.0000000 | 0.00e+00 |
| PNMA1 | 0.58 | 0.0019796 | 6.46e-04 |
| PNMAL1 | -1.26 | 0.0037973 | 1.32e-03 |
| PNN | -1.59 | 0.0000000 | 0.00e+00 |
| PNO1 | -0.73 | 0.0001061 | 2.76e-05 |
| PNP | 1.50 | 0.0000000 | 0.00e+00 |
| PNPLA2 | 1.91 | 0.0000000 | 0.00e+00 |
| PNPLA4 | -1.77 | 0.0040705 | 1.42e-03 |
| PNPLA6 | 0.72 | 0.0003643 | 1.04e-04 |
| PNPLA8 | 0.88 | 0.0004956 | 1.44e-04 |
| PNPO | -0.98 | 0.0000266 | 6.40e-06 |
| PNRC1 | 4.52 | 0.0000000 | 0.00e+00 |
| PNRC2 | 0.61 | 0.0151865 | 6.09e-03 |
| POC1A | -2.15 | 0.0000000 | 0.00e+00 |
| POC1B-GALNT4 | 1.23 | 0.0000000 | 0.00e+00 |
| POC5 | 0.90 | 0.0075133 | 2.79e-03 |
| PODXL | 3.17 | 0.0000000 | 0.00e+00 |
| POF1B | 0.89 | 0.0002627 | 7.26e-05 |
| POFUT1 | -0.39 | 0.0270403 | 1.16e-02 |
| POGZ | 0.53 | 0.0080506 | 3.02e-03 |
| POLA1 | -2.62 | 0.0000000 | 0.00e+00 |
| POLA2 | -1.64 | 0.0000000 | 0.00e+00 |
| POLD1 | -1.47 | 0.0000000 | 0.00e+00 |
| POLD2 | -1.36 | 0.0000000 | 0.00e+00 |
| POLD3 | -1.04 | 0.0005706 | 1.67e-04 |
| POLD4 | 1.51 | 0.0000000 | 0.00e+00 |
| POLDIP2 | -0.34 | 0.0052085 | 1.86e-03 |
| POLDIP3 | 0.48 | 0.0018592 | 6.03e-04 |
| POLE | -1.32 | 0.0000000 | 0.00e+00 |
| POLE2 | -3.99 | 0.0000000 | 0.00e+00 |
| POLE3 | -1.60 | 0.0000000 | 0.00e+00 |
| POLE4 | -1.52 | 0.0000027 | 6.00e-07 |
| POLG | 0.99 | 0.0000000 | 0.00e+00 |
| POLG2 | -0.86 | 0.0649947 | 3.12e-02 |
| POLL | 0.84 | 0.0013373 | 4.20e-04 |
| POLQ | -3.01 | 0.0000000 | 0.00e+00 |
| POLR1A | -0.74 | 0.0001554 | 4.17e-05 |
| POLR1B | -1.16 | 0.0000021 | 4.00e-07 |
| POLR1C | -0.54 | 0.0899116 | 4.56e-02 |
| POLR1D | 0.34 | 0.0622821 | 2.97e-02 |
| POLR1E | -1.31 | 0.0000006 | 1.00e-07 |
| POLR2A | 1.31 | 0.0000000 | 0.00e+00 |
| POLR2B | -1.71 | 0.0000000 | 0.00e+00 |
| POLR2C | -0.60 | 0.0000196 | 4.60e-06 |
| POLR2D | -0.53 | 0.0229942 | 9.64e-03 |
| POLR2E | -1.02 | 0.0000000 | 0.00e+00 |
| POLR2F | -0.88 | 0.0005736 | 1.68e-04 |
| POLR2G | -1.10 | 0.0000000 | 0.00e+00 |
| POLR2H | -0.64 | 0.0322783 | 1.41e-02 |
| POLR2I | -0.61 | 0.0019096 | 6.22e-04 |
| POLR2J | -0.48 | 0.0432907 | 1.96e-02 |
| POLR2K | -0.89 | 0.0001579 | 4.24e-05 |
| POLR2L | -1.55 | 0.0000000 | 0.00e+00 |
| POLR3B | -1.26 | 0.0041938 | 1.47e-03 |
| POLR3C | 0.74 | 0.0000992 | 2.57e-05 |
| POLR3D | 1.02 | 0.0000000 | 0.00e+00 |
| POLR3G | -3.56 | 0.0000000 | 0.00e+00 |
| POLR3H | -1.05 | 0.0000675 | 1.71e-05 |
| POLR3K | -1.88 | 0.0000000 | 0.00e+00 |
| POM121 | 1.04 | 0.0000000 | 0.00e+00 |
| POM121C | 1.28 | 0.0000000 | 0.00e+00 |
| POMGNT1 | 0.62 | 0.0001554 | 4.17e-05 |
| PON1 | 2.43 | 0.0177056 | 7.22e-03 |
| PON2 | 0.41 | 0.0000214 | 5.10e-06 |
| POP1 | -0.93 | 0.0086861 | 3.28e-03 |
| POP5 | -1.93 | 0.0000000 | 0.00e+00 |
| POP7 | -0.80 | 0.0005104 | 1.49e-04 |
| POPDC2 | 2.70 | 0.0981007 | 5.05e-02 |
| POR | 0.71 | 0.0000003 | 1.00e-07 |
| PORCN | 0.99 | 0.0445769 | 2.03e-02 |
| POT1 | -1.40 | 0.0000290 | 7.00e-06 |
| POU2F1 | 0.56 | 0.0422657 | 1.91e-02 |
| POU2F2 | 5.14 | 0.0000000 | 0.00e+00 |
| POU2F3 | 6.53 | 0.0159475 | 6.43e-03 |
| PP7080 | -0.98 | 0.0424233 | 1.92e-02 |
| PPA1 | 0.74 | 0.0000001 | 0.00e+00 |
| PPA2 | -0.48 | 0.0923254 | 4.71e-02 |
| PPAP2A | 0.51 | 0.0544079 | 2.54e-02 |
| PPAP2B | 2.47 | 0.0000000 | 0.00e+00 |
| PPAPDC1B | 0.63 | 0.0250115 | 1.06e-02 |
| PPAPDC2 | 1.37 | 0.0000001 | 0.00e+00 |
| PPARA | 1.10 | 0.0304007 | 1.32e-02 |
| PPARD | 3.70 | 0.0000000 | 0.00e+00 |
| PPAT | -1.94 | 0.0000000 | 0.00e+00 |
| PPFIA3 | -1.10 | 0.0734676 | 3.60e-02 |
| PPFIBP1 | 1.34 | 0.0000000 | 0.00e+00 |
| PPHLN1 | -0.71 | 0.0013609 | 4.29e-04 |
| PPIA | -1.16 | 0.0000000 | 0.00e+00 |
| PPIAP30 | -1.97 | 0.0720972 | 3.52e-02 |
| PPIB | 0.45 | 0.0000015 | 3.00e-07 |
| PPIC | 0.77 | 0.0381184 | 1.70e-02 |
| PPID | -1.01 | 0.0001162 | 3.05e-05 |
| PPIE | -0.64 | 0.0217654 | 9.06e-03 |
| PPIF | 0.34 | 0.0399751 | 1.80e-02 |
| PPIG | -0.82 | 0.0020131 | 6.59e-04 |
| PPIH | -1.62 | 0.0000287 | 6.90e-06 |
| PPIL1 | -0.96 | 0.0000101 | 2.30e-06 |
| PPIL3 | -0.82 | 0.0567948 | 2.67e-02 |
| PPIP5K1 | -1.33 | 0.0017760 | 5.72e-04 |
| PPIP5K2 | -1.80 | 0.0000000 | 0.00e+00 |
| PPM1B | 0.81 | 0.0000024 | 5.00e-07 |
| PPM1D | 0.56 | 0.0867423 | 4.38e-02 |
| PPM1G | -0.21 | 0.0842670 | 4.23e-02 |
| PPM1H | -1.16 | 0.0015348 | 4.88e-04 |
| PPM1K | 5.94 | 0.0000000 | 0.00e+00 |
| PPOX | -1.45 | 0.0110147 | 4.28e-03 |
| PPP1CA | -0.45 | 0.0001181 | 3.10e-05 |
| PPP1CB | -0.75 | 0.0000036 | 8.00e-07 |
| PPP1CC | -0.95 | 0.0000000 | 0.00e+00 |
| PPP1R12C | 1.08 | 0.0000000 | 0.00e+00 |
| PPP1R13L | 2.66 | 0.0000000 | 0.00e+00 |
| PPP1R14A | -1.80 | 0.0000000 | 0.00e+00 |
| PPP1R14B | -0.35 | 0.0634484 | 3.03e-02 |
| PPP1R14C | 3.84 | 0.0021335 | 7.03e-04 |
| PPP1R14D | 4.27 | 0.0000000 | 0.00e+00 |
| PPP1R15A | 4.87 | 0.0000000 | 0.00e+00 |
| PPP1R15B | 2.27 | 0.0000000 | 0.00e+00 |
| PPP1R21 | 0.88 | 0.0015507 | 4.94e-04 |
| PPP1R26 | 0.87 | 0.0000259 | 6.20e-06 |
| PPP1R37 | 0.39 | 0.0720068 | 3.52e-02 |
| PPP1R3B | 1.28 | 0.0000510 | 1.27e-05 |
| PPP1R3D | 1.31 | 0.0030824 | 1.05e-03 |
| PPP1R9A | -0.83 | 0.0134203 | 5.32e-03 |
| PPP1R9B | 0.90 | 0.0000000 | 0.00e+00 |
| PPP2CA | -0.36 | 0.0051557 | 1.84e-03 |
| PPP2CB | 0.57 | 0.0000789 | 2.01e-05 |
| PPP2R2A | 0.65 | 0.0000836 | 2.14e-05 |
| PPP2R2C | -0.50 | 0.0161038 | 6.50e-03 |
| PPP2R3C | 0.76 | 0.0075516 | 2.81e-03 |
| PPP2R5B | 2.34 | 0.0000000 | 0.00e+00 |
| PPP2R5C | -2.00 | 0.0000000 | 0.00e+00 |
| PPP2R5D | -0.47 | 0.0296230 | 1.28e-02 |
| PPP3CB | -0.68 | 0.0444049 | 2.02e-02 |
| PPP3CC | 1.73 | 0.0000000 | 0.00e+00 |
| PPP3R1 | 0.78 | 0.0000039 | 8.00e-07 |
| PPP4C | 0.59 | 0.0000306 | 7.40e-06 |
| PPP4R1 | 0.32 | 0.0587791 | 2.78e-02 |
| PPP4R1L | 2.37 | 0.0000142 | 3.30e-06 |
| PPP4R3B | -1.25 | 0.0000000 | 0.00e+00 |
| PPP5C | -0.35 | 0.0232999 | 9.80e-03 |
| PPP6C | -0.44 | 0.0751844 | 3.70e-02 |
| PPP6R1 | 0.88 | 0.0000000 | 0.00e+00 |
| PPP6R2 | 0.54 | 0.0531922 | 2.48e-02 |
| PPP6R3 | -0.77 | 0.0000002 | 0.00e+00 |
| PPRC1 | 1.15 | 0.0000000 | 0.00e+00 |
| PPT1 | -0.95 | 0.0000000 | 0.00e+00 |
| PPTC7 | 0.75 | 0.0052305 | 1.87e-03 |
| PPWD1 | -2.17 | 0.0000001 | 0.00e+00 |
| PQBP1 | 0.36 | 0.0564998 | 2.66e-02 |
| PQLC1 | 0.80 | 0.0148747 | 5.96e-03 |
| PQLC2 | 1.08 | 0.0011717 | 3.65e-04 |
| PQLC3 | -0.58 | 0.0255790 | 1.09e-02 |
| PRADC1 | -2.77 | 0.0000003 | 0.00e+00 |
| PRAF2 | 1.26 | 0.0001032 | 2.68e-05 |
| PRC1 | -2.27 | 0.0000000 | 0.00e+00 |
| PRCC | 1.13 | 0.0000000 | 0.00e+00 |
| PRDM1 | 4.01 | 0.0000000 | 0.00e+00 |
| PRDM13 | -3.58 | 0.0899791 | 4.57e-02 |
| PRDM2 | 0.75 | 0.0109297 | 4.24e-03 |
| PRDM4 | 0.59 | 0.0045697 | 1.62e-03 |
| PRDM6 | 3.25 | 0.0705565 | 3.43e-02 |
| PRDM8 | 4.95 | 0.0113022 | 4.40e-03 |
| PRDX1 | -0.38 | 0.0000077 | 1.70e-06 |
| PRDX3 | -1.60 | 0.0000000 | 0.00e+00 |
| PRDX4 | -0.66 | 0.0000067 | 1.50e-06 |
| PRDX6 | 0.81 | 0.0000000 | 0.00e+00 |
| PREB | -0.44 | 0.0112628 | 4.39e-03 |
| PREPL | -2.62 | 0.0000000 | 0.00e+00 |
| PRICKLE1 | 1.94 | 0.0007150 | 2.13e-04 |
| PRICKLE2 | 1.97 | 0.0000000 | 0.00e+00 |
| PRICKLE3 | 1.78 | 0.0000000 | 0.00e+00 |
| PRIM1 | -3.13 | 0.0000000 | 0.00e+00 |
| PRIM2 | -1.22 | 0.0017248 | 5.54e-04 |
| PRIMPOL | -2.02 | 0.0007645 | 2.29e-04 |
| PRKAA1 | -0.83 | 0.0001322 | 3.50e-05 |
| PRKAB1 | 0.77 | 0.0005605 | 1.64e-04 |
| PRKACB | -0.85 | 0.0243820 | 1.03e-02 |
| PRKAG2 | -1.09 | 0.0010576 | 3.26e-04 |
| PRKAR1A | -0.59 | 0.0000042 | 9.00e-07 |
| PRKAR1B | -1.33 | 0.0001473 | 3.93e-05 |
| PRKAR2B | -1.66 | 0.0333841 | 1.46e-02 |
| PRKCA | -0.41 | 0.0086208 | 3.26e-03 |
| PRKCD | 1.40 | 0.0000000 | 0.00e+00 |
| PRKCG | 2.58 | 0.0000426 | 1.05e-05 |
| PRKCQ-AS1 | -0.95 | 0.0657415 | 3.16e-02 |
| PRKCSH | 1.49 | 0.0000000 | 0.00e+00 |
| PRKD1 | 0.84 | 0.0526661 | 2.45e-02 |
| PRKD2 | 2.63 | 0.0000000 | 0.00e+00 |
| PRKDC | -1.58 | 0.0000000 | 0.00e+00 |
| PRKRIR | -0.87 | 0.0001966 | 5.33e-05 |
| PRKX | -1.21 | 0.0019800 | 6.47e-04 |
| PRLR | 5.53 | 0.0835040 | 4.19e-02 |
| PRMT1 | -0.44 | 0.0000709 | 1.80e-05 |
| PRMT2 | -0.58 | 0.0130945 | 5.18e-03 |
| PRMT3 | -1.47 | 0.0000000 | 0.00e+00 |
| PRMT5 | -0.57 | 0.0000163 | 3.80e-06 |
| PRMT7 | -0.68 | 0.0222793 | 9.30e-03 |
| PROCR | -0.53 | 0.0027676 | 9.34e-04 |
| PROSER2 | 2.19 | 0.0000000 | 0.00e+00 |
| PROSER3 | 0.67 | 0.0820379 | 4.10e-02 |
| PROX1 | 6.20 | 0.0283388 | 1.22e-02 |
| PRPF19 | -0.65 | 0.0000008 | 1.00e-07 |
| PRPF38B | -0.73 | 0.0096687 | 3.70e-03 |
| PRPF4 | -0.84 | 0.0000006 | 1.00e-07 |
| PRPF40A | -0.95 | 0.0000000 | 0.00e+00 |
| PRPF40B | 0.70 | 0.0129802 | 5.12e-03 |
| PRPF4B | -1.27 | 0.0000000 | 0.00e+00 |
| PRPS1 | -1.50 | 0.0000000 | 0.00e+00 |
| PRPS2 | -1.68 | 0.0000000 | 0.00e+00 |
| PRPSAP1 | -0.52 | 0.0410956 | 1.85e-02 |
| PRR11 | -2.44 | 0.0000000 | 0.00e+00 |
| PRR12 | 1.56 | 0.0000000 | 0.00e+00 |
| PRR13 | 0.87 | 0.0000000 | 0.00e+00 |
| PRR14 | 1.61 | 0.0000000 | 0.00e+00 |
| PRR15 | 3.38 | 0.0000000 | 0.00e+00 |
| PRR15L | -5.76 | 0.0886793 | 4.49e-02 |
| PRR26 | 4.26 | 0.0000001 | 0.00e+00 |
| PRR34-AS1 | -1.54 | 0.0396376 | 1.78e-02 |
| PRRC1 | -0.51 | 0.0298077 | 1.29e-02 |
| PRRC2B | 0.45 | 0.0001257 | 3.31e-05 |
| PRRC2C | 0.61 | 0.0000001 | 0.00e+00 |
| PRRG4 | 2.99 | 0.0000116 | 2.60e-06 |
| PRRX2 | 2.35 | 0.0000000 | 0.00e+00 |
| PRSS12 | 2.56 | 0.0451604 | 2.06e-02 |
| PRSS22 | 4.98 | 0.0091463 | 3.48e-03 |
| PRSS23 | 1.33 | 0.0000000 | 0.00e+00 |
| PRSS27 | 1.88 | 0.0336575 | 1.48e-02 |
| PRSS3 | 2.45 | 0.0000009 | 2.00e-07 |
| PRTFDC1 | -1.20 | 0.0340972 | 1.50e-02 |
| PRTG | -1.48 | 0.0008108 | 2.45e-04 |
| PRUNE2 | 4.95 | 0.0096565 | 3.69e-03 |
| PSAP | 1.19 | 0.0000000 | 0.00e+00 |
| PSAT1 | -1.66 | 0.0000000 | 0.00e+00 |
| PSD3 | -0.66 | 0.0105762 | 4.09e-03 |
| PSD4 | 0.86 | 0.0676196 | 3.26e-02 |
| PSEN1 | 1.31 | 0.0000000 | 0.00e+00 |
| PSEN2 | 0.86 | 0.0003811 | 1.09e-04 |
| PSIP1 | -1.62 | 0.0000000 | 0.00e+00 |
| PSKH1 | 0.54 | 0.0433242 | 1.96e-02 |
| PSMA1 | -0.31 | 0.0361464 | 1.60e-02 |
| PSMA2 | -0.80 | 0.0000001 | 0.00e+00 |
| PSMA3 | -1.06 | 0.0000000 | 0.00e+00 |
| PSMA3-AS1 | -1.09 | 0.0333831 | 1.46e-02 |
| PSMA4 | -0.24 | 0.0463887 | 2.12e-02 |
| PSMA6 | 0.49 | 0.0001542 | 4.13e-05 |
| PSMA7 | -0.54 | 0.0000025 | 5.00e-07 |
| PSMB1 | -0.69 | 0.0000105 | 2.40e-06 |
| PSMB10 | 1.98 | 0.0000000 | 0.00e+00 |
| PSMB2 | -0.29 | 0.0782168 | 3.87e-02 |
| PSMB3 | -0.62 | 0.0001694 | 4.57e-05 |
| PSMB4 | -0.77 | 0.0000000 | 0.00e+00 |
| PSMB5 | -0.75 | 0.0000000 | 0.00e+00 |
| PSMB6 | -0.40 | 0.0008702 | 2.64e-04 |
| PSMC2 | -0.81 | 0.0000002 | 0.00e+00 |
| PSMC3 | -0.39 | 0.0021566 | 7.12e-04 |
| PSMC3IP | -2.14 | 0.0000013 | 3.00e-07 |
| PSMC4 | 0.30 | 0.0027245 | 9.19e-04 |
| PSMC5 | -0.36 | 0.0082026 | 3.08e-03 |
| PSMC6 | -0.81 | 0.0000338 | 8.20e-06 |
| PSMD1 | -0.93 | 0.0000000 | 0.00e+00 |
| PSMD10 | -0.84 | 0.0000274 | 6.60e-06 |
| PSMD12 | -1.12 | 0.0000000 | 0.00e+00 |
| PSMD13 | -0.68 | 0.0000006 | 1.00e-07 |
| PSMD14 | -0.95 | 0.0000000 | 0.00e+00 |
| PSMD2 | -0.26 | 0.0230316 | 9.67e-03 |
| PSMD5 | -0.73 | 0.0000370 | 9.10e-06 |
| PSMD6 | -0.38 | 0.0593922 | 2.81e-02 |
| PSMD8 | -0.21 | 0.0490346 | 2.26e-02 |
| PSME1 | 1.92 | 0.0000000 | 0.00e+00 |
| PSME2 | 1.34 | 0.0000000 | 0.00e+00 |
| PSMF1 | 0.36 | 0.0048840 | 1.74e-03 |
| PSMG1 | -1.64 | 0.0000000 | 0.00e+00 |
| PSMG3 | -0.66 | 0.0098280 | 3.77e-03 |
| PSPC1 | -0.73 | 0.0024794 | 8.29e-04 |
| PSPH | -1.07 | 0.0003484 | 9.83e-05 |
| PSRC1 | -2.19 | 0.0000000 | 0.00e+00 |
| PSTPIP2 | 6.15 | 0.0003007 | 8.38e-05 |
| PTAFR | 5.50 | 0.0000000 | 0.00e+00 |
| PTAR1 | -0.96 | 0.0000885 | 2.28e-05 |
| PTBP2 | -1.56 | 0.0008012 | 2.41e-04 |
| PTCD1 | 0.58 | 0.0108610 | 4.21e-03 |
| PTCD3 | -1.20 | 0.0000000 | 0.00e+00 |
| PTCHD2 | 4.04 | 0.0670968 | 3.24e-02 |
| PTDSS1 | -1.13 | 0.0000000 | 0.00e+00 |
| PTGER2 | 3.45 | 0.0000000 | 0.00e+00 |
| PTGER4 | 4.36 | 0.0000000 | 0.00e+00 |
| PTGES | 1.35 | 0.0000000 | 0.00e+00 |
| PTGES3 | -0.32 | 0.0048507 | 1.72e-03 |
| PTGES3L-AARSD1 | -1.11 | 0.0000128 | 3.00e-06 |
| PTGIR | 4.72 | 0.0000000 | 0.00e+00 |
| PTGR2 | -1.26 | 0.0029610 | 1.00e-03 |
| PTGS2 | 4.34 | 0.0000000 | 0.00e+00 |
| PTH1R | -3.12 | 0.0604714 | 2.87e-02 |
| PTHLH | 3.39 | 0.0000000 | 0.00e+00 |
| PTK2B | 2.28 | 0.0000000 | 0.00e+00 |
| PTK7 | -1.97 | 0.0075411 | 2.81e-03 |
| PTMA | -0.97 | 0.0000000 | 0.00e+00 |
| PTMS | -0.46 | 0.0003770 | 1.07e-04 |
| PTP4A1 | -0.59 | 0.0000005 | 1.00e-07 |
| PTP4A2 | -0.45 | 0.0004057 | 1.16e-04 |
| PTP4A3 | 1.71 | 0.0136294 | 5.41e-03 |
| PTPN1 | 0.91 | 0.0000001 | 0.00e+00 |
| PTPN11 | -0.89 | 0.0000000 | 0.00e+00 |
| PTPN12 | 0.96 | 0.0000000 | 0.00e+00 |
| PTPN13 | -1.38 | 0.0005756 | 1.69e-04 |
| PTPN14 | 0.86 | 0.0003490 | 9.87e-05 |
| PTPN18 | -0.66 | 0.0347160 | 1.53e-02 |
| PTPN21 | 0.77 | 0.0071766 | 2.66e-03 |
| PTPN23 | 2.10 | 0.0000000 | 0.00e+00 |
| PTPN3 | -0.71 | 0.0025780 | 8.65e-04 |
| PTPN4 | -0.66 | 0.0932548 | 4.77e-02 |
| PTPN9 | 0.64 | 0.0001017 | 2.64e-05 |
| PTPRA | 1.24 | 0.0000000 | 0.00e+00 |
| PTPRB | 2.55 | 0.0780232 | 3.86e-02 |
| PTPRD | 2.74 | 0.0009099 | 2.77e-04 |
| PTPRF | 0.73 | 0.0000000 | 0.00e+00 |
| PTPRH | 4.77 | 0.0000000 | 0.00e+00 |
| PTPRJ | 0.87 | 0.0000000 | 0.00e+00 |
| PTPRK | 1.46 | 0.0000000 | 0.00e+00 |
| PTPRN2 | 3.84 | 0.0000000 | 0.00e+00 |
| PTPRS | -0.40 | 0.0388034 | 1.73e-02 |
| PTPRU | 0.70 | 0.0015520 | 4.94e-04 |
| PTRF | 0.99 | 0.0000000 | 0.00e+00 |
| PTRH2 | -0.49 | 0.0563356 | 2.65e-02 |
| PTTG1 | -2.36 | 0.0000000 | 0.00e+00 |
| PTTG1IP | 1.45 | 0.0000000 | 0.00e+00 |
| PTX3 | 7.87 | 0.0010356 | 3.19e-04 |
| PUM1 | 0.64 | 0.0000950 | 2.45e-05 |
| PUM2 | -0.60 | 0.0003758 | 1.07e-04 |
| PUS1 | -0.43 | 0.0522963 | 2.43e-02 |
| PUS7 | -0.69 | 0.0074509 | 2.77e-03 |
| PUS7L | -1.19 | 0.0090659 | 3.44e-03 |
| PUSL1 | -1.27 | 0.0151034 | 6.06e-03 |
| PVR | 1.52 | 0.0000000 | 0.00e+00 |
| PVRL1 | 0.64 | 0.0491014 | 2.26e-02 |
| PVRL2 | 2.84 | 0.0000000 | 0.00e+00 |
| PVRL3 | -1.03 | 0.0008948 | 2.72e-04 |
| PVRL4 | 2.71 | 0.0000151 | 3.50e-06 |
| PVT1 | 0.74 | 0.0229447 | 9.62e-03 |
| PWAR5 | -2.22 | 0.0400673 | 1.80e-02 |
| PWP1 | -0.46 | 0.0088540 | 3.35e-03 |
| PXDC1 | 1.79 | 0.0000000 | 0.00e+00 |
| PXDN | 1.26 | 0.0030826 | 1.05e-03 |
| PXK | 1.32 | 0.0009800 | 3.01e-04 |
| PXMP2 | -1.75 | 0.0002880 | 8.01e-05 |
| PXMP4 | -1.77 | 0.0053262 | 1.91e-03 |
| PXN | 1.03 | 0.0000000 | 0.00e+00 |
| PXN-AS1 | -1.73 | 0.0170684 | 6.93e-03 |
| PYCR1 | -1.58 | 0.0000000 | 0.00e+00 |
| PYCRL | -0.80 | 0.0760471 | 3.75e-02 |
| PYURF | -0.57 | 0.0213989 | 8.89e-03 |
| PYY | 5.83 | 0.0522626 | 2.43e-02 |
| QDPR | -0.98 | 0.0019069 | 6.20e-04 |
| QKI | 0.68 | 0.0001538 | 4.12e-05 |
| QPCT | 1.90 | 0.0000000 | 0.00e+00 |
| QPRT | -2.44 | 0.0373473 | 1.66e-02 |
| QRSL1 | -0.73 | 0.0461872 | 2.11e-02 |
| QSER1 | -1.07 | 0.0000001 | 0.00e+00 |
| QSOX1 | 1.55 | 0.0000000 | 0.00e+00 |
| QTRT1 | -0.77 | 0.0312590 | 1.36e-02 |
| R3HDM2 | 2.13 | 0.0000000 | 0.00e+00 |
| R3HDM4 | 1.72 | 0.0000000 | 0.00e+00 |
| RAB10 | -1.35 | 0.0000000 | 0.00e+00 |
| RAB11A | 0.80 | 0.0000000 | 0.00e+00 |
| RAB11B | 0.80 | 0.0000032 | 7.00e-07 |
| RAB11B-AS1 | 1.89 | 0.0765894 | 3.78e-02 |
| RAB11FIP1 | 1.46 | 0.0000000 | 0.00e+00 |
| RAB11FIP5 | 0.96 | 0.0000002 | 0.00e+00 |
| RAB12 | 0.65 | 0.0276059 | 1.18e-02 |
| RAB13 | 0.35 | 0.0973765 | 5.01e-02 |
| RAB14 | 0.38 | 0.0233409 | 9.82e-03 |
| RAB18 | -0.42 | 0.0597133 | 2.83e-02 |
| RAB1A | -0.42 | 0.0069416 | 2.56e-03 |
| RAB21 | 0.72 | 0.0018975 | 6.17e-04 |
| RAB23 | -0.99 | 0.0274190 | 1.17e-02 |
| RAB24 | 2.76 | 0.0000000 | 0.00e+00 |
| RAB26 | -4.87 | 0.0000000 | 0.00e+00 |
| RAB29 | 0.62 | 0.0335461 | 1.47e-02 |
| RAB2A | -0.60 | 0.0002863 | 7.96e-05 |
| RAB2B | 0.90 | 0.0017315 | 5.57e-04 |
| RAB31 | 1.32 | 0.0000000 | 0.00e+00 |
| RAB32 | 1.24 | 0.0000000 | 0.00e+00 |
| RAB35 | 0.66 | 0.0000007 | 1.00e-07 |
| RAB36 | -1.96 | 0.0216401 | 9.00e-03 |
| RAB37 | -3.04 | 0.0000001 | 0.00e+00 |
| RAB3B | 1.22 | 0.0000101 | 2.30e-06 |
| RAB3D | -1.94 | 0.0000001 | 0.00e+00 |
| RAB40B | -3.57 | 0.0000009 | 2.00e-07 |
| RAB43 | 2.02 | 0.0000000 | 0.00e+00 |
| RAB4A | -2.11 | 0.0000000 | 0.00e+00 |
| RAB4B | 1.86 | 0.0000000 | 0.00e+00 |
| RAB4B-EGLN2 | 1.21 | 0.0000000 | 0.00e+00 |
| RAB5A | 1.08 | 0.0000000 | 0.00e+00 |
| RAB5B | 0.99 | 0.0000000 | 0.00e+00 |
| RAB6A | 0.39 | 0.0276881 | 1.19e-02 |
| RAB7A | 0.40 | 0.0011509 | 3.58e-04 |
| RAB8B | 1.06 | 0.0000013 | 3.00e-07 |
| RAB9A | 1.58 | 0.0000000 | 0.00e+00 |
| RABAC1 | 0.66 | 0.0000041 | 9.00e-07 |
| RABEP1 | -0.40 | 0.0419123 | 1.89e-02 |
| RABEPK | -1.53 | 0.0000004 | 1.00e-07 |
| RABGEF1 | 0.92 | 0.0003481 | 9.82e-05 |
| RABGGTA | 1.33 | 0.0000048 | 1.00e-06 |
| RABGGTB | -0.52 | 0.0734835 | 3.60e-02 |
| RABL3 | -1.15 | 0.0002583 | 7.13e-05 |
| RAC1 | 0.99 | 0.0000000 | 0.00e+00 |
| RAC2 | 7.80 | 0.0012002 | 3.74e-04 |
| RAC3 | -2.30 | 0.0000064 | 1.40e-06 |
| RACGAP1 | -2.17 | 0.0000000 | 0.00e+00 |
| RACGAP1P | -2.39 | 0.0066193 | 2.43e-03 |
| RAD1 | -0.78 | 0.0135204 | 5.36e-03 |
| RAD17 | -1.57 | 0.0007839 | 2.35e-04 |
| RAD18 | -1.68 | 0.0000003 | 1.00e-07 |
| RAD21 | -0.84 | 0.0000000 | 0.00e+00 |
| RAD23A | 0.99 | 0.0000000 | 0.00e+00 |
| RAD23B | -0.27 | 0.0376898 | 1.68e-02 |
| RAD50 | -1.14 | 0.0000000 | 0.00e+00 |
| RAD51 | -2.04 | 0.0000117 | 2.70e-06 |
| RAD51AP1 | -2.61 | 0.0000000 | 0.00e+00 |
| RAD51C | -1.92 | 0.0000000 | 0.00e+00 |
| RAD51L3-RFFL | 1.52 | 0.0000000 | 0.00e+00 |
| RAD54B | -1.18 | 0.0001296 | 3.43e-05 |
| RAD54L | -2.51 | 0.0000004 | 1.00e-07 |
| RAD54L2 | 0.80 | 0.0312372 | 1.36e-02 |
| RAD9A | -0.81 | 0.0364929 | 1.62e-02 |
| RAE1 | -0.42 | 0.0494501 | 2.28e-02 |
| RAET1G | 1.14 | 0.0937755 | 4.80e-02 |
| RAET1L | 5.86 | 0.0000000 | 0.00e+00 |
| RAF1 | 0.37 | 0.0223709 | 9.35e-03 |
| RAI1 | 0.58 | 0.0001664 | 4.48e-05 |
| RAI14 | 0.78 | 0.0000001 | 0.00e+00 |
| RALA | 0.68 | 0.0016598 | 5.31e-04 |
| RALGPS2 | -0.71 | 0.0251373 | 1.07e-02 |
| RALY | 0.46 | 0.0002227 | 6.10e-05 |
| RAN | -1.22 | 0.0000000 | 0.00e+00 |
| RANBP1 | -1.88 | 0.0000000 | 0.00e+00 |
| RANBP10 | 0.93 | 0.0000820 | 2.10e-05 |
| RANBP2 | -1.28 | 0.0000000 | 0.00e+00 |
| RANBP6 | -0.99 | 0.0033849 | 1.16e-03 |
| RANGAP1 | 0.59 | 0.0000073 | 1.60e-06 |
| RAP1GAP | -3.41 | 0.0000000 | 0.00e+00 |
| RAP1GAP2 | 1.67 | 0.0000000 | 0.00e+00 |
| RAP2B | 2.14 | 0.0000000 | 0.00e+00 |
| RAPGEF1 | 1.64 | 0.0000000 | 0.00e+00 |
| RAPGEF3 | 6.06 | 0.0000000 | 0.00e+00 |
| RAPGEF6 | -0.83 | 0.0092868 | 3.54e-03 |
| RAPH1 | 1.92 | 0.0000000 | 0.00e+00 |
| RARA | 1.28 | 0.0000004 | 1.00e-07 |
| RARA-AS1 | 1.56 | 0.0224692 | 9.40e-03 |
| RARB | 0.78 | 0.0187717 | 7.70e-03 |
| RARRES1 | 3.62 | 0.0000000 | 0.00e+00 |
| RARRES3 | 5.71 | 0.0000000 | 0.00e+00 |
| RARS | -0.78 | 0.0000000 | 0.00e+00 |
| RARS2 | -1.16 | 0.0003562 | 1.01e-04 |
| RASA1 | -0.96 | 0.0013415 | 4.22e-04 |
| RASA2 | 1.57 | 0.0000000 | 0.00e+00 |
| RASA4 | 2.50 | 0.0000248 | 5.90e-06 |
| RASA4B | 2.38 | 0.0000002 | 0.00e+00 |
| RASA4CP | 4.17 | 0.0044944 | 1.59e-03 |
| RASAL2 | 0.63 | 0.0156669 | 6.30e-03 |
| RASD1 | 6.44 | 0.0000000 | 0.00e+00 |
| RASEF | 2.16 | 0.0000000 | 0.00e+00 |
| RASGEF1A | 1.48 | 0.0000031 | 7.00e-07 |
| RASGRP1 | 4.04 | 0.0000000 | 0.00e+00 |
| RASGRP3 | 6.28 | 0.0000000 | 0.00e+00 |
| RASSF3 | 1.25 | 0.0000000 | 0.00e+00 |
| RASSF4 | 1.42 | 0.0279450 | 1.20e-02 |
| RASSF5 | 4.72 | 0.0000000 | 0.00e+00 |
| RASSF8 | 1.11 | 0.0010912 | 3.38e-04 |
| RAVER1 | 1.65 | 0.0000000 | 0.00e+00 |
| RAVER2 | -1.62 | 0.0001410 | 3.75e-05 |
| RB1 | -0.92 | 0.0010460 | 3.22e-04 |
| RBBP4 | -0.64 | 0.0009264 | 2.83e-04 |
| RBBP7 | -1.12 | 0.0000000 | 0.00e+00 |
| RBBP8 | -1.33 | 0.0000007 | 1.00e-07 |
| RBCK1 | 1.67 | 0.0000000 | 0.00e+00 |
| RBFA | -0.62 | 0.0379947 | 1.69e-02 |
| RBFOX3 | 2.33 | 0.0000000 | 0.00e+00 |
| RBL1 | -2.58 | 0.0000000 | 0.00e+00 |
| RBM10 | 0.50 | 0.0025526 | 8.56e-04 |
| RBM12 | -0.61 | 0.0006155 | 1.81e-04 |
| RBM12B | -1.03 | 0.0012948 | 4.06e-04 |
| RBM14 | -0.76 | 0.0002457 | 6.77e-05 |
| RBM14-RBM4 | -0.58 | 0.0026105 | 8.76e-04 |
| RBM17 | -0.40 | 0.0534182 | 2.49e-02 |
| RBM19 | 0.44 | 0.0315846 | 1.38e-02 |
| RBM20 | 1.22 | 0.0008364 | 2.53e-04 |
| RBM22 | 0.68 | 0.0002090 | 5.69e-05 |
| RBM24 | 1.44 | 0.0000001 | 0.00e+00 |
| RBM25 | -0.94 | 0.0000006 | 1.00e-07 |
| RBM27 | -0.42 | 0.0704464 | 3.42e-02 |
| RBM28 | -0.54 | 0.0207595 | 8.60e-03 |
| RBM3 | -0.56 | 0.0000021 | 4.00e-07 |
| RBM33 | 0.76 | 0.0000581 | 1.46e-05 |
| RBM39 | -1.00 | 0.0000000 | 0.00e+00 |
| RBM4 | -0.60 | 0.0008073 | 2.43e-04 |
| RBM42 | 0.53 | 0.0000066 | 1.50e-06 |
| RBM45 | -1.42 | 0.0083444 | 3.14e-03 |
| RBM4B | -0.80 | 0.0184053 | 7.54e-03 |
| RBM5 | 0.46 | 0.0341414 | 1.50e-02 |
| RBM8A | -1.19 | 0.0000000 | 0.00e+00 |
| RBMS1 | 0.54 | 0.0124156 | 4.88e-03 |
| RBMS2 | 1.98 | 0.0000000 | 0.00e+00 |
| RBMX | -1.30 | 0.0000000 | 0.00e+00 |
| RBP1 | 5.03 | 0.0000000 | 0.00e+00 |
| RBPMS | 1.71 | 0.0000000 | 0.00e+00 |
| RBPMS2 | 1.30 | 0.0263332 | 1.12e-02 |
| RBSN | 0.93 | 0.0000635 | 1.60e-05 |
| RBX1 | -0.64 | 0.0241722 | 1.02e-02 |
| RCAN1 | 1.85 | 0.0000000 | 0.00e+00 |
| RCBTB1 | -0.83 | 0.0221366 | 9.24e-03 |
| RCC1 | -0.87 | 0.0000000 | 0.00e+00 |
| RCC2 | 0.39 | 0.0086348 | 3.26e-03 |
| RCCD1 | -1.60 | 0.0000000 | 0.00e+00 |
| RCN1 | 0.56 | 0.0000399 | 9.80e-06 |
| RCN2 | -2.02 | 0.0000000 | 0.00e+00 |
| RCOR1 | 1.60 | 0.0000000 | 0.00e+00 |
| RCSD1 | 10.12 | 0.0000049 | 1.10e-06 |
| RDH10 | 3.50 | 0.0000000 | 0.00e+00 |
| RDH11 | 1.02 | 0.0000000 | 0.00e+00 |
| RDH16 | 3.21 | 0.0737892 | 3.62e-02 |
| RDX | -1.29 | 0.0000000 | 0.00e+00 |
| REC8 | 1.79 | 0.0863321 | 4.35e-02 |
| RECQL | -1.39 | 0.0000000 | 0.00e+00 |
| RECQL4 | -2.32 | 0.0000000 | 0.00e+00 |
| REEP1 | -2.54 | 0.0681991 | 3.30e-02 |
| REEP2 | -2.22 | 0.0045219 | 1.60e-03 |
| REEP4 | -0.51 | 0.0322245 | 1.41e-02 |
| REEP5 | -0.42 | 0.0100243 | 3.85e-03 |
| REL | 1.16 | 0.0000464 | 1.15e-05 |
| RELA | 2.73 | 0.0000000 | 0.00e+00 |
| RELB | 3.91 | 0.0000000 | 0.00e+00 |
| RELL1 | 1.14 | 0.0005859 | 1.72e-04 |
| RELT | 1.60 | 0.0000003 | 1.00e-07 |
| REM1 | 7.37 | 0.0029927 | 1.02e-03 |
| REPIN1 | 0.69 | 0.0000002 | 0.00e+00 |
| RER1 | -0.30 | 0.0977429 | 5.03e-02 |
| RERE | 1.02 | 0.0000013 | 3.00e-07 |
| RET | 2.78 | 0.0014578 | 4.62e-04 |
| RETSAT | 1.17 | 0.0000000 | 0.00e+00 |
| REXO1 | 1.38 | 0.0000000 | 0.00e+00 |
| REXO2 | -1.03 | 0.0000167 | 3.90e-06 |
| RFC1 | -1.11 | 0.0000038 | 8.00e-07 |
| RFC2 | -0.95 | 0.0000032 | 7.00e-07 |
| RFC3 | -2.90 | 0.0000000 | 0.00e+00 |
| RFC4 | -2.83 | 0.0000000 | 0.00e+00 |
| RFC5 | -2.44 | 0.0000000 | 0.00e+00 |
| RFFL | 1.80 | 0.0000000 | 0.00e+00 |
| RFK | 0.43 | 0.0095801 | 3.66e-03 |
| RFNG | 0.79 | 0.0057510 | 2.08e-03 |
| RFT1 | -0.79 | 0.0090461 | 3.44e-03 |
| RFTN1 | 3.46 | 0.0000000 | 0.00e+00 |
| RFWD3 | -0.53 | 0.0015584 | 4.97e-04 |
| RFX1 | 1.54 | 0.0000095 | 2.20e-06 |
| RFX2 | 1.49 | 0.0000483 | 1.20e-05 |
| RFX3 | 1.55 | 0.0018354 | 5.94e-04 |
| RFX5 | 0.33 | 0.0639763 | 3.06e-02 |
| RFX7 | -0.86 | 0.0013368 | 4.20e-04 |
| RGAG1 | 4.85 | 0.0002566 | 7.08e-05 |
| RGMB | 0.96 | 0.0252187 | 1.07e-02 |
| RGP1 | 0.59 | 0.0837591 | 4.20e-02 |
| RGPD3 | -0.85 | 0.0979188 | 5.04e-02 |
| RGS2 | 1.06 | 0.0008989 | 2.74e-04 |
| RGS20 | 0.75 | 0.0078632 | 2.94e-03 |
| RGS22 | 5.61 | 0.0735043 | 3.61e-02 |
| RGS5 | -2.85 | 0.0342530 | 1.51e-02 |
| RGS9 | 5.71 | 0.0000000 | 0.00e+00 |
| RHBDD2 | 0.97 | 0.0000001 | 0.00e+00 |
| RHBDF1 | 1.90 | 0.0000000 | 0.00e+00 |
| RHBDF2 | 2.96 | 0.0000000 | 0.00e+00 |
| RHBDL2 | 2.28 | 0.0000119 | 2.70e-06 |
| RHCG | 6.39 | 0.0000000 | 0.00e+00 |
| RHEBL1 | 3.24 | 0.0000000 | 0.00e+00 |
| RHNO1 | -0.95 | 0.0000358 | 8.70e-06 |
| RHOB | 2.51 | 0.0000000 | 0.00e+00 |
| RHOBTB1 | -0.99 | 0.0004624 | 1.34e-04 |
| RHOBTB2 | -1.18 | 0.0000794 | 2.03e-05 |
| RHOBTB3 | -1.98 | 0.0000001 | 0.00e+00 |
| RHOC | 0.96 | 0.0000000 | 0.00e+00 |
| RHOF | 1.41 | 0.0000000 | 0.00e+00 |
| RHOG | 2.29 | 0.0000000 | 0.00e+00 |
| RHOQ | -0.42 | 0.0549387 | 2.57e-02 |
| RHOT1 | -0.73 | 0.0017457 | 5.62e-04 |
| RHOV | 0.69 | 0.0017241 | 5.54e-04 |
| RHPN1 | -1.03 | 0.0238008 | 1.00e-02 |
| RIBC2 | -2.70 | 0.0120554 | 4.72e-03 |
| RIC1 | -0.51 | 0.0733478 | 3.59e-02 |
| RIC8B | 0.53 | 0.0398185 | 1.79e-02 |
| RIF1 | -1.44 | 0.0000000 | 0.00e+00 |
| RILP | 0.93 | 0.0719698 | 3.52e-02 |
| RIMKLA | -1.66 | 0.0069198 | 2.55e-03 |
| RIMKLB | 1.44 | 0.0000000 | 0.00e+00 |
| RIN1 | 0.75 | 0.0845570 | 4.25e-02 |
| RIN2 | 0.82 | 0.0001127 | 2.95e-05 |
| RIN3 | 2.70 | 0.0000000 | 0.00e+00 |
| RINL | -1.34 | 0.0792750 | 3.93e-02 |
| RINT1 | -0.54 | 0.0657064 | 3.16e-02 |
| RIOK2 | -0.82 | 0.0288244 | 1.24e-02 |
| RIOK3 | 1.32 | 0.0000000 | 0.00e+00 |
| RIPK1 | 0.76 | 0.0036718 | 1.27e-03 |
| RIPK2 | 2.44 | 0.0000000 | 0.00e+00 |
| RIPK4 | 1.70 | 0.0000000 | 0.00e+00 |
| RIT1 | 0.56 | 0.0724081 | 3.54e-02 |
| RITA1 | -0.48 | 0.0637602 | 3.05e-02 |
| RLTPR | 2.15 | 0.0735918 | 3.61e-02 |
| RMDN1 | -0.91 | 0.0089189 | 3.38e-03 |
| RMI1 | -0.84 | 0.0313615 | 1.37e-02 |
| RMI2 | -2.10 | 0.0000002 | 0.00e+00 |
| RMND5B | 0.56 | 0.0375882 | 1.67e-02 |
| RN7SL1 | 1.99 | 0.0249154 | 1.06e-02 |
| RN7SL2 | 1.62 | 0.0522966 | 2.43e-02 |
| RNASEH1 | -0.54 | 0.0177376 | 7.24e-03 |
| RNASEH1-AS1 | -1.65 | 0.0000044 | 9.00e-07 |
| RNASEH2A | -1.78 | 0.0000000 | 0.00e+00 |
| RNASEH2B | -1.49 | 0.0000468 | 1.16e-05 |
| RNASEH2C | -1.69 | 0.0000016 | 3.00e-07 |
| RNASEK | 1.66 | 0.0000000 | 0.00e+00 |
| RNASEK-C17orf49 | 0.89 | 0.0000000 | 0.00e+00 |
| RND1 | 5.63 | 0.0000000 | 0.00e+00 |
| RNF10 | 0.90 | 0.0000000 | 0.00e+00 |
| RNF103 | 0.67 | 0.0009916 | 3.05e-04 |
| RNF103-CHMP3 | -0.57 | 0.0000501 | 1.25e-05 |
| RNF111 | 0.82 | 0.0000212 | 5.00e-06 |
| RNF114 | 1.18 | 0.0000000 | 0.00e+00 |
| RNF115 | 0.92 | 0.0000779 | 1.99e-05 |
| RNF122 | 2.77 | 0.0000000 | 0.00e+00 |
| RNF130 | -0.43 | 0.0865481 | 4.37e-02 |
| RNF139 | -1.27 | 0.0000125 | 2.90e-06 |
| RNF14 | -0.41 | 0.0798648 | 3.97e-02 |
| RNF141 | -1.34 | 0.0000253 | 6.10e-06 |
| RNF144A | 0.98 | 0.0009864 | 3.03e-04 |
| RNF144A-AS1 | 2.84 | 0.0081400 | 3.06e-03 |
| RNF144B | 2.02 | 0.0004032 | 1.16e-04 |
| RNF145 | 0.79 | 0.0000000 | 0.00e+00 |
| RNF149 | 2.03 | 0.0000000 | 0.00e+00 |
| RNF167 | -0.41 | 0.0201017 | 8.31e-03 |
| RNF169 | 0.84 | 0.0019747 | 6.44e-04 |
| RNF181 | 0.78 | 0.0000002 | 0.00e+00 |
| RNF185 | 0.85 | 0.0004193 | 1.20e-04 |
| RNF187 | 0.79 | 0.0000000 | 0.00e+00 |
| RNF19B | 4.47 | 0.0000000 | 0.00e+00 |
| RNF20 | -0.77 | 0.0004420 | 1.27e-04 |
| RNF213 | 0.99 | 0.0000000 | 0.00e+00 |
| RNF214 | 0.94 | 0.0084743 | 3.19e-03 |
| RNF216 | 0.76 | 0.0000081 | 1.80e-06 |
| RNF216P1 | 0.82 | 0.0137281 | 5.46e-03 |
| RNF217 | 1.08 | 0.0021904 | 7.24e-04 |
| RNF219 | -0.85 | 0.0294499 | 1.27e-02 |
| RNF24 | 2.97 | 0.0000000 | 0.00e+00 |
| RNF25 | 1.03 | 0.0000106 | 2.40e-06 |
| RNF26 | -0.59 | 0.0173953 | 7.08e-03 |
| RNF31 | 1.31 | 0.0000000 | 0.00e+00 |
| RNF34 | 0.48 | 0.0411631 | 1.85e-02 |
| RNF40 | 1.05 | 0.0000000 | 0.00e+00 |
| RNF41 | 0.86 | 0.0000163 | 3.80e-06 |
| RNF44 | 1.25 | 0.0000000 | 0.00e+00 |
| RNF6 | -0.59 | 0.0348891 | 1.54e-02 |
| RNFT2 | -1.59 | 0.0147088 | 5.88e-03 |
| RNGTT | -1.27 | 0.0000231 | 5.50e-06 |
| RNPEPL1 | 1.41 | 0.0000000 | 0.00e+00 |
| RNPS1 | 0.39 | 0.0047629 | 1.69e-03 |
| RNU1-1 | 2.27 | 0.0000001 | 0.00e+00 |
| RNU1-2 | 2.27 | 0.0000001 | 0.00e+00 |
| RNU1-27P | 2.27 | 0.0000001 | 0.00e+00 |
| RNU1-28P | 2.27 | 0.0000001 | 0.00e+00 |
| RNU1-3 | 2.27 | 0.0000001 | 0.00e+00 |
| RNU1-4 | 2.27 | 0.0000001 | 0.00e+00 |
| RNVU1-18 | 2.27 | 0.0000001 | 0.00e+00 |
| ROBO1 | 2.14 | 0.0000000 | 0.00e+00 |
| ROBO3 | 2.71 | 0.0000010 | 2.00e-07 |
| ROBO4 | 7.17 | 0.0044928 | 1.58e-03 |
| ROCK2 | -1.12 | 0.0000000 | 0.00e+00 |
| ROGDI | -1.36 | 0.0035713 | 1.23e-03 |
| ROMO1 | -0.34 | 0.0938373 | 4.80e-02 |
| ROR1 | -1.46 | 0.0000299 | 7.20e-06 |
| ROR1-AS1 | 5.61 | 0.0750936 | 3.69e-02 |
| RP2 | 1.05 | 0.0000325 | 7.90e-06 |
| RPA1 | -1.94 | 0.0000000 | 0.00e+00 |
| RPA2 | -0.80 | 0.0014752 | 4.68e-04 |
| RPA3 | -2.02 | 0.0000000 | 0.00e+00 |
| RPAP1 | 0.69 | 0.0293936 | 1.27e-02 |
| RPAP3 | -1.40 | 0.0000000 | 0.00e+00 |
| RPARP-AS1 | -2.55 | 0.0020822 | 6.84e-04 |
| RPE | -1.19 | 0.0000000 | 0.00e+00 |
| RPEL1 | -1.51 | 0.0003996 | 1.14e-04 |
| RPF1 | -1.04 | 0.0002095 | 5.71e-05 |
| RPF2 | -1.57 | 0.0000039 | 8.00e-07 |
| RPGRIP1L | -2.80 | 0.0000000 | 0.00e+00 |
| RPH3AL | 0.91 | 0.0077308 | 2.89e-03 |
| RPL10A | -0.66 | 0.0000000 | 0.00e+00 |
| RPL13A | 0.28 | 0.0020965 | 6.90e-04 |
| RPL13AP20 | 0.25 | 0.0638162 | 3.05e-02 |
| RPL13AP5 | 0.28 | 0.0025448 | 8.53e-04 |
| RPL14 | -0.43 | 0.0008834 | 2.68e-04 |
| RPL15 | -0.36 | 0.0000474 | 1.18e-05 |
| RPL17 | -0.57 | 0.0000073 | 1.60e-06 |
| RPL17-C18orf32 | -0.50 | 0.0001431 | 3.81e-05 |
| RPL18 | 0.23 | 0.0104254 | 4.02e-03 |
| RPL18A | 0.57 | 0.0000003 | 1.00e-07 |
| RPL22 | -0.84 | 0.0000000 | 0.00e+00 |
| RPL22L1 | -1.74 | 0.0000000 | 0.00e+00 |
| RPL23 | -0.72 | 0.0000000 | 0.00e+00 |
| RPL23A | 0.84 | 0.0000000 | 0.00e+00 |
| RPL23P8 | -1.00 | 0.0000002 | 0.00e+00 |
| RPL24 | -0.42 | 0.0001113 | 2.91e-05 |
| RPL26L1 | -1.24 | 0.0000063 | 1.40e-06 |
| RPL28 | 2.00 | 0.0000000 | 0.00e+00 |
| RPL3 | -0.77 | 0.0000000 | 0.00e+00 |
| RPL35 | -0.28 | 0.0057533 | 2.08e-03 |
| RPL35A | -0.28 | 0.0556636 | 2.61e-02 |
| RPL36 | 0.23 | 0.0431912 | 1.96e-02 |
| RPL36A-HNRNPH2 | -0.45 | 0.0040197 | 1.40e-03 |
| RPL36AL | 0.32 | 0.0312647 | 1.36e-02 |
| RPL37 | 0.34 | 0.0007708 | 2.31e-04 |
| RPL37A | 0.42 | 0.0000112 | 2.50e-06 |
| RPL38 | 0.27 | 0.0084627 | 3.19e-03 |
| RPL39L | -0.65 | 0.0178292 | 7.28e-03 |
| RPL4 | -0.71 | 0.0000000 | 0.00e+00 |
| RPL5 | -0.93 | 0.0000000 | 0.00e+00 |
| RPL6 | -0.82 | 0.0000000 | 0.00e+00 |
| RPL7 | -0.65 | 0.0000003 | 1.00e-07 |
| RPL7A | -0.26 | 0.0096659 | 3.70e-03 |
| RPLP0 | -0.40 | 0.0000003 | 1.00e-07 |
| RPLP1 | 0.26 | 0.0012797 | 4.01e-04 |
| RPLP2 | 0.60 | 0.0000000 | 0.00e+00 |
| RPP30 | -1.10 | 0.0001350 | 3.58e-05 |
| RPP38 | 0.64 | 0.0182895 | 7.48e-03 |
| RPP40 | -1.88 | 0.0002079 | 5.66e-05 |
| RPRD1A | -0.61 | 0.0103204 | 3.98e-03 |
| RPRD2 | 0.99 | 0.0000006 | 1.00e-07 |
| RPRM | 2.87 | 0.0654263 | 3.14e-02 |
| RPS11 | 0.27 | 0.0007244 | 2.16e-04 |
| RPS12 | 0.56 | 0.0000000 | 0.00e+00 |
| RPS14P3 | -0.39 | 0.0910402 | 4.63e-02 |
| RPS16 | 0.82 | 0.0000000 | 0.00e+00 |
| RPS19 | 0.64 | 0.0000000 | 0.00e+00 |
| RPS19BP1 | -0.78 | 0.0012608 | 3.95e-04 |
| RPS2 | -0.94 | 0.0000000 | 0.00e+00 |
| RPS20 | 0.26 | 0.0070423 | 2.60e-03 |
| RPS25 | 0.28 | 0.0446991 | 2.03e-02 |
| RPS27 | 0.57 | 0.0003583 | 1.02e-04 |
| RPS27L | -0.47 | 0.0024220 | 8.08e-04 |
| RPS3A | -0.56 | 0.0000444 | 1.10e-05 |
| RPS4X | -0.41 | 0.0000022 | 5.00e-07 |
| RPS4Y1 | -1.16 | 0.0000000 | 0.00e+00 |
| RPS6 | -0.42 | 0.0000002 | 0.00e+00 |
| RPS6KA2 | 4.23 | 0.0035869 | 1.24e-03 |
| RPS6KA3 | 0.29 | 0.0350296 | 1.55e-02 |
| RPS6KB1 | -0.49 | 0.0176392 | 7.20e-03 |
| RPS6KB2 | -0.43 | 0.0613592 | 2.92e-02 |
| RPS6KC1 | 1.89 | 0.0000000 | 0.00e+00 |
| RPS7 | -0.54 | 0.0000002 | 0.00e+00 |
| RPSA | -0.69 | 0.0281541 | 1.21e-02 |
| RPSAP58 | -0.85 | 0.0000000 | 0.00e+00 |
| RPTOR | 0.59 | 0.0016776 | 5.38e-04 |
| RQCD1 | -0.56 | 0.0032094 | 1.10e-03 |
| RRAD | 4.79 | 0.0000000 | 0.00e+00 |
| RRAGC | 1.54 | 0.0000000 | 0.00e+00 |
| RRAS | 0.60 | 0.0001557 | 4.18e-05 |
| RRAS2 | 0.71 | 0.0104280 | 4.02e-03 |
| RRBP1 | 1.98 | 0.0000000 | 0.00e+00 |
| RREB1 | 2.12 | 0.0000000 | 0.00e+00 |
| RRM1 | -2.42 | 0.0000000 | 0.00e+00 |
| RRM2 | -3.50 | 0.0000000 | 0.00e+00 |
| RRM2B | -0.48 | 0.0515700 | 2.39e-02 |
| RRN3 | -0.60 | 0.0215246 | 8.95e-03 |
| RRNAD1 | 1.07 | 0.0000035 | 8.00e-07 |
| RRP1 | 0.50 | 0.0192438 | 7.91e-03 |
| RRP12 | 0.88 | 0.0000005 | 1.00e-07 |
| RRP15 | -0.74 | 0.0434989 | 1.97e-02 |
| RRP1B | -0.88 | 0.0000117 | 2.70e-06 |
| RRP9 | -0.80 | 0.0042037 | 1.47e-03 |
| RRS1 | -0.68 | 0.0003334 | 9.37e-05 |
| RSAD1 | -1.08 | 0.0000035 | 7.00e-07 |
| RSAD2 | 9.43 | 0.0000000 | 0.00e+00 |
| RSBN1 | 0.96 | 0.0314682 | 1.37e-02 |
| RSL1D1 | -0.44 | 0.0001926 | 5.22e-05 |
| RSL24D1 | -1.11 | 0.0000000 | 0.00e+00 |
| RSPH3 | 1.12 | 0.0911632 | 4.64e-02 |
| RSPO3 | 0.40 | 0.0217814 | 9.07e-03 |
| RSPRY1 | 0.61 | 0.0029179 | 9.87e-04 |
| RSRC1 | -1.15 | 0.0009614 | 2.95e-04 |
| RSRC2 | 0.58 | 0.0019882 | 6.50e-04 |
| RSU1 | -0.44 | 0.0439600 | 2.00e-02 |
| RTCB | -0.30 | 0.0827540 | 4.14e-02 |
| RTEL1 | -0.66 | 0.0647417 | 3.10e-02 |
| RTEL1-TNFRSF6B | -0.60 | 0.0986403 | 5.08e-02 |
| RTKN2 | -2.67 | 0.0462670 | 2.12e-02 |
| RTN2 | 1.22 | 0.0008976 | 2.73e-04 |
| RTN4 | 0.60 | 0.0000000 | 0.00e+00 |
| RTN4IP1 | -1.62 | 0.0020336 | 6.66e-04 |
| RTP4 | 9.68 | 0.0000147 | 3.40e-06 |
| RTTN | -1.55 | 0.0000001 | 0.00e+00 |
| RUFY4 | 9.14 | 0.0000561 | 1.41e-05 |
| RUNDC1 | 0.98 | 0.0000065 | 1.40e-06 |
| RUNX1 | 2.69 | 0.0000000 | 0.00e+00 |
| RUSC2 | 1.84 | 0.0000000 | 0.00e+00 |
| RUVBL1 | -1.43 | 0.0000000 | 0.00e+00 |
| RUVBL2 | -0.84 | 0.0000000 | 0.00e+00 |
| RWDD1 | -0.80 | 0.0151034 | 6.06e-03 |
| RWDD4 | -1.24 | 0.0104176 | 4.02e-03 |
| RXRA | 0.33 | 0.0838837 | 4.21e-02 |
| RYBP | 0.53 | 0.0593474 | 2.81e-02 |
| RYR2 | 4.09 | 0.0060639 | 2.21e-03 |
| S100A10 | 0.55 | 0.0000000 | 0.00e+00 |
| S100A11 | 1.80 | 0.0000000 | 0.00e+00 |
| S100A2 | 1.42 | 0.0367492 | 1.63e-02 |
| S100A3 | -0.63 | 0.0175565 | 7.16e-03 |
| S100A4 | -2.63 | 0.0000000 | 0.00e+00 |
| S100A7 | 5.61 | 0.0750936 | 3.69e-02 |
| S100A8 | 6.49 | 0.0170469 | 6.92e-03 |
| S100A9 | 8.02 | 0.0000003 | 1.00e-07 |
| S100P | 1.79 | 0.0000000 | 0.00e+00 |
| S1PR1 | 6.14 | 0.0313259 | 1.36e-02 |
| S1PR2 | 1.68 | 0.0179721 | 7.34e-03 |
| S1PR5 | -2.11 | 0.0318366 | 1.39e-02 |
| SAA1 | 8.93 | 0.0000000 | 0.00e+00 |
| SAA2 | 8.08 | 0.0000000 | 0.00e+00 |
| SAA2-SAA4 | 7.32 | 0.0000000 | 0.00e+00 |
| SAA4 | 7.47 | 0.0024324 | 8.12e-04 |
| SAAL1 | -1.24 | 0.0000022 | 5.00e-07 |
| SAC3D1 | -1.61 | 0.0000003 | 1.00e-07 |
| SACM1L | -0.92 | 0.0061355 | 2.23e-03 |
| SACS | -1.03 | 0.0000018 | 4.00e-07 |
| SAE1 | -1.02 | 0.0000000 | 0.00e+00 |
| SALL1 | 3.43 | 0.0000014 | 3.00e-07 |
| SAMD11 | -0.69 | 0.0195821 | 8.07e-03 |
| SAMD15 | -2.83 | 0.0506298 | 2.34e-02 |
| SAMD4A | 3.03 | 0.0000000 | 0.00e+00 |
| SAMD4B | 1.85 | 0.0000000 | 0.00e+00 |
| SAMD8 | 0.93 | 0.0000109 | 2.50e-06 |
| SAMD9 | 5.12 | 0.0000000 | 0.00e+00 |
| SAMD9L | 5.43 | 0.0000000 | 0.00e+00 |
| SAMHD1 | 2.05 | 0.0000000 | 0.00e+00 |
| SAMM50 | -1.37 | 0.0000000 | 0.00e+00 |
| SAP130 | 1.03 | 0.0000100 | 2.20e-06 |
| SAP30BP | 1.06 | 0.0000000 | 0.00e+00 |
| SAPCD2 | -4.08 | 0.0000000 | 0.00e+00 |
| SAR1A | 1.94 | 0.0000000 | 0.00e+00 |
| SARM1 | 1.45 | 0.0000000 | 0.00e+00 |
| SARS | 0.63 | 0.0000150 | 3.50e-06 |
| SART1 | 0.57 | 0.0002668 | 7.39e-05 |
| SASH1 | 0.76 | 0.0418205 | 1.89e-02 |
| SASS6 | -1.82 | 0.0008594 | 2.60e-04 |
| SAT1 | 3.12 | 0.0000000 | 0.00e+00 |
| SAV1 | 2.42 | 0.0000000 | 0.00e+00 |
| SAYSD1 | -1.03 | 0.0480393 | 2.21e-02 |
| SBDS | 0.51 | 0.0028286 | 9.56e-04 |
| SBF1 | 0.86 | 0.0000001 | 0.00e+00 |
| SBK1 | 2.23 | 0.0194887 | 8.02e-03 |
| SBNO1 | -1.56 | 0.0000000 | 0.00e+00 |
| SBNO2 | 2.34 | 0.0000000 | 0.00e+00 |
| SBSN | 7.41 | 0.0000000 | 0.00e+00 |
| SCAF1 | 1.55 | 0.0000000 | 0.00e+00 |
| SCAF11 | -0.43 | 0.0148223 | 5.93e-03 |
| SCAF4 | 1.62 | 0.0000000 | 0.00e+00 |
| SCAF8 | 0.65 | 0.0050811 | 1.82e-03 |
| SCAI | -1.45 | 0.0192009 | 7.90e-03 |
| SCAMP2 | 1.11 | 0.0000000 | 0.00e+00 |
| SCAMP3 | 0.47 | 0.0545374 | 2.55e-02 |
| SCAMP4 | 1.46 | 0.0000000 | 0.00e+00 |
| SCAND1 | 0.47 | 0.0339660 | 1.49e-02 |
| SCAP | 0.45 | 0.0259857 | 1.11e-02 |
| SCARA3 | -1.02 | 0.0000000 | 0.00e+00 |
| SCARA5 | -1.33 | 0.0000796 | 2.03e-05 |
| SCARB2 | 1.00 | 0.0000000 | 0.00e+00 |
| SCARF1 | 3.95 | 0.0783729 | 3.88e-02 |
| SCARNA7 | 0.61 | 0.0669107 | 3.23e-02 |
| SCD | -0.54 | 0.0000000 | 0.00e+00 |
| SCFD1 | -1.07 | 0.0000753 | 1.91e-05 |
| SCFD2 | -1.97 | 0.0000550 | 1.38e-05 |
| SCHIP1 | 1.30 | 0.0000001 | 0.00e+00 |
| SCLT1 | -1.19 | 0.0389172 | 1.74e-02 |
| SCLY | -0.76 | 0.0137082 | 5.45e-03 |
| SCMH1 | 1.09 | 0.0001127 | 2.95e-05 |
| SCML2 | -0.86 | 0.0494466 | 2.28e-02 |
| SCN1B | 1.65 | 0.0000001 | 0.00e+00 |
| SCN3A | 4.99 | 0.0000000 | 0.00e+00 |
| SCN9A | -2.26 | 0.0000120 | 2.80e-06 |
| SCNM1 | 1.72 | 0.0000000 | 0.00e+00 |
| SCNN1B | 3.23 | 0.0003060 | 8.54e-05 |
| SCNN1G | 3.58 | 0.0062675 | 2.29e-03 |
| SCO1 | -0.66 | 0.0004065 | 1.17e-04 |
| SCO2 | 1.29 | 0.0000001 | 0.00e+00 |
| SCOC | -1.49 | 0.0000068 | 1.50e-06 |
| SCP2 | -1.16 | 0.0000119 | 2.70e-06 |
| SCPEP1 | -0.60 | 0.0315743 | 1.38e-02 |
| SCRN1 | 0.69 | 0.0000000 | 0.00e+00 |
| SCRN3 | -1.52 | 0.0006415 | 1.90e-04 |
| SCTR | 6.44 | 0.0198508 | 8.19e-03 |
| SCUBE3 | 3.17 | 0.0834423 | 4.18e-02 |
| SCYL1 | 0.73 | 0.0000014 | 3.00e-07 |
| SCYL3 | 0.86 | 0.0396963 | 1.78e-02 |
| SDC4 | 4.49 | 0.0000000 | 0.00e+00 |
| SDCBP | 1.41 | 0.0000000 | 0.00e+00 |
| SDCCAG3 | -0.84 | 0.0001704 | 4.60e-05 |
| SDF4 | 0.39 | 0.0066354 | 2.43e-03 |
| SDHAF3 | -2.01 | 0.0001095 | 2.86e-05 |
| SDHB | -0.53 | 0.0034204 | 1.18e-03 |
| SDHC | -1.12 | 0.0000000 | 0.00e+00 |
| SDHD | -0.99 | 0.0003420 | 9.64e-05 |
| SDK1 | 3.03 | 0.0000978 | 2.53e-05 |
| SDK2 | 1.84 | 0.0017417 | 5.60e-04 |
| SDPR | -1.34 | 0.0000314 | 7.60e-06 |
| SDR16C5 | 4.16 | 0.0000000 | 0.00e+00 |
| SEC11A | -0.41 | 0.0045129 | 1.59e-03 |
| SEC11C | -0.50 | 0.0871611 | 4.40e-02 |
| SEC14L1 | 0.72 | 0.0003878 | 1.11e-04 |
| SEC14L2 | 1.38 | 0.0000224 | 5.30e-06 |
| SEC14L4 | -1.92 | 0.0020471 | 6.71e-04 |
| SEC16A | 0.68 | 0.0000014 | 3.00e-07 |
| SEC23A | -0.41 | 0.0072594 | 2.69e-03 |
| SEC23IP | -0.33 | 0.0887506 | 4.50e-02 |
| SEC24A | 0.66 | 0.0085872 | 3.24e-03 |
| SEC24C | 0.52 | 0.0001233 | 3.25e-05 |
| SEC24D | 1.17 | 0.0000023 | 5.00e-07 |
| SEC31A | 0.50 | 0.0009864 | 3.03e-04 |
| SEC61A1 | 0.51 | 0.0000058 | 1.30e-06 |
| SEC63 | -0.68 | 0.0067340 | 2.47e-03 |
| SECISBP2L | 0.63 | 0.0158791 | 6.40e-03 |
| SECTM1 | 4.70 | 0.0000000 | 0.00e+00 |
| SEH1L | -1.13 | 0.0000511 | 1.27e-05 |
| SEL1L | -0.49 | 0.0026177 | 8.79e-04 |
| SELE | 8.42 | 0.0003023 | 8.43e-05 |
| SELK | 0.86 | 0.0242582 | 1.02e-02 |
| SELM | 2.44 | 0.0000000 | 0.00e+00 |
| SELT | -0.85 | 0.0003396 | 9.56e-05 |
| SEMA3A | 0.64 | 0.0094938 | 3.62e-03 |
| SEMA3B | 1.02 | 0.0000228 | 5.40e-06 |
| SEMA3D | 6.08 | 0.0003785 | 1.08e-04 |
| SEMA3E | -3.49 | 0.0018501 | 5.99e-04 |
| SEMA3F | 0.85 | 0.0601960 | 2.86e-02 |
| SEMA4B | 2.07 | 0.0000000 | 0.00e+00 |
| SEMA4C | 1.59 | 0.0000000 | 0.00e+00 |
| SEMA4G | -1.15 | 0.0017975 | 5.80e-04 |
| SEMA6B | -2.02 | 0.0000305 | 7.40e-06 |
| SEMA7A | 6.39 | 0.0000000 | 0.00e+00 |
| SENP2 | 0.49 | 0.0980345 | 5.05e-02 |
| SENP3-EIF4A1 | -0.62 | 0.0000000 | 0.00e+00 |
| SENP6 | -0.51 | 0.0539285 | 2.52e-02 |
| SEP15 | -0.49 | 0.0136492 | 5.42e-03 |
| SEPHS1 | -1.66 | 0.0000000 | 0.00e+00 |
| SEPHS2 | -0.34 | 0.0526501 | 2.45e-02 |
| SEPT10 | -0.51 | 0.0566797 | 2.67e-02 |
| SEPT11 | -0.47 | 0.0199104 | 8.22e-03 |
| SEPT2 | -1.08 | 0.0000000 | 0.00e+00 |
| SEPT4 | 4.51 | 0.0000000 | 0.00e+00 |
| SEPT5 | -1.48 | 0.0066755 | 2.45e-03 |
| SEPT5-GP1BB | -1.47 | 0.0064195 | 2.35e-03 |
| SEPT7 | -1.32 | 0.0000000 | 0.00e+00 |
| SEPT8 | 0.39 | 0.0708800 | 3.45e-02 |
| SEPT9 | 0.43 | 0.0000054 | 1.20e-06 |
| SEPW1 | -0.75 | 0.0005102 | 1.48e-04 |
| SERBP1 | -1.28 | 0.0000000 | 0.00e+00 |
| SERF2-C15ORF63 | 0.37 | 0.0296742 | 1.28e-02 |
| SERINC2 | 1.90 | 0.0000000 | 0.00e+00 |
| SERPINA1 | 5.71 | 0.0000000 | 0.00e+00 |
| SERPINA3 | 6.22 | 0.0000000 | 0.00e+00 |
| SERPINB1 | 1.29 | 0.0000000 | 0.00e+00 |
| SERPINB3 | 7.81 | 0.0000007 | 1.00e-07 |
| SERPINB4 | 9.83 | 0.0000101 | 2.30e-06 |
| SERPINB6 | -0.37 | 0.0593533 | 2.81e-02 |
| SERPINB7 | 7.18 | 0.0000000 | 0.00e+00 |
| SERPINB8 | 2.75 | 0.0000000 | 0.00e+00 |
| SERPINB9 | 1.32 | 0.0000000 | 0.00e+00 |
| SERPINE1 | 1.80 | 0.0000000 | 0.00e+00 |
| SERPINE2 | 2.12 | 0.0000000 | 0.00e+00 |
| SERPING1 | 11.54 | 0.0000001 | 0.00e+00 |
| SERPINH1 | 0.55 | 0.0000977 | 2.53e-05 |
| SERTAD1 | 1.53 | 0.0000000 | 0.00e+00 |
| SERTAD2 | 1.46 | 0.0000000 | 0.00e+00 |
| SERTAD3 | 1.19 | 0.0000000 | 0.00e+00 |
| SERTAD4-AS1 | 2.17 | 0.0984199 | 5.07e-02 |
| SESN1 | -1.05 | 0.0717550 | 3.50e-02 |
| SESN2 | 3.75 | 0.0000000 | 0.00e+00 |
| SESN3 | -1.77 | 0.0384899 | 1.72e-02 |
| SESTD1 | 0.61 | 0.0233595 | 9.83e-03 |
| SET | -0.69 | 0.0000000 | 0.00e+00 |
| SETD1A | 0.76 | 0.0008683 | 2.63e-04 |
| SETD1B | 0.94 | 0.0000440 | 1.09e-05 |
| SETD3 | -0.52 | 0.0071421 | 2.64e-03 |
| SETD5 | 0.49 | 0.0011270 | 3.49e-04 |
| SETD6 | -1.46 | 0.0097791 | 3.75e-03 |
| SETD7 | -1.52 | 0.0000000 | 0.00e+00 |
| SETD8 | -0.57 | 0.0506815 | 2.35e-02 |
| SETDB1 | 0.50 | 0.0360386 | 1.60e-02 |
| SETMAR | -1.44 | 0.0679441 | 3.28e-02 |
| SETSIP | -0.84 | 0.0131344 | 5.19e-03 |
| SEZ6L2 | 2.60 | 0.0000000 | 0.00e+00 |
| SF1 | 0.53 | 0.0000025 | 5.00e-07 |
| SF3A1 | 1.48 | 0.0000000 | 0.00e+00 |
| SF3A2 | 1.35 | 0.0000000 | 0.00e+00 |
| SF3A3 | -0.56 | 0.0020509 | 6.73e-04 |
| SF3B1 | -0.66 | 0.0000000 | 0.00e+00 |
| SF3B2 | 0.73 | 0.0000000 | 0.00e+00 |
| SF3B4 | 0.53 | 0.0006233 | 1.84e-04 |
| SF3B5 | -0.78 | 0.0000059 | 1.30e-06 |
| SF3B6 | -1.04 | 0.0000000 | 0.00e+00 |
| SFI1 | -1.09 | 0.0442174 | 2.01e-02 |
| SFRP1 | -1.57 | 0.0000001 | 0.00e+00 |
| SFRP4 | -3.08 | 0.0055345 | 1.99e-03 |
| SFSWAP | 0.46 | 0.0535297 | 2.50e-02 |
| SFT2D3 | -1.25 | 0.0169127 | 6.86e-03 |
| SFTPA2 | 7.25 | 0.0038140 | 1.32e-03 |
| SFXN1 | -0.82 | 0.0000044 | 9.00e-07 |
| SFXN2 | -1.78 | 0.0500285 | 2.31e-02 |
| SFXN4 | -1.45 | 0.0000011 | 2.00e-07 |
| SGCB | -0.58 | 0.0747794 | 3.67e-02 |
| SGCD | -2.65 | 0.0000000 | 0.00e+00 |
| SGK2 | -4.03 | 0.0000018 | 4.00e-07 |
| SGK223 | 0.51 | 0.0123978 | 4.87e-03 |
| SGK494 | -1.84 | 0.0118969 | 4.66e-03 |
| SGMS1 | 0.57 | 0.0241900 | 1.02e-02 |
| SGMS2 | 1.06 | 0.0002897 | 8.06e-05 |
| SGOL1 | -2.14 | 0.0000001 | 0.00e+00 |
| SGOL2 | -3.51 | 0.0000000 | 0.00e+00 |
| SGPL1 | 0.66 | 0.0001015 | 2.64e-05 |
| SGPP2 | 4.01 | 0.0000000 | 0.00e+00 |
| SGSH | 0.83 | 0.0175185 | 7.14e-03 |
| SH2B3 | 3.42 | 0.0000000 | 0.00e+00 |
| SH2D1B | 6.07 | 0.0000003 | 0.00e+00 |
| SH2D3A | 1.16 | 0.0001607 | 4.32e-05 |
| SH2D3C | 3.13 | 0.0901372 | 4.58e-02 |
| SH2D5 | 0.65 | 0.0660294 | 3.18e-02 |
| SH3BGRL | -0.85 | 0.0046552 | 1.65e-03 |
| SH3BGRL3 | 1.73 | 0.0000000 | 0.00e+00 |
| SH3BP2 | 1.69 | 0.0000001 | 0.00e+00 |
| SH3BP5 | -0.78 | 0.0130139 | 5.14e-03 |
| SH3BP5L | 0.90 | 0.0001483 | 3.96e-05 |
| SH3GLB1 | 0.85 | 0.0000062 | 1.40e-06 |
| SH3KBP1 | 1.79 | 0.0000000 | 0.00e+00 |
| SH3PXD2A | 1.28 | 0.0000026 | 5.00e-07 |
| SH3PXD2B | 0.93 | 0.0000064 | 1.40e-06 |
| SH3RF1 | 1.86 | 0.0000000 | 0.00e+00 |
| SH3RF3 | 4.97 | 0.0000021 | 4.00e-07 |
| SH3TC1 | 1.03 | 0.0077686 | 2.90e-03 |
| SH3YL1 | -1.41 | 0.0977381 | 5.03e-02 |
| SHANK2 | 0.55 | 0.0216763 | 9.02e-03 |
| SHANK3 | 0.84 | 0.0805934 | 4.01e-02 |
| SHB | 2.23 | 0.0000000 | 0.00e+00 |
| SHC1 | 0.31 | 0.0021489 | 7.09e-04 |
| SHC2 | 3.72 | 0.0000049 | 1.00e-06 |
| SHCBP1 | -2.59 | 0.0000000 | 0.00e+00 |
| SHF | 1.78 | 0.0435177 | 1.97e-02 |
| SHFM1 | -0.62 | 0.0047951 | 1.70e-03 |
| SHH | 1.20 | 0.0832733 | 4.17e-02 |
| SHISA5 | 0.73 | 0.0000002 | 0.00e+00 |
| SHKBP1 | 1.09 | 0.0000000 | 0.00e+00 |
| SHMT1 | -2.09 | 0.0000000 | 0.00e+00 |
| SHOX2 | 1.64 | 0.0144443 | 5.76e-03 |
| SHPK | -1.29 | 0.0001200 | 3.15e-05 |
| SHPRH | -1.40 | 0.0017634 | 5.68e-04 |
| SHROOM2 | 3.98 | 0.0000000 | 0.00e+00 |
| SHROOM3 | 2.55 | 0.0000000 | 0.00e+00 |
| SHTN1 | -0.64 | 0.0015044 | 4.78e-04 |
| SIAE | -1.31 | 0.0049997 | 1.78e-03 |
| SIAH1 | 1.07 | 0.0001098 | 2.87e-05 |
| SIAH2 | 0.77 | 0.0130362 | 5.15e-03 |
| SIGMAR1 | -0.75 | 0.0000163 | 3.80e-06 |
| SIK1 | -0.45 | 0.0014883 | 4.72e-04 |
| SIK2 | 1.18 | 0.0000159 | 3.70e-06 |
| SIK3 | 1.03 | 0.0001118 | 2.92e-05 |
| SIL1 | 1.22 | 0.0000000 | 0.00e+00 |
| SIN3A | 0.49 | 0.0043334 | 1.52e-03 |
| SIN3B | 0.76 | 0.0016068 | 5.13e-04 |
| SIPA1 | 1.29 | 0.0000011 | 2.00e-07 |
| SIPA1L1 | 0.94 | 0.0011607 | 3.61e-04 |
| SIPA1L3 | 1.11 | 0.0000000 | 0.00e+00 |
| SIRPA | 1.43 | 0.0000000 | 0.00e+00 |
| SIRPB1 | 1.35 | 0.0012410 | 3.88e-04 |
| SIRT4 | 2.42 | 0.0307987 | 1.34e-02 |
| SIRT5 | -0.94 | 0.0650183 | 3.12e-02 |
| SIRT6 | 1.79 | 0.0000000 | 0.00e+00 |
| SIRT7 | 0.98 | 0.0000164 | 3.80e-06 |
| SIVA1 | -1.81 | 0.0000000 | 0.00e+00 |
| SIX1 | 1.93 | 0.0000000 | 0.00e+00 |
| SIX4 | 1.24 | 0.0050778 | 1.81e-03 |
| SIX5 | 2.13 | 0.0000000 | 0.00e+00 |
| SKA1 | -2.64 | 0.0000062 | 1.40e-06 |
| SKA2 | -1.18 | 0.0000018 | 4.00e-07 |
| SKA3 | -3.07 | 0.0000000 | 0.00e+00 |
| SKIV2L2 | -1.55 | 0.0000000 | 0.00e+00 |
| SKP1 | -0.49 | 0.0005346 | 1.56e-04 |
| SKP2 | -2.34 | 0.0000000 | 0.00e+00 |
| SLAMF7 | 6.69 | 0.0115066 | 4.49e-03 |
| SLAMF9 | 5.97 | 0.0417361 | 1.88e-02 |
| SLBP | -1.28 | 0.0000000 | 0.00e+00 |
| SLC11A2 | 1.86 | 0.0000000 | 0.00e+00 |
| SLC12A2 | -0.43 | 0.0023496 | 7.83e-04 |
| SLC12A4 | 1.65 | 0.0000000 | 0.00e+00 |
| SLC12A6 | 0.61 | 0.0515700 | 2.39e-02 |
| SLC12A7 | 1.97 | 0.0000000 | 0.00e+00 |
| SLC13A2 | 5.27 | 0.0042845 | 1.50e-03 |
| SLC13A5 | 7.03 | 0.0062743 | 2.29e-03 |
| SLC15A3 | 9.71 | 0.0000135 | 3.10e-06 |
| SLC15A4 | 0.56 | 0.0588367 | 2.78e-02 |
| SLC16A13 | 2.62 | 0.0000000 | 0.00e+00 |
| SLC16A14 | -2.18 | 0.0365851 | 1.62e-02 |
| SLC16A2 | 2.81 | 0.0000000 | 0.00e+00 |
| SLC16A3 | 1.98 | 0.0000000 | 0.00e+00 |
| SLC16A4 | 1.02 | 0.0000001 | 0.00e+00 |
| SLC16A5 | 0.42 | 0.0887506 | 4.50e-02 |
| SLC16A6 | 2.00 | 0.0089368 | 3.39e-03 |
| SLC17A5 | 1.11 | 0.0001693 | 4.56e-05 |
| SLC19A1 | -0.86 | 0.0763762 | 3.77e-02 |
| SLC1A2 | 6.03 | 0.0384533 | 1.72e-02 |
| SLC1A3 | 10.63 | 0.0000013 | 3.00e-07 |
| SLC1A5 | 1.27 | 0.0000000 | 0.00e+00 |
| SLC20A1 | -0.81 | 0.0000101 | 2.30e-06 |
| SLC22A11 | 6.73 | 0.0107195 | 4.15e-03 |
| SLC22A18 | 0.93 | 0.0389471 | 1.74e-02 |
| SLC22A18AS | -1.90 | 0.0497641 | 2.30e-02 |
| SLC22A23 | 1.65 | 0.0000000 | 0.00e+00 |
| SLC22A3 | -0.75 | 0.0023850 | 7.95e-04 |
| SLC23A2 | 0.53 | 0.0008149 | 2.46e-04 |
| SLC25A1 | -0.34 | 0.0977600 | 5.03e-02 |
| SLC25A10 | -2.91 | 0.0000000 | 0.00e+00 |
| SLC25A11 | -1.15 | 0.0000000 | 0.00e+00 |
| SLC25A13 | -1.22 | 0.0000000 | 0.00e+00 |
| SLC25A14 | -1.79 | 0.0023145 | 7.70e-04 |
| SLC25A15 | -1.70 | 0.0000005 | 1.00e-07 |
| SLC25A16 | -1.93 | 0.0022311 | 7.39e-04 |
| SLC25A19 | -1.19 | 0.0033273 | 1.14e-03 |
| SLC25A24 | -0.71 | 0.0040895 | 1.43e-03 |
| SLC25A25 | 1.80 | 0.0000000 | 0.00e+00 |
| SLC25A25-AS1 | 1.12 | 0.0191311 | 7.87e-03 |
| SLC25A28 | 0.72 | 0.0158206 | 6.38e-03 |
| SLC25A29 | 0.81 | 0.0173853 | 7.07e-03 |
| SLC25A3 | -0.70 | 0.0000000 | 0.00e+00 |
| SLC25A32 | -1.04 | 0.0003422 | 9.64e-05 |
| SLC25A37 | 3.41 | 0.0000000 | 0.00e+00 |
| SLC25A39 | -1.35 | 0.0000000 | 0.00e+00 |
| SLC25A4 | -1.12 | 0.0069354 | 2.56e-03 |
| SLC25A44 | 0.50 | 0.0222910 | 9.31e-03 |
| SLC25A45 | 1.26 | 0.0018877 | 6.13e-04 |
| SLC25A46 | -0.93 | 0.0006505 | 1.93e-04 |
| SLC25A5 | -0.50 | 0.0000001 | 0.00e+00 |
| SLC26A11 | 1.68 | 0.0000000 | 0.00e+00 |
| SLC26A2 | -0.90 | 0.0002199 | 6.01e-05 |
| SLC26A6 | 1.35 | 0.0000150 | 3.50e-06 |
| SLC26A9 | 3.90 | 0.0000000 | 0.00e+00 |
| SLC27A2 | -1.38 | 0.0000000 | 0.00e+00 |
| SLC27A5 | -2.23 | 0.0000440 | 1.09e-05 |
| SLC29A1 | -1.18 | 0.0000005 | 1.00e-07 |
| SLC29A4 | -1.49 | 0.0033102 | 1.13e-03 |
| SLC2A1 | 0.59 | 0.0000181 | 4.20e-06 |
| SLC2A12 | 1.60 | 0.0170930 | 6.94e-03 |
| SLC2A13 | 2.24 | 0.0000000 | 0.00e+00 |
| SLC2A3 | -0.48 | 0.0252944 | 1.07e-02 |
| SLC2A4RG | -1.98 | 0.0000000 | 0.00e+00 |
| SLC2A6 | 4.00 | 0.0000000 | 0.00e+00 |
| SLC30A5 | -0.74 | 0.0009397 | 2.87e-04 |
| SLC30A6 | -0.89 | 0.0002120 | 5.78e-05 |
| SLC30A9 | -0.91 | 0.0009451 | 2.89e-04 |
| SLC31A1 | -0.42 | 0.0722835 | 3.53e-02 |
| SLC31A2 | 1.77 | 0.0000101 | 2.30e-06 |
| SLC34A2 | 6.35 | 0.0218385 | 9.10e-03 |
| SLC35A1 | -0.88 | 0.0814111 | 4.07e-02 |
| SLC35A2 | 0.74 | 0.0007751 | 2.33e-04 |
| SLC35A3 | -0.86 | 0.0663019 | 3.19e-02 |
| SLC35A5 | -1.09 | 0.0005565 | 1.63e-04 |
| SLC35B2 | -0.34 | 0.0931635 | 4.76e-02 |
| SLC35B4 | -0.80 | 0.0000454 | 1.13e-05 |
| SLC35C1 | 0.72 | 0.0077508 | 2.90e-03 |
| SLC35C2 | 0.79 | 0.0000082 | 1.80e-06 |
| SLC35D2 | 1.04 | 0.0000160 | 3.70e-06 |
| SLC35E3 | 1.28 | 0.0000103 | 2.30e-06 |
| SLC35E4 | 1.27 | 0.0078161 | 2.93e-03 |
| SLC35G1 | -2.21 | 0.0007924 | 2.38e-04 |
| SLC35G2 | 0.99 | 0.0005044 | 1.47e-04 |
| SLC37A1 | 2.87 | 0.0000000 | 0.00e+00 |
| SLC37A4 | -0.75 | 0.0387075 | 1.73e-02 |
| SLC38A1 | -0.95 | 0.0000000 | 0.00e+00 |
| SLC38A10 | 1.12 | 0.0000000 | 0.00e+00 |
| SLC38A7 | 1.08 | 0.0000669 | 1.69e-05 |
| SLC38A9 | 0.62 | 0.0499532 | 2.31e-02 |
| SLC39A1 | 0.72 | 0.0000000 | 0.00e+00 |
| SLC39A10 | -1.24 | 0.0000000 | 0.00e+00 |
| SLC39A11 | 0.90 | 0.0004201 | 1.21e-04 |
| SLC39A13 | 0.73 | 0.0168985 | 6.85e-03 |
| SLC39A14 | 1.15 | 0.0000000 | 0.00e+00 |
| SLC39A3 | -0.61 | 0.0637815 | 3.05e-02 |
| SLC39A6 | -0.75 | 0.0000814 | 2.08e-05 |
| SLC39A8 | 2.15 | 0.0000000 | 0.00e+00 |
| SLC39A9 | -0.82 | 0.0000008 | 2.00e-07 |
| SLC3A1 | -2.32 | 0.0213190 | 8.86e-03 |
| SLC3A2 | -0.45 | 0.0000135 | 3.10e-06 |
| SLC40A1 | -1.13 | 0.0002608 | 7.20e-05 |
| SLC41A1 | 1.35 | 0.0000000 | 0.00e+00 |
| SLC41A2 | 1.37 | 0.0000000 | 0.00e+00 |
| SLC43A2 | 2.86 | 0.0000000 | 0.00e+00 |
| SLC43A3 | 3.45 | 0.0002093 | 5.70e-05 |
| SLC46A1 | -0.64 | 0.0098395 | 3.77e-03 |
| SLC47A1 | -1.56 | 0.0000208 | 4.90e-06 |
| SLC4A11 | 1.31 | 0.0003484 | 9.84e-05 |
| SLC4A2 | 0.24 | 0.0413971 | 1.87e-02 |
| SLC4A4 | -2.69 | 0.0003154 | 8.83e-05 |
| SLC4A5 | 2.26 | 0.0046223 | 1.64e-03 |
| SLC4A7 | -1.06 | 0.0000003 | 1.00e-07 |
| SLC4A8 | -3.32 | 0.0596653 | 2.83e-02 |
| SLC50A1 | 0.58 | 0.0437211 | 1.99e-02 |
| SLC52A2 | 0.91 | 0.0000000 | 0.00e+00 |
| SLC52A3 | 3.90 | 0.0000000 | 0.00e+00 |
| SLC5A3 | -0.65 | 0.0984199 | 5.07e-02 |
| SLC5A5 | 3.39 | 0.0495348 | 2.28e-02 |
| SLC5A6 | -1.42 | 0.0000000 | 0.00e+00 |
| SLC5A9 | -5.90 | 0.0687493 | 3.33e-02 |
| SLC6A12 | 7.40 | 0.0000000 | 0.00e+00 |
| SLC6A14 | 5.69 | 0.0000000 | 0.00e+00 |
| SLC6A15 | 1.91 | 0.0013555 | 4.27e-04 |
| SLC6A17 | 1.33 | 0.0348486 | 1.54e-02 |
| SLC6A20 | 5.97 | 0.0423108 | 1.91e-02 |
| SLC6A8 | 1.01 | 0.0553181 | 2.59e-02 |
| SLC6A9 | 1.92 | 0.0000035 | 7.00e-07 |
| SLC7A1 | 1.27 | 0.0000000 | 0.00e+00 |
| SLC7A11 | -2.24 | 0.0000000 | 0.00e+00 |
| SLC7A2 | 3.13 | 0.0000000 | 0.00e+00 |
| SLC7A5 | -0.76 | 0.0000000 | 0.00e+00 |
| SLC7A5P2 | -0.80 | 0.0327664 | 1.43e-02 |
| SLC7A8 | 4.52 | 0.0010811 | 3.34e-04 |
| SLC8A2 | 4.56 | 0.0000402 | 9.90e-06 |
| SLC8B1 | 1.69 | 0.0000000 | 0.00e+00 |
| SLC9A1 | 2.65 | 0.0000000 | 0.00e+00 |
| SLC9A2 | -1.54 | 0.0018847 | 6.12e-04 |
| SLC9A3R1 | -0.55 | 0.0044804 | 1.58e-03 |
| SLC9A3R2 | -0.68 | 0.0006293 | 1.86e-04 |
| SLC9A7 | 0.85 | 0.0001663 | 4.48e-05 |
| SLC9A8 | 1.12 | 0.0000002 | 0.00e+00 |
| SLCO2B1 | 1.99 | 0.0000000 | 0.00e+00 |
| SLCO3A1 | 2.83 | 0.0000000 | 0.00e+00 |
| SLCO4A1 | 2.24 | 0.0000000 | 0.00e+00 |
| SLCO5A1 | 6.13 | 0.0000002 | 0.00e+00 |
| SLF1 | -2.54 | 0.0000000 | 0.00e+00 |
| SLF2 | -1.06 | 0.0014704 | 4.66e-04 |
| SLFN5 | 3.71 | 0.0000000 | 0.00e+00 |
| SLFNL1-AS1 | -1.71 | 0.0639161 | 3.06e-02 |
| SLIRP | -1.03 | 0.0000087 | 1.90e-06 |
| SLIT1 | 2.81 | 0.0767244 | 3.79e-02 |
| SLITRK6 | -6.04 | 0.0000477 | 1.18e-05 |
| SLMAP | 1.18 | 0.0000000 | 0.00e+00 |
| SLMO2-ATP5E | -0.48 | 0.0001256 | 3.31e-05 |
| SLPI | 3.24 | 0.0000000 | 0.00e+00 |
| SLTM | -0.52 | 0.0038217 | 1.33e-03 |
| SLU7 | 0.47 | 0.0195808 | 8.07e-03 |
| SLX1A | 1.06 | 0.0000000 | 0.00e+00 |
| SLX1A-SULT1A3 | 0.61 | 0.0002784 | 7.73e-05 |
| SLX1B | 1.06 | 0.0000000 | 0.00e+00 |
| SLX1B-SULT1A4 | 0.61 | 0.0002784 | 7.73e-05 |
| SMAD1 | 0.75 | 0.0651582 | 3.13e-02 |
| SMAD2 | -0.75 | 0.0000018 | 4.00e-07 |
| SMAD3 | 0.92 | 0.0000000 | 0.00e+00 |
| SMAD4 | 0.46 | 0.0510990 | 2.37e-02 |
| SMAD5 | -0.60 | 0.0218916 | 9.12e-03 |
| SMAD6 | -0.78 | 0.0023223 | 7.73e-04 |
| SMAGP | 0.77 | 0.0462986 | 2.12e-02 |
| SMARCA1 | -0.37 | 0.0336868 | 1.48e-02 |
| SMARCA2 | -0.60 | 0.0507275 | 2.35e-02 |
| SMARCA5 | 0.30 | 0.0685735 | 3.32e-02 |
| SMARCAD1 | -1.73 | 0.0000000 | 0.00e+00 |
| SMARCAL1 | 0.57 | 0.0157741 | 6.36e-03 |
| SMARCB1 | -0.55 | 0.0572790 | 2.70e-02 |
| SMARCC2 | 0.38 | 0.0111032 | 4.32e-03 |
| SMARCD1 | 0.44 | 0.0071713 | 2.65e-03 |
| SMARCD3 | -1.34 | 0.0025084 | 8.39e-04 |
| SMARCE1 | -0.85 | 0.0000001 | 0.00e+00 |
| SMC1A | -0.88 | 0.0000000 | 0.00e+00 |
| SMC2 | -3.31 | 0.0000000 | 0.00e+00 |
| SMC3 | -1.67 | 0.0000000 | 0.00e+00 |
| SMC4 | -2.62 | 0.0000000 | 0.00e+00 |
| SMC5 | -1.39 | 0.0000028 | 6.00e-07 |
| SMC6 | -1.22 | 0.0000000 | 0.00e+00 |
| SMCHD1 | -1.11 | 0.0000017 | 4.00e-07 |
| SMCO2 | 4.61 | 0.0214952 | 8.94e-03 |
| SMCR8 | 0.77 | 0.0000022 | 4.00e-07 |
| SMG1 | -0.33 | 0.0360828 | 1.60e-02 |
| SMG5 | 1.07 | 0.0000000 | 0.00e+00 |
| SMG6 | 0.85 | 0.0000283 | 6.80e-06 |
| SMG7 | 1.08 | 0.0000000 | 0.00e+00 |
| SMG9 | 0.88 | 0.0000168 | 3.90e-06 |
| SMIM11 | -1.26 | 0.0103397 | 3.99e-03 |
| SMIM15 | -0.61 | 0.0072347 | 2.68e-03 |
| SMIM20 | -0.93 | 0.0337558 | 1.48e-02 |
| SMIM4 | -1.33 | 0.0333943 | 1.46e-02 |
| SMIM6 | -1.58 | 0.0549254 | 2.57e-02 |
| SMNDC1 | 0.79 | 0.0000454 | 1.13e-05 |
| SMO | 0.62 | 0.0057166 | 2.06e-03 |
| SMOC1 | 1.55 | 0.0000000 | 0.00e+00 |
| SMOX | 1.63 | 0.0000000 | 0.00e+00 |
| SMPD1 | 2.83 | 0.0000000 | 0.00e+00 |
| SMTN | 1.69 | 0.0000000 | 0.00e+00 |
| SMU1 | -0.45 | 0.0203990 | 8.44e-03 |
| SMURF1 | 1.82 | 0.0000000 | 0.00e+00 |
| SMURF2 | 0.62 | 0.0137325 | 5.46e-03 |
| SMYD2 | -1.10 | 0.0003611 | 1.02e-04 |
| SMYD5 | -0.55 | 0.0049842 | 1.78e-03 |
| SNAP25 | 0.58 | 0.0087176 | 3.30e-03 |
| SNAPC4 | 0.92 | 0.0006437 | 1.90e-04 |
| SNAR-B1 | 1.73 | 0.0044902 | 1.58e-03 |
| SNAR-B2 | 1.73 | 0.0044902 | 1.58e-03 |
| SNCG | -1.69 | 0.0302487 | 1.31e-02 |
| SND1 | 0.46 | 0.0000518 | 1.29e-05 |
| SNHG1 | -0.74 | 0.0004905 | 1.42e-04 |
| SNHG10 | -1.43 | 0.0064829 | 2.38e-03 |
| SNHG12 | 0.87 | 0.0144039 | 5.74e-03 |
| SNHG15 | 1.59 | 0.0000000 | 0.00e+00 |
| SNHG16 | 0.27 | 0.0885626 | 4.48e-02 |
| SNHG19 | -2.18 | 0.0000000 | 0.00e+00 |
| SNHG21 | -2.22 | 0.0111132 | 4.32e-03 |
| SNHG3 | -1.29 | 0.0000019 | 4.00e-07 |
| SNHG4 | -2.56 | 0.0000416 | 1.03e-05 |
| SNHG6 | 0.44 | 0.0691739 | 3.35e-02 |
| SNHG7 | -0.46 | 0.0658477 | 3.17e-02 |
| SNHG9 | -1.69 | 0.0033740 | 1.16e-03 |
| SNIP1 | 0.56 | 0.0823005 | 4.12e-02 |
| SNPH | 4.66 | 0.0000000 | 0.00e+00 |
| SNRK | 0.99 | 0.0073923 | 2.74e-03 |
| SNRNP200 | -0.69 | 0.0000000 | 0.00e+00 |
| SNRNP25 | -2.51 | 0.0000000 | 0.00e+00 |
| SNRNP27 | 0.61 | 0.0115534 | 4.51e-03 |
| SNRNP40 | -0.93 | 0.0000846 | 2.17e-05 |
| SNRNP48 | -1.00 | 0.0374056 | 1.66e-02 |
| SNRNP70 | -0.47 | 0.0000827 | 2.12e-05 |
| SNRPA | -0.40 | 0.0224829 | 9.41e-03 |
| SNRPB | -0.45 | 0.0000152 | 3.50e-06 |
| SNRPC | -0.76 | 0.0003121 | 8.73e-05 |
| SNRPD1 | -1.23 | 0.0000000 | 0.00e+00 |
| SNRPD2 | -0.72 | 0.0000000 | 0.00e+00 |
| SNRPD3 | -0.90 | 0.0000001 | 0.00e+00 |
| SNRPE | -1.79 | 0.0000000 | 0.00e+00 |
| SNRPF | -1.23 | 0.0000000 | 0.00e+00 |
| SNRPG | -1.19 | 0.0000000 | 0.00e+00 |
| SNTA1 | -1.18 | 0.0091045 | 3.46e-03 |
| SNTB2 | 1.24 | 0.0000000 | 0.00e+00 |
| SNU13 | -0.79 | 0.0000940 | 2.43e-05 |
| SNUPN | -0.71 | 0.0286802 | 1.23e-02 |
| SNX11 | 1.16 | 0.0000000 | 0.00e+00 |
| SNX13 | -0.90 | 0.0019342 | 6.30e-04 |
| SNX15 | 0.92 | 0.0000318 | 7.70e-06 |
| SNX16 | 1.40 | 0.0793965 | 3.94e-02 |
| SNX18 | 0.93 | 0.0006559 | 1.94e-04 |
| SNX19 | 0.61 | 0.0011739 | 3.65e-04 |
| SNX2 | -0.46 | 0.0711437 | 3.47e-02 |
| SNX25 | -0.79 | 0.0993137 | 5.12e-02 |
| SNX29 | 1.20 | 0.0036778 | 1.27e-03 |
| SNX30 | -0.81 | 0.0964831 | 4.95e-02 |
| SNX33 | 1.81 | 0.0000000 | 0.00e+00 |
| SNX5 | -1.06 | 0.0000000 | 0.00e+00 |
| SNX6 | -0.47 | 0.0068461 | 2.52e-03 |
| SNX9 | 0.92 | 0.0000002 | 0.00e+00 |
| SOCS1 | 5.82 | 0.0000013 | 3.00e-07 |
| SOCS2 | 1.88 | 0.0000000 | 0.00e+00 |
| SOCS3 | 4.73 | 0.0000000 | 0.00e+00 |
| SOCS4 | -1.36 | 0.0000040 | 9.00e-07 |
| SOCS5 | -0.93 | 0.0001321 | 3.50e-05 |
| SOCS6 | -0.81 | 0.0000428 | 1.06e-05 |
| SOD1 | -0.95 | 0.0000000 | 0.00e+00 |
| SOD2 | 4.94 | 0.0000000 | 0.00e+00 |
| SORBS1 | 1.93 | 0.0005477 | 1.60e-04 |
| SORBS3 | 0.71 | 0.0000394 | 9.70e-06 |
| SORCS2 | 3.31 | 0.0000000 | 0.00e+00 |
| SORL1 | -2.30 | 0.0000000 | 0.00e+00 |
| SOX12 | -0.87 | 0.0001334 | 3.54e-05 |
| SOX13 | 2.42 | 0.0000000 | 0.00e+00 |
| SOX2 | 1.93 | 0.0000000 | 0.00e+00 |
| SOX4 | 2.18 | 0.0000000 | 0.00e+00 |
| SOX7 | 2.20 | 0.0886565 | 4.49e-02 |
| SOX9 | 1.05 | 0.0002109 | 5.75e-05 |
| SP1 | 0.30 | 0.0849193 | 4.27e-02 |
| SP100 | 2.36 | 0.0000000 | 0.00e+00 |
| SP110 | 3.68 | 0.0000000 | 0.00e+00 |
| SP140 | 4.79 | 0.0003638 | 1.03e-04 |
| SP140L | 2.81 | 0.0000000 | 0.00e+00 |
| SP2 | 1.68 | 0.0000000 | 0.00e+00 |
| SP3 | -0.54 | 0.0022766 | 7.56e-04 |
| SP6 | 1.63 | 0.0013480 | 4.24e-04 |
| SP8 | 5.00 | 0.0000017 | 3.00e-07 |
| SPA17 | -1.53 | 0.0043892 | 1.54e-03 |
| SPAG16 | -1.79 | 0.0015487 | 4.93e-04 |
| SPAG5 | -2.55 | 0.0000000 | 0.00e+00 |
| SPAG7 | 0.64 | 0.0002034 | 5.53e-05 |
| SPAG9 | 1.97 | 0.0000000 | 0.00e+00 |
| SPANXN5 | 4.13 | 0.0600794 | 2.85e-02 |
| SPARC | -2.58 | 0.0262728 | 1.12e-02 |
| SPATA13 | 0.73 | 0.0227307 | 9.52e-03 |
| SPATA2 | 1.33 | 0.0000087 | 2.00e-06 |
| SPATA20 | -1.26 | 0.0000000 | 0.00e+00 |
| SPATA2L | 0.96 | 0.0313482 | 1.36e-02 |
| SPATA5 | -1.30 | 0.0033399 | 1.15e-03 |
| SPATA6L | 2.08 | 0.0010576 | 3.26e-04 |
| SPATS2L | 1.23 | 0.0000000 | 0.00e+00 |
| SPC24 | -3.39 | 0.0000000 | 0.00e+00 |
| SPC25 | -3.24 | 0.0000006 | 1.00e-07 |
| SPCS1 | -0.57 | 0.0071275 | 2.63e-03 |
| SPDEF | -1.27 | 0.0031183 | 1.06e-03 |
| SPDL1 | -1.24 | 0.0000001 | 0.00e+00 |
| SPECC1 | 0.56 | 0.0044476 | 1.57e-03 |
| SPECC1L-ADORA2A | 2.82 | 0.0000000 | 0.00e+00 |
| SPEN | 1.05 | 0.0000000 | 0.00e+00 |
| SPG20 | -0.52 | 0.0393742 | 1.76e-02 |
| SPG7 | 0.46 | 0.0130078 | 5.13e-03 |
| SPHAR | -3.42 | 0.0045586 | 1.61e-03 |
| SPHK1 | 3.74 | 0.0000000 | 0.00e+00 |
| SPICE1 | -0.68 | 0.0775915 | 3.84e-02 |
| SPIN1 | -0.78 | 0.0004247 | 1.22e-04 |
| SPINT1 | 5.13 | 0.0000000 | 0.00e+00 |
| SPINT2 | 0.44 | 0.0000015 | 3.00e-07 |
| SPIRE1 | 1.04 | 0.0000147 | 3.40e-06 |
| SPNS1 | 0.59 | 0.0071170 | 2.63e-03 |
| SPNS2 | 5.07 | 0.0000000 | 0.00e+00 |
| SPOCD1 | 3.44 | 0.0002111 | 5.75e-05 |
| SPOCK1 | 4.89 | 0.0000000 | 0.00e+00 |
| SPOP | 0.38 | 0.0914741 | 4.66e-02 |
| SPP1 | 0.26 | 0.0244853 | 1.03e-02 |
| SPPL3 | 1.10 | 0.0000000 | 0.00e+00 |
| SPR | -0.68 | 0.0000056 | 1.20e-06 |
| SPRED2 | 1.53 | 0.0000000 | 0.00e+00 |
| SPRED3 | 2.02 | 0.0003021 | 8.42e-05 |
| SPRTN | -1.22 | 0.0008097 | 2.44e-04 |
| SPRY1 | 2.87 | 0.0000009 | 2.00e-07 |
| SPRY2 | 1.98 | 0.0000000 | 0.00e+00 |
| SPRY4 | 2.90 | 0.0000000 | 0.00e+00 |
| SPRYD3 | 0.85 | 0.0000040 | 9.00e-07 |
| SPRYD4 | -0.99 | 0.0210724 | 8.75e-03 |
| SPSB1 | 1.18 | 0.0000000 | 0.00e+00 |
| SPTAN1 | 0.30 | 0.0108917 | 4.22e-03 |
| SPTBN1 | 0.54 | 0.0000000 | 0.00e+00 |
| SPTBN5 | 4.24 | 0.0002640 | 7.30e-05 |
| SPTSSA | 0.40 | 0.0501449 | 2.32e-02 |
| SPX | 3.08 | 0.0000000 | 0.00e+00 |
| SQLE | 1.33 | 0.0000000 | 0.00e+00 |
| SQRDL | 2.93 | 0.0000000 | 0.00e+00 |
| SRBD1 | -1.32 | 0.0003285 | 9.23e-05 |
| SRC | 1.09 | 0.0000000 | 0.00e+00 |
| SRCAP | 0.61 | 0.0000945 | 2.44e-05 |
| SRD5A1 | -1.67 | 0.0007912 | 2.38e-04 |
| SREBF1 | 0.34 | 0.0059999 | 2.18e-03 |
| SREBF2 | 1.46 | 0.0000000 | 0.00e+00 |
| SREK1 | -0.85 | 0.0001482 | 3.96e-05 |
| SRF | 2.38 | 0.0000000 | 0.00e+00 |
| SRGAP2 | 0.96 | 0.0000012 | 2.00e-07 |
| SRGAP2C | 0.64 | 0.0916435 | 4.67e-02 |
| SRGN | 1.84 | 0.0000000 | 0.00e+00 |
| SRM | -0.78 | 0.0026728 | 9.00e-04 |
| SRP14 | -0.32 | 0.0507746 | 2.35e-02 |
| SRP72 | -0.57 | 0.0001493 | 3.99e-05 |
| SRP9 | -1.83 | 0.0000000 | 0.00e+00 |
| SRPK2 | 0.76 | 0.0000270 | 6.50e-06 |
| SRPR | 0.81 | 0.0000001 | 0.00e+00 |
| SRPRB | -0.87 | 0.0003083 | 8.61e-05 |
| SRRM1 | 0.42 | 0.0299107 | 1.29e-02 |
| SRRM2 | 0.24 | 0.0210417 | 8.73e-03 |
| SRRT | -0.49 | 0.0051469 | 1.84e-03 |
| SRSF1 | -1.85 | 0.0000000 | 0.00e+00 |
| SRSF10 | -1.45 | 0.0000000 | 0.00e+00 |
| SRSF11 | -0.55 | 0.0025255 | 8.46e-04 |
| SRSF12 | 1.65 | 0.0471744 | 2.16e-02 |
| SRSF2 | -1.19 | 0.0000000 | 0.00e+00 |
| SRSF3 | -0.97 | 0.0000000 | 0.00e+00 |
| SRSF4 | 0.64 | 0.0003185 | 8.92e-05 |
| SRSF5 | -0.45 | 0.0045873 | 1.62e-03 |
| SRSF6 | -1.19 | 0.0000000 | 0.00e+00 |
| SRSF7 | -1.31 | 0.0000000 | 0.00e+00 |
| SRSF8 | -0.96 | 0.0018084 | 5.84e-04 |
| SRSF9 | -0.30 | 0.0417851 | 1.89e-02 |
| SRXN1 | -1.14 | 0.0000000 | 0.00e+00 |
| SS18L1 | -0.78 | 0.0813160 | 4.06e-02 |
| SSB | -1.39 | 0.0000000 | 0.00e+00 |
| SSBP1 | -0.68 | 0.0008870 | 2.70e-04 |
| SSBP3 | 0.78 | 0.0414114 | 1.87e-02 |
| SSBP4 | -1.04 | 0.0002769 | 7.68e-05 |
| SSC5D | 4.24 | 0.0002680 | 7.42e-05 |
| SSH1 | 1.44 | 0.0000000 | 0.00e+00 |
| SSH2 | 2.02 | 0.0000000 | 0.00e+00 |
| SSNA1 | -0.55 | 0.0074652 | 2.77e-03 |
| SSR1 | -0.63 | 0.0004527 | 1.31e-04 |
| SSR2 | 0.40 | 0.0020143 | 6.59e-04 |
| SSR4 | 0.44 | 0.0011245 | 3.48e-04 |
| SSRP1 | -1.19 | 0.0000000 | 0.00e+00 |
| SSTR2 | 4.72 | 0.0000126 | 2.90e-06 |
| SSUH2 | 3.91 | 0.0002101 | 5.72e-05 |
| SSX2IP | -2.00 | 0.0000000 | 0.00e+00 |
| ST20 | 1.31 | 0.0006233 | 1.84e-04 |
| ST20-MTHFS | 0.51 | 0.0135327 | 5.37e-03 |
| ST3GAL1 | 3.32 | 0.0000000 | 0.00e+00 |
| ST3GAL2 | 1.45 | 0.0000000 | 0.00e+00 |
| ST3GAL4 | 2.34 | 0.0000000 | 0.00e+00 |
| ST3GAL4-AS1 | 1.99 | 0.0004724 | 1.37e-04 |
| ST5 | 2.31 | 0.0000000 | 0.00e+00 |
| ST6GAL2 | -3.70 | 0.0000000 | 0.00e+00 |
| ST6GALNAC6 | 0.79 | 0.0075354 | 2.80e-03 |
| ST7 | 0.56 | 0.0739462 | 3.63e-02 |
| STAG1 | -0.82 | 0.0057397 | 2.07e-03 |
| STAG2 | -0.51 | 0.0007923 | 2.38e-04 |
| STAMBP | 0.50 | 0.0039208 | 1.36e-03 |
| STAMBPL1 | 1.70 | 0.0000000 | 0.00e+00 |
| STAP2 | 3.52 | 0.0352621 | 1.56e-02 |
| STARD10 | 2.12 | 0.0000000 | 0.00e+00 |
| STARD13 | 2.69 | 0.0000000 | 0.00e+00 |
| STARD3 | 0.71 | 0.0002166 | 5.92e-05 |
| STARD4 | 1.40 | 0.0000000 | 0.00e+00 |
| STARD7 | -1.09 | 0.0000000 | 0.00e+00 |
| STARD9 | -1.67 | 0.0068248 | 2.51e-03 |
| STAT1 | 1.40 | 0.0000000 | 0.00e+00 |
| STAT2 | 3.49 | 0.0000000 | 0.00e+00 |
| STAT3 | 1.48 | 0.0000000 | 0.00e+00 |
| STAT4 | -1.33 | 0.0072932 | 2.70e-03 |
| STAT5A | 5.01 | 0.0000000 | 0.00e+00 |
| STAT5B | 1.32 | 0.0000000 | 0.00e+00 |
| STAT6 | 1.17 | 0.0000000 | 0.00e+00 |
| STAU1 | 0.68 | 0.0000009 | 2.00e-07 |
| STAU2 | -0.86 | 0.0009904 | 3.04e-04 |
| STC1 | 2.45 | 0.0000000 | 0.00e+00 |
| STC2 | 1.49 | 0.0000000 | 0.00e+00 |
| STEAP1 | 0.83 | 0.0031436 | 1.07e-03 |
| STEAP3 | -1.00 | 0.0001278 | 3.38e-05 |
| STEAP4 | 6.14 | 0.0311640 | 1.35e-02 |
| STIL | -1.60 | 0.0000001 | 0.00e+00 |
| STIM1 | 0.90 | 0.0000008 | 2.00e-07 |
| STIM2 | 0.72 | 0.0471728 | 2.16e-02 |
| STIP1 | -0.89 | 0.0000000 | 0.00e+00 |
| STK10 | 2.46 | 0.0000000 | 0.00e+00 |
| STK11IP | 0.53 | 0.0775597 | 3.84e-02 |
| STK17A | 1.54 | 0.0000000 | 0.00e+00 |
| STK24 | 0.93 | 0.0000000 | 0.00e+00 |
| STK26 | -1.60 | 0.0000827 | 2.11e-05 |
| STK3 | 0.61 | 0.0343089 | 1.51e-02 |
| STK32B | -2.32 | 0.0000000 | 0.00e+00 |
| STK32C | -0.85 | 0.0294555 | 1.27e-02 |
| STK35 | 0.46 | 0.0329287 | 1.44e-02 |
| STK38L | 0.79 | 0.0070333 | 2.60e-03 |
| STK4 | -0.49 | 0.0488044 | 2.24e-02 |
| STK40 | 1.46 | 0.0000006 | 1.00e-07 |
| STMN1 | -2.11 | 0.0000000 | 0.00e+00 |
| STMN3 | -1.63 | 0.0001085 | 2.83e-05 |
| STMN4 | 5.83 | 0.0517380 | 2.40e-02 |
| STOM | 1.01 | 0.0000000 | 0.00e+00 |
| STOML1 | 2.76 | 0.0000000 | 0.00e+00 |
| STOML2 | -0.84 | 0.0000000 | 0.00e+00 |
| STOX2 | -1.81 | 0.0102022 | 3.93e-03 |
| STRA13 | -1.78 | 0.0000000 | 0.00e+00 |
| STRA6 | 1.61 | 0.0000000 | 0.00e+00 |
| STRAP | -1.07 | 0.0000000 | 0.00e+00 |
| STRBP | -1.40 | 0.0000001 | 0.00e+00 |
| STRIP1 | 0.76 | 0.0021748 | 7.18e-04 |
| STRIP2 | 2.01 | 0.0000000 | 0.00e+00 |
| STRN | -0.41 | 0.0723156 | 3.54e-02 |
| STRN4 | 0.79 | 0.0000000 | 0.00e+00 |
| STS | 3.64 | 0.0000000 | 0.00e+00 |
| STT3A | -0.53 | 0.0017284 | 5.56e-04 |
| STT3B | -0.42 | 0.0012527 | 3.92e-04 |
| STUB1 | -0.71 | 0.0007988 | 2.41e-04 |
| STX10 | -0.70 | 0.0157001 | 6.32e-03 |
| STX11 | 4.34 | 0.0000000 | 0.00e+00 |
| STX12 | 1.08 | 0.0001413 | 3.76e-05 |
| STX16 | 0.37 | 0.0667886 | 3.22e-02 |
| STX18 | 0.72 | 0.0061797 | 2.25e-03 |
| STX1A | 0.69 | 0.0021896 | 7.23e-04 |
| STX2 | -1.28 | 0.0000065 | 1.40e-06 |
| STX3 | 1.31 | 0.0000002 | 0.00e+00 |
| STX4 | 1.94 | 0.0000000 | 0.00e+00 |
| STX5 | 1.26 | 0.0000000 | 0.00e+00 |
| STXBP1 | 0.57 | 0.0130936 | 5.18e-03 |
| STXBP2 | 1.64 | 0.0000000 | 0.00e+00 |
| STXBP4 | -1.79 | 0.0001542 | 4.13e-05 |
| STXBP5-AS1 | 4.13 | 0.0579864 | 2.74e-02 |
| STYK1 | 3.98 | 0.0010181 | 3.13e-04 |
| STYX | -0.81 | 0.0612866 | 2.92e-02 |
| SUCLA2 | -0.94 | 0.0019659 | 6.41e-04 |
| SUCLG1 | -0.77 | 0.0000243 | 5.80e-06 |
| SUCLG2 | -0.99 | 0.0000114 | 2.60e-06 |
| SUCLG2-AS1 | -1.84 | 0.0794078 | 3.94e-02 |
| SUFU | 0.78 | 0.0330824 | 1.45e-02 |
| SUGP1 | 1.02 | 0.0000231 | 5.50e-06 |
| SUGP2 | -0.70 | 0.0011371 | 3.53e-04 |
| SULF1 | 5.83 | 0.0515073 | 2.39e-02 |
| SULT1C2P1 | 7.95 | 0.0008573 | 2.60e-04 |
| SUMF1 | -1.45 | 0.0001271 | 3.36e-05 |
| SUMO1 | -0.52 | 0.0025581 | 8.58e-04 |
| SUN2 | 0.58 | 0.0025386 | 8.50e-04 |
| SUOX | -1.25 | 0.0006726 | 2.00e-04 |
| SUPT16H | -1.48 | 0.0000000 | 0.00e+00 |
| SUPT5H | 1.16 | 0.0000000 | 0.00e+00 |
| SUPT6H | 1.00 | 0.0000000 | 0.00e+00 |
| SUPT7L | 0.47 | 0.0095027 | 3.62e-03 |
| SURF2 | -0.78 | 0.0354630 | 1.57e-02 |
| SUSD1 | 0.66 | 0.0660858 | 3.18e-02 |
| SUSD2 | 1.40 | 0.0000000 | 0.00e+00 |
| SUSD4 | 3.27 | 0.0200760 | 8.29e-03 |
| SUSD6 | 2.94 | 0.0000000 | 0.00e+00 |
| SUV39H1 | -1.32 | 0.0000106 | 2.40e-06 |
| SUV39H2 | -1.33 | 0.0006963 | 2.07e-04 |
| SUV420H2 | -1.01 | 0.0696910 | 3.38e-02 |
| SUZ12 | -0.52 | 0.0148090 | 5.93e-03 |
| SVEP1 | -1.53 | 0.0842902 | 4.24e-02 |
| SVIL | 2.28 | 0.0000000 | 0.00e+00 |
| SVIP | -1.21 | 0.0793219 | 3.94e-02 |
| SWAP70 | 1.41 | 0.0000000 | 0.00e+00 |
| SWI5 | -1.12 | 0.0035226 | 1.21e-03 |
| SYBU | -1.94 | 0.0008051 | 2.43e-04 |
| SYCP2 | -2.93 | 0.0054700 | 1.97e-03 |
| SYDE1 | 0.78 | 0.0050143 | 1.79e-03 |
| SYMPK | 1.07 | 0.0000000 | 0.00e+00 |
| SYNCRIP | -1.04 | 0.0000000 | 0.00e+00 |
| SYNE2 | -0.52 | 0.0073572 | 2.73e-03 |
| SYNE3 | 4.12 | 0.0000000 | 0.00e+00 |
| SYNGR2 | 0.63 | 0.0000002 | 0.00e+00 |
| SYNGR3 | 2.36 | 0.0000000 | 0.00e+00 |
| SYNJ2BP | -0.63 | 0.0288567 | 1.24e-02 |
| SYNJ2BP-COX16 | -1.01 | 0.0001401 | 3.73e-05 |
| SYNM | -0.93 | 0.0000013 | 3.00e-07 |
| SYNPO | 2.33 | 0.0000000 | 0.00e+00 |
| SYNPO2 | 1.24 | 0.0000012 | 3.00e-07 |
| SYNRG | 0.63 | 0.0238113 | 1.00e-02 |
| SYPL1 | -0.43 | 0.0307080 | 1.33e-02 |
| SYS1 | 1.11 | 0.0000002 | 0.00e+00 |
| SYS1-DBNDD2 | 0.56 | 0.0424233 | 1.92e-02 |
| SYT1 | 1.31 | 0.0000000 | 0.00e+00 |
| SYT13 | 1.72 | 0.0000000 | 0.00e+00 |
| SYT3 | 2.32 | 0.0898677 | 4.56e-02 |
| SYT6 | -2.85 | 0.0439772 | 2.00e-02 |
| SYTL4 | 0.83 | 0.0848823 | 4.27e-02 |
| SYVN1 | 1.77 | 0.0000000 | 0.00e+00 |
| SZT2 | 1.07 | 0.0000017 | 3.00e-07 |
| TAB2 | 1.56 | 0.0000000 | 0.00e+00 |
| TAB3 | 0.69 | 0.0086861 | 3.28e-03 |
| TAC3 | 10.74 | 0.0000009 | 2.00e-07 |
| TACC1 | -0.76 | 0.0000799 | 2.04e-05 |
| TACC3 | -2.28 | 0.0000000 | 0.00e+00 |
| TADA2B | 0.82 | 0.0050023 | 1.78e-03 |
| TADA3 | 0.46 | 0.0046181 | 1.63e-03 |
| TAF10 | 0.49 | 0.0041871 | 1.47e-03 |
| TAF15 | -1.61 | 0.0000000 | 0.00e+00 |
| TAF1B | -0.90 | 0.0073930 | 2.75e-03 |
| TAF1D | 0.67 | 0.0073074 | 2.71e-03 |
| TAF2 | -0.59 | 0.0237081 | 9.99e-03 |
| TAF3 | 0.65 | 0.0913660 | 4.66e-02 |
| TAF7 | 0.42 | 0.0046431 | 1.64e-03 |
| TAF8 | 0.60 | 0.0724836 | 3.55e-02 |
| TAF9 | -0.71 | 0.0090775 | 3.45e-03 |
| TAF9B | -2.04 | 0.0000002 | 0.00e+00 |
| TAGLN2 | -0.60 | 0.0000000 | 0.00e+00 |
| TALDO1 | -0.62 | 0.0000000 | 0.00e+00 |
| TANC2 | 1.55 | 0.0000000 | 0.00e+00 |
| TANK | 0.99 | 0.0000038 | 8.00e-07 |
| TAOK1 | -0.50 | 0.0008455 | 2.56e-04 |
| TAOK2 | 0.83 | 0.0000001 | 0.00e+00 |
| TAOK3 | 1.23 | 0.0000000 | 0.00e+00 |
| TAPBP | 2.83 | 0.0000000 | 0.00e+00 |
| TAPBPL | 4.42 | 0.0000000 | 0.00e+00 |
| TARBP1 | -0.97 | 0.0011416 | 3.54e-04 |
| TARBP2 | -0.77 | 0.0032343 | 1.11e-03 |
| TARDBP | -1.11 | 0.0000000 | 0.00e+00 |
| TARS | -0.36 | 0.0093793 | 3.57e-03 |
| TARS2 | -0.55 | 0.0256455 | 1.09e-02 |
| TATDN1 | -1.10 | 0.0016643 | 5.33e-04 |
| TATDN2 | 0.52 | 0.0030868 | 1.05e-03 |
| TAX1BP3 | 0.57 | 0.0000202 | 4.80e-06 |
| TBC1D10A | 2.66 | 0.0000000 | 0.00e+00 |
| TBC1D10B | 1.04 | 0.0000001 | 0.00e+00 |
| TBC1D12 | 1.44 | 0.0000562 | 1.41e-05 |
| TBC1D13 | 0.50 | 0.0724466 | 3.54e-02 |
| TBC1D14 | -1.27 | 0.0000000 | 0.00e+00 |
| TBC1D15 | -0.67 | 0.0224602 | 9.39e-03 |
| TBC1D17 | 0.98 | 0.0000534 | 1.34e-05 |
| TBC1D22B | 2.02 | 0.0000000 | 0.00e+00 |
| TBC1D25 | 1.77 | 0.0000000 | 0.00e+00 |
| TBC1D2B | 1.61 | 0.0000000 | 0.00e+00 |
| TBC1D31 | -1.94 | 0.0000253 | 6.10e-06 |
| TBC1D4 | -1.24 | 0.0004376 | 1.26e-04 |
| TBC1D5 | -0.95 | 0.0000069 | 1.50e-06 |
| TBC1D8 | 0.26 | 0.0861538 | 4.34e-02 |
| TBC1D9 | 0.72 | 0.0077279 | 2.88e-03 |
| TBCB | -0.49 | 0.0000634 | 1.60e-05 |
| TBCC | 0.97 | 0.0013218 | 4.15e-04 |
| TBCD | -1.17 | 0.0000001 | 0.00e+00 |
| TBCE | -0.83 | 0.0001876 | 5.08e-05 |
| TBKBP1 | 1.53 | 0.0000617 | 1.55e-05 |
| TBL1X | -0.53 | 0.0240671 | 1.02e-02 |
| TBL2 | 0.43 | 0.0492505 | 2.27e-02 |
| TBL3 | 0.45 | 0.0338598 | 1.49e-02 |
| TBRG1 | 0.49 | 0.0837865 | 4.21e-02 |
| TBX2 | 2.58 | 0.0000000 | 0.00e+00 |
| TBX21 | 5.69 | 0.0644894 | 3.09e-02 |
| TBX3 | 1.71 | 0.0000000 | 0.00e+00 |
| TCEAL3 | 1.24 | 0.0294861 | 1.27e-02 |
| TCEANC2 | -1.05 | 0.0850498 | 4.28e-02 |
| TCEB2 | -0.38 | 0.0105499 | 4.08e-03 |
| TCEB3 | 1.04 | 0.0000000 | 0.00e+00 |
| TCERG1 | -0.95 | 0.0000019 | 4.00e-07 |
| TCF20 | 1.03 | 0.0002429 | 6.68e-05 |
| TCF7 | 2.53 | 0.0000000 | 0.00e+00 |
| TCF7L1 | 1.98 | 0.0000049 | 1.10e-06 |
| TCF7L2 | 1.28 | 0.0000002 | 0.00e+00 |
| TCFL5 | -1.32 | 0.0068103 | 2.51e-03 |
| TCIRG1 | 1.51 | 0.0000000 | 0.00e+00 |
| TCN1 | 4.32 | 0.0000000 | 0.00e+00 |
| TCN2 | 3.21 | 0.0000000 | 0.00e+00 |
| TCOF1 | 0.38 | 0.0057123 | 2.06e-03 |
| TCP1 | -0.87 | 0.0000000 | 0.00e+00 |
| TCP11L2 | 2.40 | 0.0000000 | 0.00e+00 |
| TCTA | -0.73 | 0.0580192 | 2.74e-02 |
| TCTN1 | -0.97 | 0.0190227 | 7.82e-03 |
| TCTN2 | -1.06 | 0.0758305 | 3.74e-02 |
| TCTN3 | -0.66 | 0.0028337 | 9.58e-04 |
| TDG | -0.56 | 0.0341976 | 1.50e-02 |
| TDP1 | -1.23 | 0.0008082 | 2.44e-04 |
| TDP2 | 0.41 | 0.0191047 | 7.85e-03 |
| TDRD3 | -0.85 | 0.0902182 | 4.58e-02 |
| TDRD7 | 2.78 | 0.0000000 | 0.00e+00 |
| TEAD1 | -0.62 | 0.0000053 | 1.10e-06 |
| TEAD3 | 1.26 | 0.0034430 | 1.18e-03 |
| TEAD4 | 0.57 | 0.0069416 | 2.56e-03 |
| TECPR1 | 0.90 | 0.0018767 | 6.09e-04 |
| TEKT4P2 | 1.29 | 0.0225856 | 9.45e-03 |
| TEP1 | 2.58 | 0.0000000 | 0.00e+00 |
| TERF2 | 0.97 | 0.0000138 | 3.20e-06 |
| TERF2IP | 0.82 | 0.0000875 | 2.25e-05 |
| TERT | -5.38 | 0.0000028 | 6.00e-07 |
| TES | 0.72 | 0.0000280 | 6.70e-06 |
| TESC | 0.71 | 0.0000003 | 1.00e-07 |
| TESK1 | 1.49 | 0.0000000 | 0.00e+00 |
| TESK2 | 2.34 | 0.0253412 | 1.08e-02 |
| TEX15 | -4.32 | 0.0440886 | 2.00e-02 |
| TEX2 | -0.85 | 0.0020027 | 6.55e-04 |
| TEX261 | -1.25 | 0.0000000 | 0.00e+00 |
| TEX9 | -1.97 | 0.0620406 | 2.96e-02 |
| TF | 4.46 | 0.0000000 | 0.00e+00 |
| TFAM | -1.26 | 0.0000000 | 0.00e+00 |
| TFAP2C | 1.33 | 0.0000000 | 0.00e+00 |
| TFAP4 | -1.07 | 0.0130855 | 5.17e-03 |
| TFB2M | -2.02 | 0.0000000 | 0.00e+00 |
| TFDP1 | -1.31 | 0.0000000 | 0.00e+00 |
| TFDP2 | -1.24 | 0.0000440 | 1.09e-05 |
| TFE3 | 1.23 | 0.0000000 | 0.00e+00 |
| TFF1 | 4.01 | 0.0000000 | 0.00e+00 |
| TFG | 0.52 | 0.0001189 | 3.12e-05 |
| TFIP11 | 0.91 | 0.0000015 | 3.00e-07 |
| TFPI2 | 1.61 | 0.0000000 | 0.00e+00 |
| TFRC | -1.56 | 0.0000000 | 0.00e+00 |
| TG | 4.64 | 0.0198508 | 8.19e-03 |
| TGFA | 1.09 | 0.0018906 | 6.14e-04 |
| TGFB1 | 2.19 | 0.0000000 | 0.00e+00 |
| TGFB1I1 | 1.53 | 0.0000010 | 2.00e-07 |
| TGFBI | 1.23 | 0.0000000 | 0.00e+00 |
| TGFBR3 | -0.88 | 0.0372736 | 1.66e-02 |
| TGFBRAP1 | -0.57 | 0.0208266 | 8.63e-03 |
| TGIF1 | 1.01 | 0.0000000 | 0.00e+00 |
| TGIF2 | 1.40 | 0.0001704 | 4.59e-05 |
| TGM2 | 3.09 | 0.0000000 | 0.00e+00 |
| THADA | -0.88 | 0.0000600 | 1.51e-05 |
| THAP10 | -1.19 | 0.0612097 | 2.91e-02 |
| THAP2 | 1.29 | 0.0251663 | 1.07e-02 |
| THAP5 | -1.37 | 0.0003476 | 9.81e-05 |
| THAP9 | -1.37 | 0.0825343 | 4.13e-02 |
| THBD | 2.40 | 0.0000000 | 0.00e+00 |
| THBS1 | 1.05 | 0.0000000 | 0.00e+00 |
| THBS3 | 0.63 | 0.0971378 | 4.99e-02 |
| THEM4 | -0.99 | 0.0107567 | 4.16e-03 |
| THEM6 | -1.04 | 0.0013913 | 4.39e-04 |
| THEMIS2 | 6.04 | 0.0000000 | 0.00e+00 |
| THNSL1 | -2.24 | 0.0001388 | 3.69e-05 |
| THOC1 | -1.12 | 0.0021817 | 7.20e-04 |
| THOC2 | -0.39 | 0.0287086 | 1.24e-02 |
| THOC3 | -1.94 | 0.0000000 | 0.00e+00 |
| THOC6 | -1.00 | 0.0007275 | 2.17e-04 |
| THOC7 | -0.94 | 0.0026276 | 8.83e-04 |
| THOP1 | -0.53 | 0.0080905 | 3.04e-03 |
| THRA | -1.61 | 0.0000004 | 1.00e-07 |
| THRAP3 | -0.47 | 0.0005503 | 1.61e-04 |
| THSD4 | 1.01 | 0.0000000 | 0.00e+00 |
| THSD7A | -1.23 | 0.0000085 | 1.90e-06 |
| THUMPD1 | -0.50 | 0.0557984 | 2.62e-02 |
| THUMPD3 | -0.46 | 0.0196981 | 8.12e-03 |
| THUMPD3-AS1 | -1.62 | 0.0430951 | 1.95e-02 |
| THYN1 | -1.49 | 0.0052901 | 1.90e-03 |
| TIA1 | -0.99 | 0.0000992 | 2.57e-05 |
| TIAM2 | 3.36 | 0.0000091 | 2.10e-06 |
| TICAM1 | 4.14 | 0.0000000 | 0.00e+00 |
| TICRR | -1.81 | 0.0000000 | 0.00e+00 |
| TIFA | 1.91 | 0.0000000 | 0.00e+00 |
| TIGD1 | -1.34 | 0.0436339 | 1.98e-02 |
| TIGD2 | -1.22 | 0.0814111 | 4.06e-02 |
| TIMELESS | -1.77 | 0.0000000 | 0.00e+00 |
| TIMM10 | -1.83 | 0.0000000 | 0.00e+00 |
| TIMM13 | -1.21 | 0.0000001 | 0.00e+00 |
| TIMM17A | -0.85 | 0.0000008 | 2.00e-07 |
| TIMM21 | -3.11 | 0.0000000 | 0.00e+00 |
| TIMM22 | -0.80 | 0.0891887 | 4.52e-02 |
| TIMM23 | -0.91 | 0.0000001 | 0.00e+00 |
| TIMM50 | -0.77 | 0.0000001 | 0.00e+00 |
| TIMM8A | -1.63 | 0.0014369 | 4.55e-04 |
| TIMM8B | -0.90 | 0.0019893 | 6.50e-04 |
| TIMMDC1 | -0.85 | 0.0000758 | 1.93e-05 |
| TIMP1 | 2.04 | 0.0000000 | 0.00e+00 |
| TIMP2 | 1.22 | 0.0000000 | 0.00e+00 |
| TIMP3 | 3.89 | 0.0016088 | 5.14e-04 |
| TIMP4 | -1.85 | 0.0023739 | 7.91e-04 |
| TINAGL1 | 3.25 | 0.0217322 | 9.05e-03 |
| TINF2 | 1.35 | 0.0000000 | 0.00e+00 |
| TIPARP | 3.19 | 0.0000000 | 0.00e+00 |
| TIPIN | -1.71 | 0.0000185 | 4.30e-06 |
| TJAP1 | 1.54 | 0.0000000 | 0.00e+00 |
| TJP2 | 1.90 | 0.0000000 | 0.00e+00 |
| TJP3 | 2.55 | 0.0000000 | 0.00e+00 |
| TK1 | -1.91 | 0.0000000 | 0.00e+00 |
| TKT | -0.52 | 0.0000000 | 0.00e+00 |
| TLDC1 | 1.98 | 0.0000000 | 0.00e+00 |
| TLE1 | 2.09 | 0.0000000 | 0.00e+00 |
| TLE3 | 2.68 | 0.0000000 | 0.00e+00 |
| TLE4 | 0.68 | 0.0126463 | 4.98e-03 |
| TLK2 | 0.73 | 0.0009643 | 2.96e-04 |
| TLL1 | 5.83 | 0.0522626 | 2.43e-02 |
| TLN1 | 1.30 | 0.0000000 | 0.00e+00 |
| TLR2 | 6.83 | 0.0087355 | 3.31e-03 |
| TLR3 | 2.02 | 0.0126654 | 4.99e-03 |
| TM2D2 | -0.38 | 0.0897016 | 4.55e-02 |
| TM4SF1 | -0.29 | 0.0017924 | 5.78e-04 |
| TM4SF18 | -2.13 | 0.0000000 | 0.00e+00 |
| TM4SF20 | -3.22 | 0.0000000 | 0.00e+00 |
| TM4SF4 | -2.71 | 0.0000000 | 0.00e+00 |
| TM7SF3 | -1.71 | 0.0000000 | 0.00e+00 |
| TM9SF1 | 0.44 | 0.0111556 | 4.34e-03 |
| TM9SF2 | -0.36 | 0.0415098 | 1.87e-02 |
| TM9SF3 | -0.49 | 0.0003742 | 1.06e-04 |
| TM9SF4 | 0.99 | 0.0000000 | 0.00e+00 |
| TMA7 | -0.91 | 0.0014098 | 4.45e-04 |
| TMBIM1 | 2.16 | 0.0000000 | 0.00e+00 |
| TMBIM6 | 0.39 | 0.0000116 | 2.60e-06 |
| TMC6 | 1.73 | 0.0000000 | 0.00e+00 |
| TMC8 | 3.23 | 0.0000000 | 0.00e+00 |
| TMCC1-AS1 | -2.28 | 0.0218055 | 9.08e-03 |
| TMCC3 | 1.82 | 0.0079967 | 3.00e-03 |
| TMCO1 | -0.42 | 0.0205624 | 8.51e-03 |
| TMCO3 | -1.26 | 0.0000012 | 2.00e-07 |
| TMED3 | -0.55 | 0.0000893 | 2.30e-05 |
| TMED9 | 0.47 | 0.0001190 | 3.13e-05 |
| TMEM104 | 0.82 | 0.0015500 | 4.94e-04 |
| TMEM106A | 4.70 | 0.0000000 | 0.00e+00 |
| TMEM106B | -1.53 | 0.0000000 | 0.00e+00 |
| TMEM106C | -0.93 | 0.0000000 | 0.00e+00 |
| TMEM107 | -1.85 | 0.0015730 | 5.02e-04 |
| TMEM109 | 0.55 | 0.0008018 | 2.42e-04 |
| TMEM110 | -0.78 | 0.0693234 | 3.36e-02 |
| TMEM115 | 0.98 | 0.0000004 | 1.00e-07 |
| TMEM116 | -1.73 | 0.0072594 | 2.69e-03 |
| TMEM123 | -0.24 | 0.0657415 | 3.16e-02 |
| TMEM126B | -0.98 | 0.0025221 | 8.44e-04 |
| TMEM127 | 1.12 | 0.0000000 | 0.00e+00 |
| TMEM132A | 3.99 | 0.0000000 | 0.00e+00 |
| TMEM132B | 6.44 | 0.0185731 | 7.61e-03 |
| TMEM133 | 1.43 | 0.0683320 | 3.31e-02 |
| TMEM138 | -0.71 | 0.0154091 | 6.19e-03 |
| TMEM139 | 1.70 | 0.0118813 | 4.65e-03 |
| TMEM140 | 5.96 | 0.0000000 | 0.00e+00 |
| TMEM141 | -0.82 | 0.0004172 | 1.20e-04 |
| TMEM143 | -1.77 | 0.0128391 | 5.06e-03 |
| TMEM147 | -0.57 | 0.0000304 | 7.30e-06 |
| TMEM14A | -2.00 | 0.0000429 | 1.06e-05 |
| TMEM14B | -1.55 | 0.0000000 | 0.00e+00 |
| TMEM14C | -0.91 | 0.0000023 | 5.00e-07 |
| TMEM150A | 1.39 | 0.0013462 | 4.24e-04 |
| TMEM150B | 4.09 | 0.0621421 | 2.96e-02 |
| TMEM154 | 6.87 | 0.0081890 | 3.08e-03 |
| TMEM156 | 2.02 | 0.0000000 | 0.00e+00 |
| TMEM158 | 4.36 | 0.0000000 | 0.00e+00 |
| TMEM159 | 3.06 | 0.0000000 | 0.00e+00 |
| TMEM160 | -1.38 | 0.0001560 | 4.19e-05 |
| TMEM163 | 5.69 | 0.0644894 | 3.09e-02 |
| TMEM165 | 1.37 | 0.0000000 | 0.00e+00 |
| TMEM167A | -0.74 | 0.0009260 | 2.83e-04 |
| TMEM167B | 0.93 | 0.0095968 | 3.67e-03 |
| TMEM168 | -2.44 | 0.0000002 | 0.00e+00 |
| TMEM170A | 0.71 | 0.0573582 | 2.71e-02 |
| TMEM171 | 3.93 | 0.0000000 | 0.00e+00 |
| TMEM173 | 5.61 | 0.0000000 | 0.00e+00 |
| TMEM177 | -1.32 | 0.0007567 | 2.26e-04 |
| TMEM179B | 1.05 | 0.0000004 | 1.00e-07 |
| TMEM18 | -0.56 | 0.0728760 | 3.57e-02 |
| TMEM181 | -1.60 | 0.0000010 | 2.00e-07 |
| TMEM183A | -0.61 | 0.0112183 | 4.37e-03 |
| TMEM184A | -1.11 | 0.0003447 | 9.72e-05 |
| TMEM184B | 1.65 | 0.0000000 | 0.00e+00 |
| TMEM184C | -0.54 | 0.0901954 | 4.58e-02 |
| TMEM189 | 1.68 | 0.0000000 | 0.00e+00 |
| TMEM189-UBE2V1 | 0.53 | 0.0002341 | 6.43e-05 |
| TMEM19 | -2.00 | 0.0000000 | 0.00e+00 |
| TMEM190 | 6.30 | 0.0249539 | 1.06e-02 |
| TMEM2 | 0.25 | 0.0578778 | 2.73e-02 |
| TMEM200C | 7.51 | 0.0022179 | 7.34e-04 |
| TMEM203 | -0.44 | 0.0998729 | 5.15e-02 |
| TMEM209 | -0.79 | 0.0333472 | 1.46e-02 |
| TMEM214 | 0.73 | 0.0000025 | 5.00e-07 |
| TMEM217 | 3.81 | 0.0997294 | 5.15e-02 |
| TMEM218 | -1.61 | 0.0004265 | 1.23e-04 |
| TMEM219 | 0.85 | 0.0000252 | 6.00e-06 |
| TMEM220-AS1 | 5.53 | 0.0806408 | 4.02e-02 |
| TMEM223 | -0.81 | 0.0337408 | 1.48e-02 |
| TMEM229B | 2.85 | 0.0155962 | 6.27e-03 |
| TMEM230 | -0.42 | 0.0187311 | 7.68e-03 |
| TMEM231 | -1.11 | 0.0767686 | 3.79e-02 |
| TMEM237 | -2.19 | 0.0000000 | 0.00e+00 |
| TMEM239 | 8.09 | 0.0006426 | 1.90e-04 |
| TMEM242 | -0.83 | 0.0746991 | 3.67e-02 |
| TMEM248 | 0.28 | 0.0896005 | 4.55e-02 |
| TMEM256 | -1.07 | 0.0000485 | 1.20e-05 |
| TMEM259 | 0.56 | 0.0001319 | 3.49e-05 |
| TMEM261 | -1.39 | 0.0000000 | 0.00e+00 |
| TMEM265 | 3.46 | 0.0000000 | 0.00e+00 |
| TMEM30A | -0.40 | 0.0100084 | 3.84e-03 |
| TMEM39A | 0.56 | 0.0380637 | 1.70e-02 |
| TMEM40 | 4.66 | 0.0000006 | 1.00e-07 |
| TMEM41A | 1.05 | 0.0000158 | 3.70e-06 |
| TMEM43 | 0.56 | 0.0003571 | 1.01e-04 |
| TMEM44 | 1.37 | 0.0004019 | 1.15e-04 |
| TMEM47 | 2.09 | 0.0006824 | 2.03e-04 |
| TMEM5 | -1.17 | 0.0000242 | 5.80e-06 |
| TMEM50A | 0.71 | 0.0000210 | 5.00e-06 |
| TMEM51 | 1.69 | 0.0000000 | 0.00e+00 |
| TMEM51-AS1 | 2.40 | 0.0037602 | 1.30e-03 |
| TMEM52B | 9.19 | 0.0000483 | 1.20e-05 |
| TMEM53 | 1.74 | 0.0011628 | 3.62e-04 |
| TMEM55B | 0.66 | 0.0014556 | 4.61e-04 |
| TMEM56 | -2.16 | 0.0000000 | 0.00e+00 |
| TMEM56-RWDD3 | -1.54 | 0.0058390 | 2.11e-03 |
| TMEM57 | 0.88 | 0.0073879 | 2.74e-03 |
| TMEM59 | 0.52 | 0.0041131 | 1.44e-03 |
| TMEM62 | 1.48 | 0.0000001 | 0.00e+00 |
| TMEM63A | 1.51 | 0.0000005 | 1.00e-07 |
| TMEM63B | 1.68 | 0.0000000 | 0.00e+00 |
| TMEM64 | -1.53 | 0.0000116 | 2.60e-06 |
| TMEM65 | -0.50 | 0.0963991 | 4.95e-02 |
| TMEM70 | -0.89 | 0.0075498 | 2.81e-03 |
| TMEM86A | 7.53 | 0.0021399 | 7.05e-04 |
| TMEM87A | 0.48 | 0.0554250 | 2.60e-02 |
| TMEM87B | 0.76 | 0.0031873 | 1.09e-03 |
| TMEM8A | 1.29 | 0.0000000 | 0.00e+00 |
| TMEM8B | 1.92 | 0.0011462 | 3.56e-04 |
| TMEM9 | -0.36 | 0.0756462 | 3.73e-02 |
| TMEM92 | 2.02 | 0.0000000 | 0.00e+00 |
| TMEM98 | -0.82 | 0.0084760 | 3.19e-03 |
| TMEM99 | -1.56 | 0.0030366 | 1.03e-03 |
| TMOD1 | 3.40 | 0.0000000 | 0.00e+00 |
| TMOD3 | -0.36 | 0.0031165 | 1.06e-03 |
| TMPO | -1.27 | 0.0000000 | 0.00e+00 |
| TMPO-AS1 | -1.51 | 0.0067102 | 2.47e-03 |
| TMPRSS2 | 7.17 | 0.0045032 | 1.59e-03 |
| TMPRSS3 | 6.09 | 0.0003665 | 1.04e-04 |
| TMPRSS4 | 7.12 | 0.0050146 | 1.79e-03 |
| TMSB10 | 1.23 | 0.0000000 | 0.00e+00 |
| TMSB4X | -1.00 | 0.0000000 | 0.00e+00 |
| TMTC3 | -1.34 | 0.0000006 | 1.00e-07 |
| TMTC4 | -0.92 | 0.0228628 | 9.58e-03 |
| TMUB1 | 0.69 | 0.0173492 | 7.05e-03 |
| TMUB2 | 0.87 | 0.0000054 | 1.20e-06 |
| TMX1 | -0.96 | 0.0000003 | 1.00e-07 |
| TMX2 | -0.83 | 0.0000000 | 0.00e+00 |
| TMX2-CTNND1 | 0.72 | 0.0000000 | 0.00e+00 |
| TMX3 | -1.27 | 0.0000032 | 7.00e-07 |
| TMX4 | -0.86 | 0.0007942 | 2.39e-04 |
| TNC | 3.75 | 0.0000171 | 4.00e-06 |
| TNFAIP1 | 2.12 | 0.0000000 | 0.00e+00 |
| TNFAIP2 | 3.70 | 0.0000000 | 0.00e+00 |
| TNFAIP3 | 7.14 | 0.0000000 | 0.00e+00 |
| TNFAIP6 | 5.55 | 0.0000000 | 0.00e+00 |
| TNFAIP8 | 1.41 | 0.0000000 | 0.00e+00 |
| TNFAIP8L2-SCNM1 | 1.70 | 0.0000000 | 0.00e+00 |
| TNFAIP8L3 | 3.05 | 0.0000122 | 2.80e-06 |
| TNFRSF10A | 1.57 | 0.0000000 | 0.00e+00 |
| TNFRSF10B | 1.66 | 0.0000000 | 0.00e+00 |
| TNFRSF10D | -1.28 | 0.0000235 | 5.60e-06 |
| TNFRSF11A | 1.12 | 0.0036296 | 1.25e-03 |
| TNFRSF12A | 1.19 | 0.0000000 | 0.00e+00 |
| TNFRSF14 | 7.28 | 0.0036749 | 1.27e-03 |
| TNFRSF1B | 6.95 | 0.0000000 | 0.00e+00 |
| TNFRSF21 | 1.40 | 0.0000000 | 0.00e+00 |
| TNFRSF8 | 6.20 | 0.0000000 | 0.00e+00 |
| TNFRSF9 | 5.96 | 0.0000000 | 0.00e+00 |
| TNFSF10 | 6.24 | 0.0000000 | 0.00e+00 |
| TNFSF12-TNFSF13 | 1.05 | 0.0666908 | 3.21e-02 |
| TNFSF13 | 1.75 | 0.0151353 | 6.07e-03 |
| TNFSF13B | 4.82 | 0.0000071 | 1.60e-06 |
| TNFSF14 | 6.65 | 0.0129194 | 5.09e-03 |
| TNFSF15 | 1.51 | 0.0001124 | 2.94e-05 |
| TNFSF4 | 2.91 | 0.0600171 | 2.85e-02 |
| TNFSF8 | 5.61 | 0.0724836 | 3.55e-02 |
| TNFSF9 | 1.73 | 0.0000000 | 0.00e+00 |
| TNIP1 | 4.28 | 0.0000000 | 0.00e+00 |
| TNIP2 | 1.98 | 0.0000000 | 0.00e+00 |
| TNIP3 | 4.85 | 0.0000000 | 0.00e+00 |
| TNKS1BP1 | 2.37 | 0.0000000 | 0.00e+00 |
| TNNT2 | 6.49 | 0.0170469 | 6.92e-03 |
| TNPO1 | -0.70 | 0.0000000 | 0.00e+00 |
| TNPO3 | -0.72 | 0.0000538 | 1.34e-05 |
| TNRC18 | 1.61 | 0.0000000 | 0.00e+00 |
| TNRC6C | 1.05 | 0.0596341 | 2.82e-02 |
| TNRC6C-AS1 | 2.58 | 0.0722667 | 3.53e-02 |
| TNS1 | 2.18 | 0.0000000 | 0.00e+00 |
| TNS2 | -1.55 | 0.0000018 | 4.00e-07 |
| TNS3 | 0.41 | 0.0000192 | 4.50e-06 |
| TNS4 | 0.53 | 0.0003933 | 1.13e-04 |
| TOB2 | 1.44 | 0.0000000 | 0.00e+00 |
| TOE1 | -0.95 | 0.0107994 | 4.18e-03 |
| TOLLIP | 0.69 | 0.0001910 | 5.17e-05 |
| TOM1 | 2.40 | 0.0000000 | 0.00e+00 |
| TOM1L2 | 1.68 | 0.0000000 | 0.00e+00 |
| TOMM20 | -1.07 | 0.0000000 | 0.00e+00 |
| TOMM22 | -0.94 | 0.0000050 | 1.10e-06 |
| TOMM40 | -1.11 | 0.0000000 | 0.00e+00 |
| TOMM5 | -1.32 | 0.0000000 | 0.00e+00 |
| TOMM6 | -0.69 | 0.0000728 | 1.85e-05 |
| TOMM70A | -0.59 | 0.0001325 | 3.51e-05 |
| TONSL | -2.21 | 0.0000000 | 0.00e+00 |
| TOP1 | 0.58 | 0.0001508 | 4.03e-05 |
| TOP1MT | -0.53 | 0.0598925 | 2.84e-02 |
| TOP2A | -3.92 | 0.0000000 | 0.00e+00 |
| TOP2B | -1.03 | 0.0000000 | 0.00e+00 |
| TOP3B | 0.58 | 0.0950249 | 4.87e-02 |
| TOPBP1 | -2.30 | 0.0000000 | 0.00e+00 |
| TOR1B | 1.13 | 0.0000000 | 0.00e+00 |
| TOR3A | -1.64 | 0.0000000 | 0.00e+00 |
| TOR4A | 1.74 | 0.0000000 | 0.00e+00 |
| TOX3 | 1.30 | 0.0763535 | 3.77e-02 |
| TOX4 | 0.45 | 0.0094732 | 3.61e-03 |
| TP53BP2 | 1.67 | 0.0000000 | 0.00e+00 |
| TP53I11 | 0.69 | 0.0000227 | 5.40e-06 |
| TP53I13 | 0.39 | 0.0924825 | 4.72e-02 |
| TP53I3 | -0.46 | 0.0031968 | 1.09e-03 |
| TP53INP1 | 1.30 | 0.0025164 | 8.42e-04 |
| TP53INP2 | 4.28 | 0.0000000 | 0.00e+00 |
| TP53RK | 0.82 | 0.0033912 | 1.16e-03 |
| TP53TG1 | -1.10 | 0.0791471 | 3.92e-02 |
| TP63 | 1.52 | 0.0002386 | 6.56e-05 |
| TPBG | 2.31 | 0.0000000 | 0.00e+00 |
| TPCN1 | 0.36 | 0.0730418 | 3.58e-02 |
| TPCN2 | 0.75 | 0.0108826 | 4.22e-03 |
| TPD52 | -0.58 | 0.0028027 | 9.47e-04 |
| TPD52L1 | -0.96 | 0.0020033 | 6.55e-04 |
| TPD52L2 | 0.56 | 0.0000037 | 8.00e-07 |
| TPGS1 | 1.06 | 0.0148337 | 5.94e-03 |
| TPGS2 | -0.75 | 0.0003913 | 1.12e-04 |
| TPI1 | -1.09 | 0.0000000 | 0.00e+00 |
| TPI1P2 | -1.17 | 0.0061280 | 2.23e-03 |
| TPI1P3 | -0.92 | 0.0042884 | 1.51e-03 |
| TPK1 | 3.01 | 0.0000034 | 7.00e-07 |
| TPM1 | 0.95 | 0.0000000 | 0.00e+00 |
| TPM2 | -1.31 | 0.0000000 | 0.00e+00 |
| TPM3 | -0.28 | 0.0111361 | 4.33e-03 |
| TPM4 | 0.27 | 0.0097139 | 3.72e-03 |
| TPP1 | 1.24 | 0.0000000 | 0.00e+00 |
| TPP2 | -0.85 | 0.0013853 | 4.37e-04 |
| TPPP | -1.83 | 0.0080175 | 3.01e-03 |
| TPPP3 | -2.55 | 0.0016741 | 5.36e-04 |
| TPR | -1.14 | 0.0000000 | 0.00e+00 |
| TPRA1 | 1.81 | 0.0000000 | 0.00e+00 |
| TPRKB | -1.54 | 0.0000001 | 0.00e+00 |
| TPRN | 1.30 | 0.0007541 | 2.26e-04 |
| TPST2 | 0.94 | 0.0484965 | 2.23e-02 |
| TPT1 | 0.74 | 0.0000000 | 0.00e+00 |
| TPX2 | -1.89 | 0.0000000 | 0.00e+00 |
| TRA2A | -0.80 | 0.0002619 | 7.24e-05 |
| TRA2B | -1.24 | 0.0000000 | 0.00e+00 |
| TRABD2A | 1.17 | 0.0928263 | 4.74e-02 |
| TRADD | 1.23 | 0.0000002 | 0.00e+00 |
| TRAF1 | 6.01 | 0.0000000 | 0.00e+00 |
| TRAF2 | 0.74 | 0.0000039 | 8.00e-07 |
| TRAF3 | 0.64 | 0.0004206 | 1.21e-04 |
| TRAF3IP2 | 2.93 | 0.0000000 | 0.00e+00 |
| TRAF4 | 1.33 | 0.0000000 | 0.00e+00 |
| TRAF5 | -0.93 | 0.0532816 | 2.48e-02 |
| TRAF6 | 0.94 | 0.0044363 | 1.56e-03 |
| TRAF7 | 0.56 | 0.0000598 | 1.51e-05 |
| TRAFD1 | 1.82 | 0.0000000 | 0.00e+00 |
| TRAIP | -1.94 | 0.0000938 | 2.42e-05 |
| TRAK2 | -0.92 | 0.0002538 | 7.00e-05 |
| TRAM1 | -0.70 | 0.0000000 | 0.00e+00 |
| TRAM2 | -0.50 | 0.0452630 | 2.07e-02 |
| TRANK1 | 5.04 | 0.0000000 | 0.00e+00 |
| TRAP1 | -1.61 | 0.0000000 | 0.00e+00 |
| TRAPPC1 | -0.41 | 0.0174998 | 7.13e-03 |
| TRAPPC11 | -0.57 | 0.0619785 | 2.95e-02 |
| TRAPPC2L | -1.61 | 0.0000000 | 0.00e+00 |
| TRAPPC8 | -0.57 | 0.0638162 | 3.05e-02 |
| TRAPPC9 | 0.65 | 0.0233919 | 9.85e-03 |
| TRDMT1 | -1.77 | 0.0233094 | 9.80e-03 |
| TRERF1 | 3.05 | 0.0000000 | 0.00e+00 |
| TREX1 | 2.06 | 0.0000000 | 0.00e+00 |
| TRIAP1 | -1.06 | 0.0000057 | 1.20e-06 |
| TRIB1 | 3.76 | 0.0000000 | 0.00e+00 |
| TRIB2 | 4.51 | 0.0000570 | 1.43e-05 |
| TRIB3 | 1.67 | 0.0000000 | 0.00e+00 |
| TRIM11 | 1.70 | 0.0000000 | 0.00e+00 |
| TRIM14 | 1.60 | 0.0000000 | 0.00e+00 |
| TRIM16 | -0.21 | 0.0659119 | 3.17e-02 |
| TRIM16L | -0.55 | 0.0000034 | 7.00e-07 |
| TRIM21 | 2.90 | 0.0000000 | 0.00e+00 |
| TRIM22 | 7.05 | 0.0000000 | 0.00e+00 |
| TRIM25 | 2.57 | 0.0000000 | 0.00e+00 |
| TRIM29 | 2.53 | 0.0000000 | 0.00e+00 |
| TRIM31 | 8.14 | 0.0005657 | 1.66e-04 |
| TRIM35 | 0.97 | 0.0001058 | 2.75e-05 |
| TRIM36 | 2.51 | 0.0117685 | 4.60e-03 |
| TRIM38 | 2.25 | 0.0000000 | 0.00e+00 |
| TRIM4 | 0.64 | 0.0116871 | 4.57e-03 |
| TRIM44 | 0.47 | 0.0045129 | 1.59e-03 |
| TRIM47 | 1.27 | 0.0000000 | 0.00e+00 |
| TRIM5 | 1.47 | 0.0000000 | 0.00e+00 |
| TRIM56 | 1.35 | 0.0000000 | 0.00e+00 |
| TRIM59 | -1.64 | 0.0002044 | 5.56e-05 |
| TRIM6 | -1.38 | 0.0000006 | 1.00e-07 |
| TRIM6-TRIM34 | -0.57 | 0.0911874 | 4.65e-02 |
| TRIM66 | -1.42 | 0.0081106 | 3.05e-03 |
| TRIM69 | 6.53 | 0.0155876 | 6.27e-03 |
| TRIM7 | -0.77 | 0.0572374 | 2.70e-02 |
| TRIM8 | 3.45 | 0.0000000 | 0.00e+00 |
| TRIML2 | 0.76 | 0.0007299 | 2.18e-04 |
| TRIO | 1.03 | 0.0000000 | 0.00e+00 |
| TRIP10 | 1.72 | 0.0000000 | 0.00e+00 |
| TRIP11 | -0.86 | 0.0039812 | 1.39e-03 |
| TRIP12 | 0.26 | 0.0426510 | 1.93e-02 |
| TRIP13 | -2.78 | 0.0000000 | 0.00e+00 |
| TRIP4 | 0.95 | 0.0000044 | 1.00e-06 |
| TRIQK | -1.71 | 0.0028397 | 9.60e-04 |
| TRMT10A | -1.50 | 0.0353727 | 1.56e-02 |
| TRMT10C | -0.66 | 0.0296492 | 1.28e-02 |
| TRMT11 | -0.85 | 0.0565974 | 2.66e-02 |
| TRMT112 | -1.11 | 0.0000000 | 0.00e+00 |
| TRMT5 | -0.75 | 0.0120530 | 4.72e-03 |
| TRMT6 | -0.57 | 0.0159681 | 6.44e-03 |
| TRMT61B | -1.47 | 0.0000668 | 1.69e-05 |
| TRNP1 | -0.97 | 0.0000000 | 0.00e+00 |
| TRNT1 | -0.93 | 0.0071956 | 2.66e-03 |
| TROAP | -1.40 | 0.0000003 | 1.00e-07 |
| TRPA1 | 0.66 | 0.0953839 | 4.89e-02 |
| TRPC4AP | 0.66 | 0.0000014 | 3.00e-07 |
| TRPM4 | 1.66 | 0.0000000 | 0.00e+00 |
| TRPM7 | -0.46 | 0.0174401 | 7.10e-03 |
| TRPS1 | 3.09 | 0.0963469 | 4.94e-02 |
| TRUB1 | -0.89 | 0.0022016 | 7.28e-04 |
| TRUB2 | -0.82 | 0.0001068 | 2.78e-05 |
| TSC2 | 0.78 | 0.0000446 | 1.10e-05 |
| TSC22D1 | 0.89 | 0.0000000 | 0.00e+00 |
| TSC22D2 | 1.44 | 0.0000029 | 6.00e-07 |
| TSC22D3 | 1.55 | 0.0000000 | 0.00e+00 |
| TSC22D4 | 2.48 | 0.0000000 | 0.00e+00 |
| TSEN15 | -1.15 | 0.0000549 | 1.37e-05 |
| TSEN2 | -2.24 | 0.0000005 | 1.00e-07 |
| TSEN54 | -0.72 | 0.0041011 | 1.43e-03 |
| TSG101 | 0.31 | 0.0770095 | 3.81e-02 |
| TSHZ3 | 1.97 | 0.0001253 | 3.30e-05 |
| TSKU | 0.65 | 0.0000000 | 0.00e+00 |
| TSN | -1.41 | 0.0000000 | 0.00e+00 |
| TSNAX | -0.45 | 0.0504330 | 2.33e-02 |
| TSPAN13 | -1.83 | 0.0000000 | 0.00e+00 |
| TSPAN14 | 0.39 | 0.0018021 | 5.82e-04 |
| TSPAN15 | 0.71 | 0.0000006 | 1.00e-07 |
| TSPAN18 | 2.87 | 0.0001020 | 2.65e-05 |
| TSPAN3 | 0.57 | 0.0000003 | 1.00e-07 |
| TSPAN31 | 1.14 | 0.0000010 | 2.00e-07 |
| TSPAN6 | -1.03 | 0.0005695 | 1.67e-04 |
| TSPAN7 | -2.62 | 0.0431872 | 1.96e-02 |
| TSPAN9 | 1.19 | 0.0000002 | 0.00e+00 |
| TSPYL5 | 0.51 | 0.0734676 | 3.60e-02 |
| TSR1 | -0.48 | 0.0011890 | 3.70e-04 |
| TSSC4 | 0.60 | 0.0025183 | 8.43e-04 |
| TSTA3 | -0.54 | 0.0966676 | 4.96e-02 |
| TTC1 | -0.45 | 0.0209702 | 8.70e-03 |
| TTC12 | -1.70 | 0.0118555 | 4.64e-03 |
| TTC13 | -0.76 | 0.0338598 | 1.49e-02 |
| TTC14 | -1.41 | 0.0002214 | 6.06e-05 |
| TTC17 | 0.56 | 0.0021108 | 6.95e-04 |
| TTC21B | -1.37 | 0.0003767 | 1.07e-04 |
| TTC26 | -0.90 | 0.0364585 | 1.62e-02 |
| TTC27 | -0.71 | 0.0078908 | 2.95e-03 |
| TTC28 | 2.23 | 0.0000000 | 0.00e+00 |
| TTC3 | -2.07 | 0.0000000 | 0.00e+00 |
| TTC30A | -1.51 | 0.0019738 | 6.44e-04 |
| TTC30B | -1.53 | 0.0107575 | 4.17e-03 |
| TTC37 | -1.54 | 0.0000000 | 0.00e+00 |
| TTC38 | -0.84 | 0.0266298 | 1.14e-02 |
| TTC39A | 1.93 | 0.0000001 | 0.00e+00 |
| TTC39B | 2.87 | 0.0000000 | 0.00e+00 |
| TTC7B | 1.47 | 0.0000087 | 2.00e-06 |
| TTC8 | -0.69 | 0.0232999 | 9.80e-03 |
| TTC9 | 1.66 | 0.0000000 | 0.00e+00 |
| TTC9B | 7.09 | 0.0053193 | 1.91e-03 |
| TTF2 | -0.75 | 0.0031369 | 1.07e-03 |
| TTI1 | -0.53 | 0.0343092 | 1.51e-02 |
| TTK | -2.71 | 0.0000000 | 0.00e+00 |
| TTL | -0.74 | 0.0006198 | 1.83e-04 |
| TTLL12 | -0.50 | 0.0686086 | 3.32e-02 |
| TTLL3 | 1.11 | 0.0070174 | 2.59e-03 |
| TTLL4 | 0.46 | 0.0567364 | 2.67e-02 |
| TTLL5 | -0.82 | 0.0205495 | 8.50e-03 |
| TTLL6 | 0.86 | 0.0122479 | 4.81e-03 |
| TTYH2 | 2.19 | 0.0000116 | 2.60e-06 |
| TTYH3 | 1.02 | 0.0000000 | 0.00e+00 |
| TUB | -2.80 | 0.0000000 | 0.00e+00 |
| TUBA1A | -1.59 | 0.0000000 | 0.00e+00 |
| TUBA1B | -2.06 | 0.0000000 | 0.00e+00 |
| TUBA1C | -1.39 | 0.0000000 | 0.00e+00 |
| TUBA4A | -0.62 | 0.0000001 | 0.00e+00 |
| TUBA8 | 2.93 | 0.0248015 | 1.05e-02 |
| TUBB3 | -2.04 | 0.0000000 | 0.00e+00 |
| TUBB4A | -1.56 | 0.0000117 | 2.70e-06 |
| TUBB4B | -1.71 | 0.0000000 | 0.00e+00 |
| TUBB6 | -0.38 | 0.0062675 | 2.29e-03 |
| TUBG1 | -0.97 | 0.0000000 | 0.00e+00 |
| TUBG2 | -0.55 | 0.0230605 | 9.68e-03 |
| TUBGCP3 | -1.42 | 0.0000129 | 3.00e-06 |
| TUBGCP5 | -1.22 | 0.0008977 | 2.73e-04 |
| TUFM | -0.72 | 0.0000000 | 0.00e+00 |
| TUFT1 | 1.79 | 0.0000000 | 0.00e+00 |
| TUSC1 | 1.04 | 0.0287476 | 1.24e-02 |
| TUSC3 | -1.07 | 0.0000026 | 6.00e-07 |
| TUT1 | 0.86 | 0.0004944 | 1.44e-04 |
| TVP23C-CDRT4 | 0.98 | 0.0008854 | 2.69e-04 |
| TWF1 | -1.02 | 0.0000000 | 0.00e+00 |
| TWF2 | 0.35 | 0.0684502 | 3.31e-02 |
| TWIST2 | 2.02 | 0.0000000 | 0.00e+00 |
| TWISTNB | -0.57 | 0.0602711 | 2.86e-02 |
| TXLNG | -0.93 | 0.0004429 | 1.28e-04 |
| TXLNGY | -1.57 | 0.0003770 | 1.07e-04 |
| TXN | -1.35 | 0.0000000 | 0.00e+00 |
| TXN2 | -0.92 | 0.0000102 | 2.30e-06 |
| TXNDC11 | 0.48 | 0.0398726 | 1.79e-02 |
| TXNDC12 | -0.44 | 0.0136811 | 5.44e-03 |
| TXNDC17 | -0.93 | 0.0000072 | 1.60e-06 |
| TXNDC5 | -1.02 | 0.0000000 | 0.00e+00 |
| TXNDC9 | -0.97 | 0.0021234 | 6.99e-04 |
| TXNIP | 5.12 | 0.0000000 | 0.00e+00 |
| TXNL4A | -0.74 | 0.0009397 | 2.87e-04 |
| TXNRD1 | 0.38 | 0.0000004 | 1.00e-07 |
| TXNRD2 | -0.93 | 0.0048507 | 1.73e-03 |
| TYK2 | 1.24 | 0.0000000 | 0.00e+00 |
| TYMP | 4.71 | 0.0000000 | 0.00e+00 |
| TYMS | -2.74 | 0.0000000 | 0.00e+00 |
| TYMSOS | -2.58 | 0.0491756 | 2.26e-02 |
| TYSND1 | 0.77 | 0.0001289 | 3.41e-05 |
| TYW3 | -0.97 | 0.0050075 | 1.79e-03 |
| TYW5 | 0.77 | 0.0088810 | 3.37e-03 |
| U2AF2 | -0.77 | 0.0000000 | 0.00e+00 |
| U2SURP | -1.19 | 0.0000000 | 0.00e+00 |
| UACA | -1.14 | 0.0000003 | 1.00e-07 |
| UAP1L1 | -0.85 | 0.0891113 | 4.52e-02 |
| UBA1 | 0.71 | 0.0000000 | 0.00e+00 |
| UBA2 | -0.83 | 0.0000000 | 0.00e+00 |
| UBA5 | -0.68 | 0.0396262 | 1.77e-02 |
| UBA7 | 5.48 | 0.0000000 | 0.00e+00 |
| UBAC1 | -1.00 | 0.0000435 | 1.07e-05 |
| UBALD1 | 2.15 | 0.0000000 | 0.00e+00 |
| UBALD2 | 2.29 | 0.0000000 | 0.00e+00 |
| UBAP1 | 1.63 | 0.0000000 | 0.00e+00 |
| UBAP2 | 0.61 | 0.0016470 | 5.27e-04 |
| UBAP2L | 0.34 | 0.0190368 | 7.82e-03 |
| UBASH3A | 6.53 | 0.0161724 | 6.54e-03 |
| UBASH3B | 0.52 | 0.0031583 | 1.08e-03 |
| UBB | 0.55 | 0.0000000 | 0.00e+00 |
| UBC | 1.72 | 0.0000000 | 0.00e+00 |
| UBD | 8.33 | 0.0003872 | 1.11e-04 |
| UBE2B | 0.98 | 0.0002156 | 5.89e-05 |
| UBE2C | -2.32 | 0.0000000 | 0.00e+00 |
| UBE2D1 | 0.61 | 0.0707394 | 3.44e-02 |
| UBE2D2 | 0.35 | 0.0227457 | 9.53e-03 |
| UBE2D3 | 0.66 | 0.0000000 | 0.00e+00 |
| UBE2E1 | 1.25 | 0.0000000 | 0.00e+00 |
| UBE2E2 | 1.72 | 0.0000000 | 0.00e+00 |
| UBE2E3 | -0.85 | 0.0038906 | 1.35e-03 |
| UBE2G2 | -0.53 | 0.0526945 | 2.45e-02 |
| UBE2H | 2.21 | 0.0000000 | 0.00e+00 |
| UBE2I | -0.61 | 0.0001066 | 2.78e-05 |
| UBE2L6 | 3.97 | 0.0000000 | 0.00e+00 |
| UBE2M | 0.44 | 0.0014442 | 4.57e-04 |
| UBE2MP1 | 0.61 | 0.0653264 | 3.14e-02 |
| UBE2N | -0.61 | 0.0003241 | 9.10e-05 |
| UBE2O | 0.82 | 0.0000007 | 1.00e-07 |
| UBE2Q1 | 0.46 | 0.0079004 | 2.96e-03 |
| UBE2Q2 | -0.74 | 0.0042904 | 1.51e-03 |
| UBE2R2 | 0.81 | 0.0000001 | 0.00e+00 |
| UBE2S | -1.12 | 0.0000000 | 0.00e+00 |
| UBE2T | -2.02 | 0.0000000 | 0.00e+00 |
| UBE2V2 | -0.82 | 0.0003890 | 1.11e-04 |
| UBE2Z | 1.45 | 0.0000000 | 0.00e+00 |
| UBE3A | -0.77 | 0.0020471 | 6.71e-04 |
| UBE3C | -0.68 | 0.0000171 | 4.00e-06 |
| UBE4B | -1.01 | 0.0008051 | 2.43e-04 |
| UBL3 | -0.94 | 0.0401372 | 1.80e-02 |
| UBL4A | -0.46 | 0.0177714 | 7.25e-03 |
| UBL5 | -0.44 | 0.0272112 | 1.17e-02 |
| UBN1 | 0.52 | 0.0076538 | 2.85e-03 |
| UBN2 | 0.57 | 0.0810496 | 4.04e-02 |
| UBOX5 | 1.54 | 0.0000034 | 7.00e-07 |
| UBQLN1 | 0.32 | 0.0074294 | 2.76e-03 |
| UBQLN2 | 0.50 | 0.0341069 | 1.50e-02 |
| UBR2 | 0.66 | 0.0100335 | 3.86e-03 |
| UBR3 | -0.69 | 0.0090664 | 3.45e-03 |
| UBR4 | 1.07 | 0.0000000 | 0.00e+00 |
| UBR7 | -1.58 | 0.0000078 | 1.70e-06 |
| UBTD2 | 1.10 | 0.0000443 | 1.10e-05 |
| UBTF | -1.23 | 0.0000000 | 0.00e+00 |
| UBXN1 | 0.65 | 0.0000114 | 2.60e-06 |
| UBXN2B | -0.64 | 0.0347157 | 1.53e-02 |
| UBXN4 | -0.29 | 0.0767686 | 3.79e-02 |
| UBXN6 | 0.62 | 0.0009090 | 2.77e-04 |
| UBXN8 | -1.35 | 0.0099149 | 3.80e-03 |
| UCHL1 | -0.82 | 0.0000000 | 0.00e+00 |
| UCHL5 | -1.40 | 0.0000006 | 1.00e-07 |
| UCK2 | -0.33 | 0.0296395 | 1.28e-02 |
| UCKL1 | 0.59 | 0.0424233 | 1.92e-02 |
| UCN2 | 2.38 | 0.0007791 | 2.34e-04 |
| UCP2 | -0.65 | 0.0203459 | 8.41e-03 |
| UFD1L | -0.37 | 0.0612866 | 2.92e-02 |
| UFL1 | -1.55 | 0.0000076 | 1.70e-06 |
| UGCG | 1.76 | 0.0000000 | 0.00e+00 |
| UGDH | -1.36 | 0.0000000 | 0.00e+00 |
| UGGT1 | -0.34 | 0.0235286 | 9.91e-03 |
| UGGT2 | -0.90 | 0.0179965 | 7.35e-03 |
| UGP2 | -0.47 | 0.0026477 | 8.91e-04 |
| UGT1A1 | -1.52 | 0.0026541 | 8.94e-04 |
| UGT1A10 | -1.54 | 0.0030528 | 1.04e-03 |
| UGT1A3 | -1.57 | 0.0022925 | 7.62e-04 |
| UGT1A4 | -1.53 | 0.0029214 | 9.89e-04 |
| UGT1A5 | -1.55 | 0.0030182 | 1.03e-03 |
| UGT1A6 | -1.55 | 0.0019577 | 6.38e-04 |
| UGT1A7 | -1.41 | 0.0038513 | 1.34e-03 |
| UGT1A8 | -1.55 | 0.0024495 | 8.18e-04 |
| UGT1A9 | -1.84 | 0.0000665 | 1.68e-05 |
| UHMK1 | -0.39 | 0.0444049 | 2.02e-02 |
| UHRF1 | -2.17 | 0.0000000 | 0.00e+00 |
| UHRF1BP1 | 1.04 | 0.0000001 | 0.00e+00 |
| ULBP2 | 2.93 | 0.0000009 | 2.00e-07 |
| ULK1 | 1.22 | 0.0000000 | 0.00e+00 |
| ULK3 | -0.91 | 0.0002668 | 7.38e-05 |
| UMPS | -1.23 | 0.0000000 | 0.00e+00 |
| UNC119B | -0.97 | 0.0001479 | 3.95e-05 |
| UNC13A | 1.93 | 0.0000000 | 0.00e+00 |
| UNC13B | -0.58 | 0.0344227 | 1.52e-02 |
| UNC13D | -0.75 | 0.0552215 | 2.59e-02 |
| UNC5A | 3.32 | 0.0175290 | 7.15e-03 |
| UNC5B | 5.35 | 0.0000000 | 0.00e+00 |
| UNG | -2.06 | 0.0000000 | 0.00e+00 |
| UNK | 1.18 | 0.0000000 | 0.00e+00 |
| UNKL | -1.38 | 0.0000034 | 7.00e-07 |
| UPB1 | 6.53 | 0.0156707 | 6.31e-03 |
| UPF1 | 0.66 | 0.0000339 | 8.20e-06 |
| UPF2 | -0.51 | 0.0367492 | 1.63e-02 |
| UPK1A-AS1 | 4.04 | 0.0672812 | 3.25e-02 |
| UPK1B | -1.98 | 0.0000000 | 0.00e+00 |
| UPP1 | 0.36 | 0.0886565 | 4.49e-02 |
| UQCC1 | -1.29 | 0.0000016 | 3.00e-07 |
| UQCC2 | -0.94 | 0.0022560 | 7.49e-04 |
| UQCC3 | -1.39 | 0.0000001 | 0.00e+00 |
| UQCR11 | -0.54 | 0.0073259 | 2.72e-03 |
| UQCRB | -0.78 | 0.0000338 | 8.20e-06 |
| UQCRC2 | -0.91 | 0.0000000 | 0.00e+00 |
| UQCRFS1 | -0.87 | 0.0000000 | 0.00e+00 |
| UQCRH | -0.87 | 0.0000001 | 0.00e+00 |
| UQCRHL | -0.66 | 0.0074715 | 2.78e-03 |
| UQCRQ | -1.09 | 0.0000000 | 0.00e+00 |
| URB1 | -0.49 | 0.0442069 | 2.01e-02 |
| URB2 | -1.05 | 0.0006558 | 1.94e-04 |
| URGCP | 1.01 | 0.0000014 | 3.00e-07 |
| URGCP-MRPS24 | -0.37 | 0.0351824 | 1.55e-02 |
| URI1 | -1.17 | 0.0000000 | 0.00e+00 |
| URM1 | -0.53 | 0.0108610 | 4.21e-03 |
| UROD | -0.79 | 0.0070249 | 2.59e-03 |
| UROS | -0.66 | 0.0194910 | 8.02e-03 |
| USB1 | 0.62 | 0.0008364 | 2.53e-04 |
| USF1 | 1.56 | 0.0000000 | 0.00e+00 |
| USF2 | 0.42 | 0.0019181 | 6.25e-04 |
| USMG5 | -0.65 | 0.0004973 | 1.45e-04 |
| USP1 | -1.09 | 0.0000468 | 1.16e-05 |
| USP11 | 1.64 | 0.0000000 | 0.00e+00 |
| USP12 | 0.51 | 0.0352583 | 1.56e-02 |
| USP13 | -0.66 | 0.0348162 | 1.54e-02 |
| USP14 | -0.46 | 0.0187107 | 7.67e-03 |
| USP16 | -0.64 | 0.0136034 | 5.40e-03 |
| USP18 | 3.05 | 0.0000000 | 0.00e+00 |
| USP19 | 0.48 | 0.0118008 | 4.61e-03 |
| USP20 | 0.81 | 0.0291256 | 1.25e-02 |
| USP21 | 0.57 | 0.0759043 | 3.74e-02 |
| USP22 | -0.48 | 0.0000280 | 6.70e-06 |
| USP24 | -0.63 | 0.0035732 | 1.23e-03 |
| USP3 | 0.69 | 0.0000204 | 4.80e-06 |
| USP30 | 0.87 | 0.0241317 | 1.02e-02 |
| USP31 | 0.75 | 0.0026238 | 8.82e-04 |
| USP32 | 0.38 | 0.0445943 | 2.03e-02 |
| USP33 | 0.41 | 0.0738103 | 3.62e-02 |
| USP34 | -0.93 | 0.0000000 | 0.00e+00 |
| USP36 | 0.62 | 0.0003621 | 1.03e-04 |
| USP37 | -0.74 | 0.0308216 | 1.34e-02 |
| USP39 | -0.66 | 0.0000130 | 3.00e-06 |
| USP40 | -1.15 | 0.0000000 | 0.00e+00 |
| USP42 | 1.15 | 0.0000692 | 1.75e-05 |
| USP43 | 3.19 | 0.0000000 | 0.00e+00 |
| USP46 | -1.21 | 0.0070843 | 2.62e-03 |
| USP48 | -0.54 | 0.0627512 | 3.00e-02 |
| USP5 | -0.73 | 0.0000000 | 0.00e+00 |
| USP53 | 0.80 | 0.0001760 | 4.75e-05 |
| USP54 | 1.22 | 0.0000002 | 0.00e+00 |
| USP7 | -0.74 | 0.0000023 | 5.00e-07 |
| USP9X | -0.40 | 0.0023052 | 7.67e-04 |
| USP9Y | -0.90 | 0.0109436 | 4.25e-03 |
| UTP11L | -0.99 | 0.0012868 | 4.03e-04 |
| UTP15 | -1.83 | 0.0000001 | 0.00e+00 |
| UTP18 | -0.59 | 0.0007647 | 2.29e-04 |
| UTP20 | -1.65 | 0.0000000 | 0.00e+00 |
| UTY | 0.85 | 0.0099304 | 3.81e-03 |
| UVRAG | 0.84 | 0.0031322 | 1.07e-03 |
| UXS1 | 1.50 | 0.0000000 | 0.00e+00 |
| VAC14 | 0.81 | 0.0000554 | 1.39e-05 |
| VAMP1 | -1.94 | 0.0000289 | 7.00e-06 |
| VAMP2 | 1.32 | 0.0000001 | 0.00e+00 |
| VAMP7 | -0.97 | 0.0077970 | 2.92e-03 |
| VAMP8 | 0.75 | 0.0071248 | 2.63e-03 |
| VANGL1 | -1.05 | 0.0000874 | 2.25e-05 |
| VANGL2 | 4.46 | 0.0000000 | 0.00e+00 |
| VASH1 | -1.04 | 0.0824340 | 4.13e-02 |
| VASH2 | -1.21 | 0.0359198 | 1.59e-02 |
| VASP | 1.79 | 0.0000000 | 0.00e+00 |
| VAT1 | 0.38 | 0.0033861 | 1.16e-03 |
| VAV1 | 0.97 | 0.0308288 | 1.34e-02 |
| VAV2 | 0.60 | 0.0020866 | 6.86e-04 |
| VAV3 | -2.02 | 0.0000003 | 0.00e+00 |
| VBP1 | -1.26 | 0.0000000 | 0.00e+00 |
| VCAM1 | 6.69 | 0.0116449 | 4.55e-03 |
| VCL | 1.12 | 0.0000000 | 0.00e+00 |
| VCP | -0.17 | 0.0910402 | 4.63e-02 |
| VDAC2 | 0.67 | 0.0000010 | 2.00e-07 |
| VDAC3 | -1.64 | 0.0000000 | 0.00e+00 |
| VDR | 3.41 | 0.0000000 | 0.00e+00 |
| VEGFA | 2.01 | 0.0000000 | 0.00e+00 |
| VEGFC | 2.42 | 0.0000000 | 0.00e+00 |
| VEZF1 | 0.94 | 0.0000000 | 0.00e+00 |
| VEZT | 0.60 | 0.0000168 | 3.90e-06 |
| VGLL4 | 1.02 | 0.0000003 | 1.00e-07 |
| VHL | 0.76 | 0.0000542 | 1.35e-05 |
| VIM | 1.13 | 0.0000000 | 0.00e+00 |
| VIMP | 1.16 | 0.0000000 | 0.00e+00 |
| VKORC1 | -0.59 | 0.0073260 | 2.72e-03 |
| VKORC1L1 | -0.64 | 0.0011836 | 3.69e-04 |
| VMA21 | -1.34 | 0.0000000 | 0.00e+00 |
| VMP1 | -0.30 | 0.0148369 | 5.94e-03 |
| VNN1 | 3.82 | 0.0003986 | 1.14e-04 |
| VNN2 | 5.44 | 0.0909112 | 4.63e-02 |
| VNN3 | 7.61 | 0.0017901 | 5.78e-04 |
| VOPP1 | -0.37 | 0.0573582 | 2.70e-02 |
| VPS11 | 0.59 | 0.0661660 | 3.18e-02 |
| VPS13A | -1.13 | 0.0000003 | 1.00e-07 |
| VPS13C | -0.91 | 0.0000185 | 4.40e-06 |
| VPS16 | 1.40 | 0.0000000 | 0.00e+00 |
| VPS18 | 1.24 | 0.0000000 | 0.00e+00 |
| VPS25 | -1.46 | 0.0000000 | 0.00e+00 |
| VPS26B | -0.84 | 0.0014049 | 4.44e-04 |
| VPS28 | 0.78 | 0.0000118 | 2.70e-06 |
| VPS33B | 0.62 | 0.0086141 | 3.25e-03 |
| VPS35 | -0.64 | 0.0000008 | 2.00e-07 |
| VPS36 | -0.70 | 0.0214169 | 8.90e-03 |
| VPS37A | 0.67 | 0.0013938 | 4.40e-04 |
| VPS37B | 1.54 | 0.0000000 | 0.00e+00 |
| VPS37C | 1.11 | 0.0000129 | 3.00e-06 |
| VPS45 | -1.59 | 0.0000000 | 0.00e+00 |
| VPS4A | 0.29 | 0.0564560 | 2.65e-02 |
| VPS4B | -0.52 | 0.0700656 | 3.40e-02 |
| VPS50 | -1.12 | 0.0095193 | 3.63e-03 |
| VPS51 | 0.38 | 0.0593336 | 2.81e-02 |
| VPS72 | 0.44 | 0.0461872 | 2.11e-02 |
| VPS9D1 | 3.29 | 0.0000000 | 0.00e+00 |
| VPS9D1-AS1 | -1.59 | 0.0049852 | 1.78e-03 |
| VRK1 | -1.96 | 0.0000002 | 0.00e+00 |
| VRK3 | 1.49 | 0.0000000 | 0.00e+00 |
| VSIG1 | -2.86 | 0.0666349 | 3.21e-02 |
| VSTM2L | 2.12 | 0.0009035 | 2.75e-04 |
| VTA1 | -0.97 | 0.0000453 | 1.12e-05 |
| VTI1B | -0.62 | 0.0049130 | 1.75e-03 |
| VTN | 3.65 | 0.0000000 | 0.00e+00 |
| VWA1 | -0.83 | 0.0085300 | 3.22e-03 |
| VWDE | -3.87 | 0.0607711 | 2.89e-02 |
| WAC | 0.55 | 0.0000719 | 1.83e-05 |
| WARS | 4.42 | 0.0000000 | 0.00e+00 |
| WASF2 | 1.90 | 0.0000000 | 0.00e+00 |
| WASH1 | 0.77 | 0.0722835 | 3.53e-02 |
| WASH2P | 1.11 | 0.0055233 | 1.99e-03 |
| WASH3P | 0.92 | 0.0150367 | 6.03e-03 |
| WASH7P | 1.10 | 0.0076349 | 2.85e-03 |
| WASL | 0.76 | 0.0002877 | 8.00e-05 |
| WBP1 | -0.60 | 0.0120772 | 4.73e-03 |
| WBP11 | 0.33 | 0.0562798 | 2.64e-02 |
| WBP1L | 1.07 | 0.0000000 | 0.00e+00 |
| WBP2 | 2.02 | 0.0000000 | 0.00e+00 |
| WBSCR22 | -0.60 | 0.0006916 | 2.05e-04 |
| WBSCR27 | 0.83 | 0.0551168 | 2.58e-02 |
| WDFY2 | 1.26 | 0.0000362 | 8.90e-06 |
| WDHD1 | -3.69 | 0.0000000 | 0.00e+00 |
| WDR1 | 0.30 | 0.0026477 | 8.91e-04 |
| WDR11 | -0.88 | 0.0001172 | 3.07e-05 |
| WDR12 | -1.52 | 0.0000000 | 0.00e+00 |
| WDR13 | 0.45 | 0.0463326 | 2.12e-02 |
| WDR17 | -1.93 | 0.0051505 | 1.84e-03 |
| WDR24 | 0.77 | 0.0367878 | 1.64e-02 |
| WDR25 | 2.22 | 0.0000000 | 0.00e+00 |
| WDR26 | 1.03 | 0.0000000 | 0.00e+00 |
| WDR3 | -0.96 | 0.0000127 | 2.90e-06 |
| WDR34 | -2.50 | 0.0000000 | 0.00e+00 |
| WDR35 | -1.97 | 0.0000000 | 0.00e+00 |
| WDR36 | -1.31 | 0.0000007 | 1.00e-07 |
| WDR4 | -0.74 | 0.0249226 | 1.06e-02 |
| WDR43 | -0.74 | 0.0000028 | 6.00e-07 |
| WDR45 | 1.56 | 0.0000000 | 0.00e+00 |
| WDR5 | -0.91 | 0.0000043 | 9.00e-07 |
| WDR54 | -1.98 | 0.0000000 | 0.00e+00 |
| WDR61 | -1.02 | 0.0000027 | 6.00e-07 |
| WDR62 | -2.22 | 0.0000000 | 0.00e+00 |
| WDR73 | -0.66 | 0.0135375 | 5.37e-03 |
| WDR75 | -1.13 | 0.0000000 | 0.00e+00 |
| WDR76 | -1.87 | 0.0000000 | 0.00e+00 |
| WDR77 | -0.88 | 0.0000756 | 1.92e-05 |
| WDR81 | 0.74 | 0.0173403 | 7.05e-03 |
| WDR82 | -0.33 | 0.0709942 | 3.46e-02 |
| WDR89 | -1.15 | 0.0167262 | 6.77e-03 |
| WDR90 | -1.13 | 0.0123940 | 4.87e-03 |
| WDR92 | -1.22 | 0.0021490 | 7.09e-04 |
| WDTC1 | 1.39 | 0.0000000 | 0.00e+00 |
| WFDC21P | 3.40 | 0.0000020 | 4.00e-07 |
| WFDC5 | 5.97 | 0.0417361 | 1.88e-02 |
| WHAMM | 1.62 | 0.0000000 | 0.00e+00 |
| WHSC1 | -1.39 | 0.0000000 | 0.00e+00 |
| WIBG | 0.58 | 0.0238866 | 1.01e-02 |
| WIPF1 | 0.90 | 0.0022551 | 7.48e-04 |
| WIPF2 | 1.31 | 0.0000000 | 0.00e+00 |
| WIPI1 | 1.57 | 0.0000014 | 3.00e-07 |
| WIPI2 | 0.32 | 0.0791471 | 3.93e-02 |
| WISP1 | 4.19 | 0.0041237 | 1.44e-03 |
| WISP2 | -3.55 | 0.0000000 | 0.00e+00 |
| WIZ | 1.08 | 0.0000553 | 1.39e-05 |
| WLS | 0.78 | 0.0711988 | 3.47e-02 |
| WNK1 | 0.67 | 0.0000000 | 0.00e+00 |
| WNT4 | 5.17 | 0.0000000 | 0.00e+00 |
| WNT6 | 3.54 | 0.0095065 | 3.63e-03 |
| WNT7A | 6.14 | 0.0321172 | 1.40e-02 |
| WNT7B | 0.83 | 0.0000000 | 0.00e+00 |
| WNT9A | 2.87 | 0.0065935 | 2.42e-03 |
| WRAP53 | -1.23 | 0.0000008 | 2.00e-07 |
| WRB | -2.65 | 0.0000000 | 0.00e+00 |
| WRN | -1.09 | 0.0026189 | 8.80e-04 |
| WSB1 | 0.60 | 0.0052511 | 1.88e-03 |
| WT1 | 2.00 | 0.0001120 | 2.93e-05 |
| WTAP | 1.64 | 0.0000000 | 0.00e+00 |
| WTIP | 0.56 | 0.0842902 | 4.24e-02 |
| WWC1 | 0.99 | 0.0000000 | 0.00e+00 |
| WWC2-AS2 | 2.91 | 0.0610203 | 2.90e-02 |
| WWOX | -0.98 | 0.0704721 | 3.43e-02 |
| WWP2 | 0.94 | 0.0000030 | 6.00e-07 |
| WWTR1 | -0.43 | 0.0078049 | 2.92e-03 |
| XAB2 | 0.96 | 0.0000007 | 1.00e-07 |
| XAF1 | 12.49 | 0.0000000 | 0.00e+00 |
| XAGE1B | 0.41 | 0.0835040 | 4.19e-02 |
| XAGE1E | 0.41 | 0.0835040 | 4.19e-02 |
| XBP1 | 2.21 | 0.0000000 | 0.00e+00 |
| XDH | 3.76 | 0.0000000 | 0.00e+00 |
| XIAP | 0.68 | 0.0013794 | 4.35e-04 |
| XIRP1 | 7.78 | 0.0012948 | 4.06e-04 |
| XKR8 | 1.51 | 0.0007066 | 2.10e-04 |
| XPA | 0.99 | 0.0053116 | 1.91e-03 |
| XPNPEP1 | 0.37 | 0.0498899 | 2.30e-02 |
| XPNPEP3 | -0.98 | 0.0038538 | 1.34e-03 |
| XPO1 | -1.14 | 0.0000000 | 0.00e+00 |
| XPO4 | -1.39 | 0.0000008 | 2.00e-07 |
| XPO6 | 0.60 | 0.0000410 | 1.01e-05 |
| XPO7 | -1.09 | 0.0000000 | 0.00e+00 |
| XPOT | -0.43 | 0.0035707 | 1.23e-03 |
| XRCC1 | 0.50 | 0.0183503 | 7.51e-03 |
| XRCC2 | -3.39 | 0.0000000 | 0.00e+00 |
| XRCC3 | -1.85 | 0.0000000 | 0.00e+00 |
| XRCC4 | -1.02 | 0.0450304 | 2.05e-02 |
| XRCC5 | -1.34 | 0.0000000 | 0.00e+00 |
| XRCC6 | -1.09 | 0.0000000 | 0.00e+00 |
| XRCC6BP1 | -1.87 | 0.0885134 | 4.48e-02 |
| XRN2 | -0.56 | 0.0000103 | 2.30e-06 |
| XYLB | -1.94 | 0.0038280 | 1.33e-03 |
| XYLT1 | 1.39 | 0.0093707 | 3.57e-03 |
| YAP1 | 0.75 | 0.0000013 | 3.00e-07 |
| YARS | 0.57 | 0.0000196 | 4.60e-06 |
| YBX1 | -1.11 | 0.0000000 | 0.00e+00 |
| YBX3 | 1.19 | 0.0000000 | 0.00e+00 |
| YBX3P1 | 1.13 | 0.0026963 | 9.09e-04 |
| YEATS2 | 0.63 | 0.0106314 | 4.11e-03 |
| YEATS4 | -0.97 | 0.0033955 | 1.17e-03 |
| YES1 | -0.67 | 0.0055385 | 1.99e-03 |
| YIF1B | -0.37 | 0.0639247 | 3.06e-02 |
| YIPF3 | 0.68 | 0.0002267 | 6.21e-05 |
| YLPM1 | 0.55 | 0.0020027 | 6.55e-04 |
| YME1L1 | -0.37 | 0.0031435 | 1.07e-03 |
| YPEL2 | 2.10 | 0.0000000 | 0.00e+00 |
| YPEL3 | 1.92 | 0.0000216 | 5.10e-06 |
| YPEL5 | 1.14 | 0.0000000 | 0.00e+00 |
| YRDC | 0.56 | 0.0834777 | 4.19e-02 |
| YTHDC2 | -1.48 | 0.0000000 | 0.00e+00 |
| YTHDF1 | 0.46 | 0.0110134 | 4.28e-03 |
| YWHAE | -0.58 | 0.0000001 | 0.00e+00 |
| YWHAG | -0.23 | 0.0534803 | 2.49e-02 |
| YWHAQ | -0.72 | 0.0000000 | 0.00e+00 |
| YY1AP1 | 1.36 | 0.0000000 | 0.00e+00 |
| ZADH2 | -0.91 | 0.0099074 | 3.80e-03 |
| ZAK | -0.50 | 0.0013317 | 4.18e-04 |
| ZBED5 | -1.00 | 0.0000430 | 1.06e-05 |
| ZBED5-AS1 | -1.40 | 0.0464488 | 2.13e-02 |
| ZBP1 | 7.03 | 0.0061949 | 2.26e-03 |
| ZBTB10 | 2.55 | 0.0000000 | 0.00e+00 |
| ZBTB11 | -0.77 | 0.0123084 | 4.83e-03 |
| ZBTB14 | -1.25 | 0.0133613 | 5.29e-03 |
| ZBTB17 | 1.44 | 0.0000001 | 0.00e+00 |
| ZBTB2 | 0.77 | 0.0170489 | 6.92e-03 |
| ZBTB21 | 0.65 | 0.0495759 | 2.29e-02 |
| ZBTB32 | 8.28 | 0.0000000 | 0.00e+00 |
| ZBTB39 | 0.65 | 0.0291870 | 1.26e-02 |
| ZBTB4 | 1.07 | 0.0000000 | 0.00e+00 |
| ZBTB43 | 1.24 | 0.0000469 | 1.16e-05 |
| ZBTB44 | -0.85 | 0.0313367 | 1.36e-02 |
| ZBTB45 | 0.61 | 0.0781138 | 3.87e-02 |
| ZBTB46 | 4.00 | 0.0000000 | 0.00e+00 |
| ZBTB49 | 1.12 | 0.0983376 | 5.06e-02 |
| ZBTB5 | 0.67 | 0.0101198 | 3.89e-03 |
| ZBTB7A | 1.86 | 0.0000000 | 0.00e+00 |
| ZBTB7B | 0.86 | 0.0000002 | 0.00e+00 |
| ZC2HC1A | -1.07 | 0.0851530 | 4.29e-02 |
| ZC3H12A | 3.73 | 0.0000000 | 0.00e+00 |
| ZC3H12C | 2.02 | 0.0000000 | 0.00e+00 |
| ZC3H13 | -0.95 | 0.0005520 | 1.61e-04 |
| ZC3H14 | -1.01 | 0.0000000 | 0.00e+00 |
| ZC3H15 | -0.60 | 0.0007351 | 2.19e-04 |
| ZC3H18 | 0.55 | 0.0004058 | 1.16e-04 |
| ZC3H3 | 2.07 | 0.0000000 | 0.00e+00 |
| ZC3H7A | -0.51 | 0.0730418 | 3.58e-02 |
| ZC3H7B | 0.89 | 0.0000000 | 0.00e+00 |
| ZC3H8 | -1.27 | 0.0077405 | 2.89e-03 |
| ZC3HAV1 | 3.39 | 0.0000000 | 0.00e+00 |
| ZC3HAV1L | -1.42 | 0.0220640 | 9.20e-03 |
| ZCCHC11 | -0.75 | 0.0320134 | 1.40e-02 |
| ZCCHC14 | -0.80 | 0.0666908 | 3.21e-02 |
| ZCCHC2 | 0.70 | 0.0767121 | 3.79e-02 |
| ZCCHC3 | 0.40 | 0.0585653 | 2.77e-02 |
| ZCCHC6 | 0.64 | 0.0083479 | 3.14e-03 |
| ZCCHC7 | 0.65 | 0.0282471 | 1.21e-02 |
| ZCRB1 | -0.62 | 0.0131443 | 5.20e-03 |
| ZDHHC1 | 1.51 | 0.0024138 | 8.05e-04 |
| ZDHHC12 | -0.87 | 0.0004539 | 1.31e-04 |
| ZDHHC14 | 1.46 | 0.0085717 | 3.23e-03 |
| ZDHHC16 | -0.42 | 0.0827638 | 4.14e-02 |
| ZDHHC17 | -0.94 | 0.0060759 | 2.21e-03 |
| ZDHHC18 | 0.52 | 0.0303698 | 1.32e-02 |
| ZDHHC20 | -0.74 | 0.0042100 | 1.48e-03 |
| ZDHHC21 | -1.13 | 0.0129004 | 5.09e-03 |
| ZDHHC5 | 0.76 | 0.0000000 | 0.00e+00 |
| ZDHHC6 | -0.49 | 0.0552439 | 2.59e-02 |
| ZDHHC7 | -0.68 | 0.0006966 | 2.07e-04 |
| ZDHHC9 | 1.73 | 0.0000000 | 0.00e+00 |
| ZEB2 | 2.46 | 0.0000000 | 0.00e+00 |
| ZER1 | 1.11 | 0.0000009 | 2.00e-07 |
| ZFAND2A | 1.76 | 0.0000000 | 0.00e+00 |
| ZFAND3 | 1.38 | 0.0000000 | 0.00e+00 |
| ZFAND5 | 0.75 | 0.0000000 | 0.00e+00 |
| ZFAND6 | 0.90 | 0.0000000 | 0.00e+00 |
| ZFAS1 | 1.39 | 0.0000000 | 0.00e+00 |
| ZFAT | 0.84 | 0.0525366 | 2.44e-02 |
| ZFHX2 | 4.48 | 0.0000000 | 0.00e+00 |
| ZFHX3 | 0.77 | 0.0000215 | 5.10e-06 |
| ZFP36 | 3.83 | 0.0000000 | 0.00e+00 |
| ZFP36L1 | 2.85 | 0.0000000 | 0.00e+00 |
| ZFP36L2 | 2.60 | 0.0000000 | 0.00e+00 |
| ZFP62 | -1.20 | 0.0159155 | 6.42e-03 |
| ZFP64 | 0.93 | 0.0000939 | 2.43e-05 |
| ZFP91 | 0.48 | 0.0009304 | 2.84e-04 |
| ZFP91-CNTF | 0.65 | 0.0017779 | 5.73e-04 |
| ZFPL1 | 1.19 | 0.0000007 | 1.00e-07 |
| ZFPM1 | 1.07 | 0.0145494 | 5.81e-03 |
| ZFR | -0.50 | 0.0009035 | 2.75e-04 |
| ZFYVE1 | 2.23 | 0.0000000 | 0.00e+00 |
| ZFYVE16 | -0.98 | 0.0001296 | 3.43e-05 |
| ZFYVE19 | 0.73 | 0.0091299 | 3.47e-03 |
| ZFYVE26 | 0.61 | 0.0134323 | 5.33e-03 |
| ZFYVE27 | 1.05 | 0.0000000 | 0.00e+00 |
| ZFYVE28 | 1.32 | 0.0312372 | 1.36e-02 |
| ZGPAT | 0.77 | 0.0034407 | 1.18e-03 |
| ZGRF1 | -2.84 | 0.0000577 | 1.45e-05 |
| ZHX1-C8orf76 | 0.57 | 0.0793866 | 3.94e-02 |
| ZHX2 | 3.29 | 0.0000000 | 0.00e+00 |
| ZHX3 | -1.58 | 0.0075698 | 2.82e-03 |
| ZMAT2 | -0.42 | 0.0361281 | 1.60e-02 |
| ZMAT3 | -1.33 | 0.0221959 | 9.27e-03 |
| ZMIZ1 | 1.26 | 0.0000000 | 0.00e+00 |
| ZMIZ1-AS1 | 4.04 | 0.0670968 | 3.24e-02 |
| ZMIZ2 | 2.64 | 0.0000000 | 0.00e+00 |
| ZMPSTE24 | -0.48 | 0.0258873 | 1.10e-02 |
| ZMYM1 | -1.32 | 0.0112131 | 4.36e-03 |
| ZMYM5 | 1.10 | 0.0104687 | 4.04e-03 |
| ZMYM6NB | 1.76 | 0.0000001 | 0.00e+00 |
| ZMYND11 | -1.17 | 0.0000000 | 0.00e+00 |
| ZMYND19 | -0.66 | 0.0304007 | 1.32e-02 |
| ZMYND8 | 1.05 | 0.0000000 | 0.00e+00 |
| ZNF101 | 1.23 | 0.0027896 | 9.42e-04 |
| ZNF114 | -1.81 | 0.0529579 | 2.46e-02 |
| ZNF131 | -0.75 | 0.0403894 | 1.82e-02 |
| ZNF133 | 1.38 | 0.0000002 | 0.00e+00 |
| ZNF134 | 0.87 | 0.0239937 | 1.01e-02 |
| ZNF140 | 1.07 | 0.0027761 | 9.37e-04 |
| ZNF142 | 0.99 | 0.0000912 | 2.35e-05 |
| ZNF146 | -1.18 | 0.0000000 | 0.00e+00 |
| ZNF148 | -0.64 | 0.0561853 | 2.64e-02 |
| ZNF155 | 1.48 | 0.0040818 | 1.43e-03 |
| ZNF180 | -0.91 | 0.0396376 | 1.78e-02 |
| ZNF181 | -0.95 | 0.0617707 | 2.94e-02 |
| ZNF185 | 0.69 | 0.0180304 | 7.37e-03 |
| ZNF205 | 1.12 | 0.0011401 | 3.54e-04 |
| ZNF207 | 0.50 | 0.0000024 | 5.00e-07 |
| ZNF212 | 1.16 | 0.0008758 | 2.66e-04 |
| ZNF213 | 1.74 | 0.0000001 | 0.00e+00 |
| ZNF215 | -2.34 | 0.0615695 | 2.93e-02 |
| ZNF217 | 1.45 | 0.0000000 | 0.00e+00 |
| ZNF222 | 1.65 | 0.0040065 | 1.40e-03 |
| ZNF248 | -1.81 | 0.0002161 | 5.91e-05 |
| ZNF25 | -2.08 | 0.0063839 | 2.34e-03 |
| ZNF251 | 0.92 | 0.0708526 | 3.45e-02 |
| ZNF260 | -0.70 | 0.0711369 | 3.47e-02 |
| ZNF263 | 0.57 | 0.0057045 | 2.06e-03 |
| ZNF264 | 1.29 | 0.0006696 | 1.99e-04 |
| ZNF267 | 1.19 | 0.0000050 | 1.10e-06 |
| ZNF274 | 1.42 | 0.0000000 | 0.00e+00 |
| ZNF277 | 0.87 | 0.0049212 | 1.75e-03 |
| ZNF280A | 1.80 | 0.0231021 | 9.70e-03 |
| ZNF280D | -0.74 | 0.0345812 | 1.52e-02 |
| ZNF282 | 1.57 | 0.0000000 | 0.00e+00 |
| ZNF286A | -0.80 | 0.0254500 | 1.08e-02 |
| ZNF296 | 1.67 | 0.0046739 | 1.66e-03 |
| ZNF3 | 0.52 | 0.0461872 | 2.11e-02 |
| ZNF302 | -1.34 | 0.0000280 | 6.70e-06 |
| ZNF316 | 1.21 | 0.0000020 | 4.00e-07 |
| ZNF317 | 0.64 | 0.0057316 | 2.07e-03 |
| ZNF319 | 1.59 | 0.0000000 | 0.00e+00 |
| ZNF324 | 1.20 | 0.0010956 | 3.39e-04 |
| ZNF326 | -0.97 | 0.0001687 | 4.55e-05 |
| ZNF331 | -1.04 | 0.0003928 | 1.12e-04 |
| ZNF335 | 1.43 | 0.0000000 | 0.00e+00 |
| ZNF34 | 0.94 | 0.0818063 | 4.09e-02 |
| ZNF350 | 1.00 | 0.0345181 | 1.52e-02 |
| ZNF354A | -1.36 | 0.0092784 | 3.53e-03 |
| ZNF358 | 1.10 | 0.0009199 | 2.81e-04 |
| ZNF367 | -3.14 | 0.0000000 | 0.00e+00 |
| ZNF384 | 0.53 | 0.0059639 | 2.17e-03 |
| ZNF385A | 3.53 | 0.0000000 | 0.00e+00 |
| ZNF385C | 4.16 | 0.0006325 | 1.87e-04 |
| ZNF394 | 1.33 | 0.0000010 | 2.00e-07 |
| ZNF398 | 0.95 | 0.0013639 | 4.30e-04 |
| ZNF408 | 1.28 | 0.0000336 | 8.20e-06 |
| ZNF410 | 0.61 | 0.0005229 | 1.52e-04 |
| ZNF414 | 1.32 | 0.0230605 | 9.68e-03 |
| ZNF416 | 1.14 | 0.0168961 | 6.85e-03 |
| ZNF431 | -1.61 | 0.0905319 | 4.60e-02 |
| ZNF436 | 1.25 | 0.0002632 | 7.28e-05 |
| ZNF444 | 0.96 | 0.0001726 | 4.66e-05 |
| ZNF445 | -0.83 | 0.0466070 | 2.14e-02 |
| ZNF462 | 1.91 | 0.0000000 | 0.00e+00 |
| ZNF467 | 2.26 | 0.0000011 | 2.00e-07 |
| ZNF488 | -3.01 | 0.0000466 | 1.15e-05 |
| ZNF496 | 0.82 | 0.0056332 | 2.03e-03 |
| ZNF503 | 2.28 | 0.0000000 | 0.00e+00 |
| ZNF507 | -1.08 | 0.0000001 | 0.00e+00 |
| ZNF512 | -1.22 | 0.0000007 | 1.00e-07 |
| ZNF513 | 1.36 | 0.0000000 | 0.00e+00 |
| ZNF514 | -1.12 | 0.0558607 | 2.62e-02 |
| ZNF518A | -0.93 | 0.0008568 | 2.59e-04 |
| ZNF519 | -2.58 | 0.0923812 | 4.72e-02 |
| ZNF524 | 1.05 | 0.0008658 | 2.63e-04 |
| ZNF526 | 1.24 | 0.0000000 | 0.00e+00 |
| ZNF543 | 1.01 | 0.0897087 | 4.55e-02 |
| ZNF557 | 1.06 | 0.0530936 | 2.47e-02 |
| ZNF567 | -0.84 | 0.0741527 | 3.64e-02 |
| ZNF569 | -1.35 | 0.0270380 | 1.16e-02 |
| ZNF574 | 1.33 | 0.0000000 | 0.00e+00 |
| ZNF579 | 2.06 | 0.0000000 | 0.00e+00 |
| ZNF580 | 0.91 | 0.0029476 | 9.99e-04 |
| ZNF581 | 1.33 | 0.0000000 | 0.00e+00 |
| ZNF592 | 1.46 | 0.0000000 | 0.00e+00 |
| ZNF598 | 0.77 | 0.0000085 | 1.90e-06 |
| ZNF609 | 0.89 | 0.0000000 | 0.00e+00 |
| ZNF618 | -0.50 | 0.0479810 | 2.20e-02 |
| ZNF622 | 0.87 | 0.0000059 | 1.30e-06 |
| ZNF628 | 2.42 | 0.0000000 | 0.00e+00 |
| ZNF638 | -1.14 | 0.0000000 | 0.00e+00 |
| ZNF641 | 2.54 | 0.0000000 | 0.00e+00 |
| ZNF646 | 2.08 | 0.0000000 | 0.00e+00 |
| ZNF652 | -0.84 | 0.0422637 | 1.91e-02 |
| ZNF653 | 1.42 | 0.0049100 | 1.75e-03 |
| ZNF654 | 1.11 | 0.0460808 | 2.11e-02 |
| ZNF655 | 1.05 | 0.0000000 | 0.00e+00 |
| ZNF664 | -1.11 | 0.0000000 | 0.00e+00 |
| ZNF668 | 1.26 | 0.0000167 | 3.90e-06 |
| ZNF672 | 0.79 | 0.0006007 | 1.77e-04 |
| ZNF675 | -1.62 | 0.0590000 | 2.79e-02 |
| ZNF680 | -1.63 | 0.0501293 | 2.32e-02 |
| ZNF687 | 1.57 | 0.0000000 | 0.00e+00 |
| ZNF688 | 1.20 | 0.0111802 | 4.35e-03 |
| ZNF689 | 0.85 | 0.0061770 | 2.25e-03 |
| ZNF692 | -0.77 | 0.0666341 | 3.21e-02 |
| ZNF697 | 4.14 | 0.0000000 | 0.00e+00 |
| ZNF703 | 1.32 | 0.0000002 | 0.00e+00 |
| ZNF706 | 0.59 | 0.0147212 | 5.89e-03 |
| ZNF707 | 1.35 | 0.0041777 | 1.46e-03 |
| ZNF710 | 2.92 | 0.0000000 | 0.00e+00 |
| ZNF714 | -1.94 | 0.0265642 | 1.13e-02 |
| ZNF740 | 0.49 | 0.0221171 | 9.23e-03 |
| ZNF746 | 1.53 | 0.0000000 | 0.00e+00 |
| ZNF747 | 1.24 | 0.0013553 | 4.27e-04 |
| ZNF749 | -1.04 | 0.0608450 | 2.89e-02 |
| ZNF764 | 1.27 | 0.0017297 | 5.56e-04 |
| ZNF766 | 0.85 | 0.0041248 | 1.44e-03 |
| ZNF768 | 0.44 | 0.0399467 | 1.79e-02 |
| ZNF770 | -0.71 | 0.0062501 | 2.28e-03 |
| ZNF777 | 1.35 | 0.0000000 | 0.00e+00 |
| ZNF783 | 0.83 | 0.0562497 | 2.64e-02 |
| ZNF784 | 1.21 | 0.0031369 | 1.07e-03 |
| ZNF787 | 0.88 | 0.0000000 | 0.00e+00 |
| ZNF79 | 1.21 | 0.0015294 | 4.86e-04 |
| ZNF792 | 1.15 | 0.0163115 | 6.59e-03 |
| ZNF821 | 2.03 | 0.0000151 | 3.50e-06 |
| ZNF827 | 1.08 | 0.0109101 | 4.23e-03 |
| ZNF862 | 0.84 | 0.0711617 | 3.47e-02 |
| ZNF865 | 2.14 | 0.0000000 | 0.00e+00 |
| ZNFX1 | 3.62 | 0.0000000 | 0.00e+00 |
| ZNHIT6 | -0.91 | 0.0096687 | 3.70e-03 |
| ZNRF1 | 1.22 | 0.0000152 | 3.50e-06 |
| ZNRF2 | 0.77 | 0.0137088 | 5.45e-03 |
| ZRANB1 | 0.52 | 0.0336957 | 1.48e-02 |
| ZRANB2 | -1.21 | 0.0000130 | 3.00e-06 |
| ZRANB3 | -1.79 | 0.0027613 | 9.31e-04 |
| ZSCAN21 | 0.82 | 0.0600152 | 2.84e-02 |
| ZSCAN25 | 0.63 | 0.0932601 | 4.77e-02 |
| ZSWIM1 | 1.06 | 0.0448712 | 2.04e-02 |
| ZSWIM3 | 1.43 | 0.0516395 | 2.40e-02 |
| ZSWIM4 | 4.11 | 0.0000000 | 0.00e+00 |
| ZSWIM6 | 1.13 | 0.0000011 | 2.00e-07 |
| ZSWIM8 | 2.03 | 0.0000000 | 0.00e+00 |
| ZW10 | -0.88 | 0.0059499 | 2.16e-03 |
| ZWILCH | -2.42 | 0.0000000 | 0.00e+00 |
| ZWINT | -2.61 | 0.0000000 | 0.00e+00 |
| ZYG11B | -0.68 | 0.0705296 | 3.43e-02 |
| ZYX | 2.82 | 0.0000000 | 0.00e+00 |
| ZZZ3 | -0.94 | 0.0046890 | 1.66e-03 |
Pathway analysis of the differntial expressed gene
library(clusterProfiler)
library(enrichplot)
deseq_res_eg <- bitr(deseq2_res[which(deseq2_res$p.adjust<0.1),"GeneID"],fromType = "SYMBOL", toType = "ENTREZID",OrgDb = "org.Hs.eg.db" )
ego <- enrichKEGG(deseq_res_eg$ENTREZID,organism = "hsa",pAdjustMethod = "none", pvalueCutoff = 0.05,minGSSize = 3)
#enrichMap(ego,vertex.label.font = 2,n = 24)
emapplot(ego )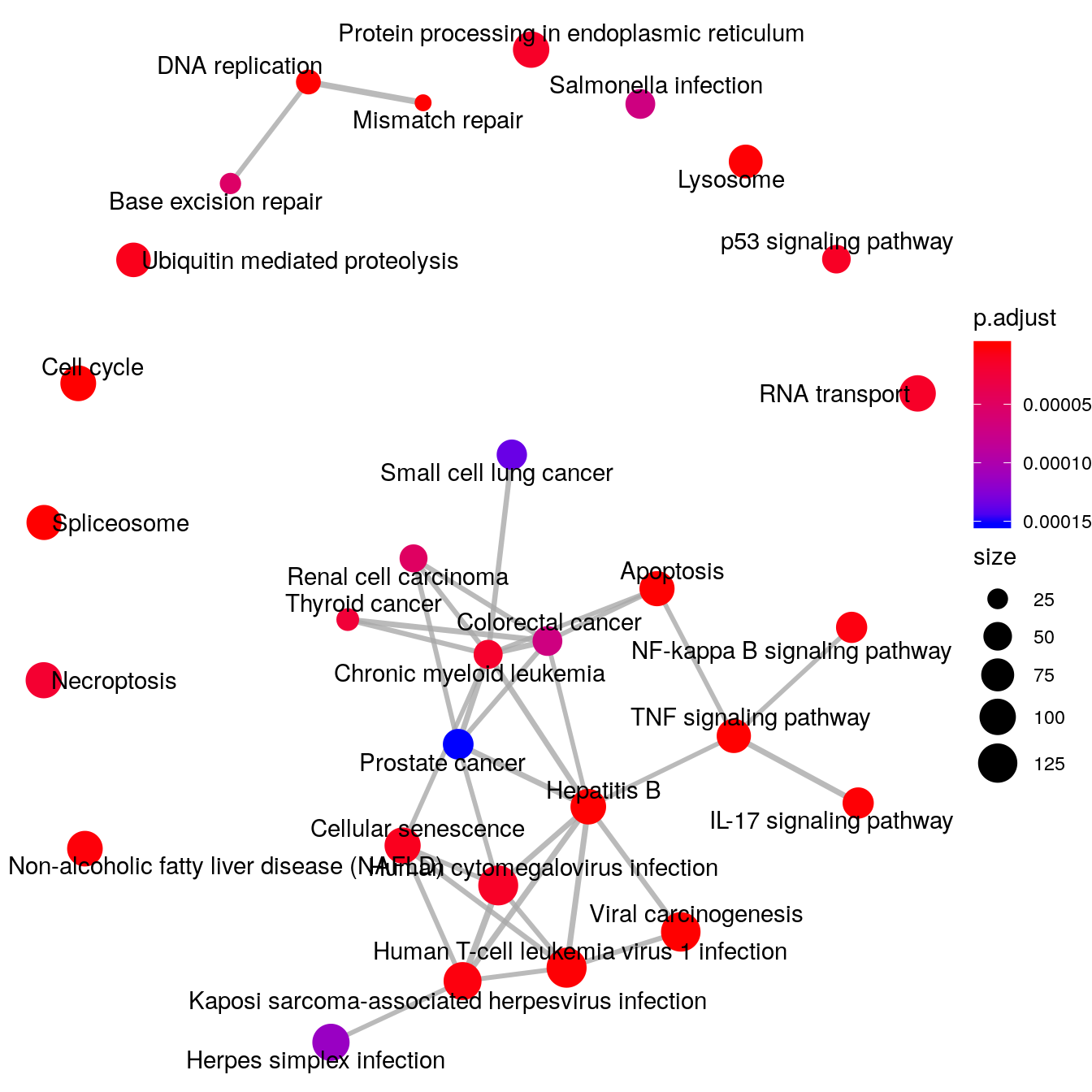
Hela host cell line
Count reads map to hg38 genome
library(m6Amonster)
samplenames <-c("Hela1","Hela2","RSVinfect1","RSVinfect2")
gtf <- "~/Database/genome/hg38/hg38_UCSC.gtf"
RSV.Hela <- countReads(samplenames = paste(samplenames,"alignTohg38",sep = "." ),
gtf = gtf,
bamFolder = "/home/zijiezhang/RSV/2017/alignToGRCh38_bam",
outputDir = "/home/zijiezhang/RSV/2017",
modification = "m6A",
threads = 40
)Call peaks
RSV.Hela <- callPeakFisher(RSV.Hela,threads = 20)
save(RSV.Hela,file = "~/RSV/2017/RSV.Hela_infected.RData")Report peaks for Ctl and Infected samples
load("~/RSV/2017/RSV.Hela_infected.RData")
CtlPeak <- reportConsistentPeak(RSV.Hela,samplenames = c("Hela1.alignTohg38","Hela2.alignTohg38"),threads = 15)Reporting peak concsistent in all samples for
Hela1.alignTohg38 Hela2.alignTohg38
Hyper-thread registered: TRUE
Using 15 thread(s) to report merged report...
Time used to report peaks: 1.48280185858409 mins... InfectedPeak <- reportConsistentPeak(RSV.Hela,samplenames = c("RSVinfect1.alignTohg38","RSVinfect2.alignTohg38"), threads = 15 )Reporting peak concsistent in all samples for
RSVinfect1.alignTohg38 RSVinfect2.alignTohg38
Hyper-thread registered: TRUE
Using 15 thread(s) to report merged report...
Time used to report peaks: 1.52456891934077 mins... Plot distribution of peaks on the meta-gene
plotMetaGeneMulti(list("Ctl"=CtlPeak, "RSV infected" = InfectedPeak), "~/Database/genome/hg38/hg38_UCSC.gtf")[1] "Converting BED12 to GRangesList"
[1] "It may take a few minutes"
[1] "Converting BED12 to GRangesList"
[1] "It may take a few minutes"
[1] "total 58259 transcripts extracted ..."
[1] "total 42543 transcripts left after ambiguity filter ..."
[1] "total 21293 mRNAs left after component length filter ..."
[1] "total 7986 ncRNAs left after ncRNA length filter ..."
[1] "Building Guitar Coordinates. It may take a few minutes ..."
[1] "Guitar Coordinates Built ..."
[1] "Using provided Guitar Coordinates"
[1] "resolving ambiguious features ..."
[1] "no figure saved ..."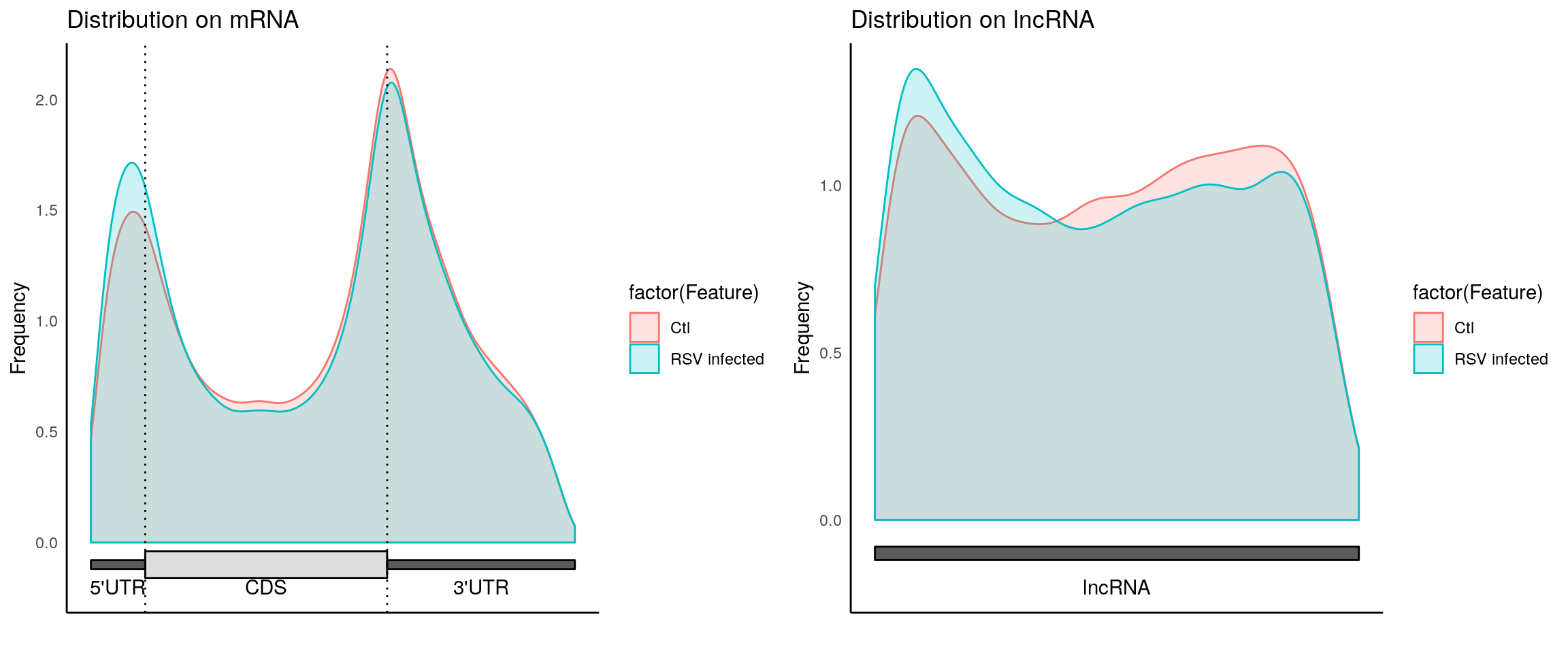
Show Ctl peaks distribution on Pie chart
peakDistribution(peak = CtlPeak, gtf = "~/Database/genome/hg38/hg38_UCSC.gtf" )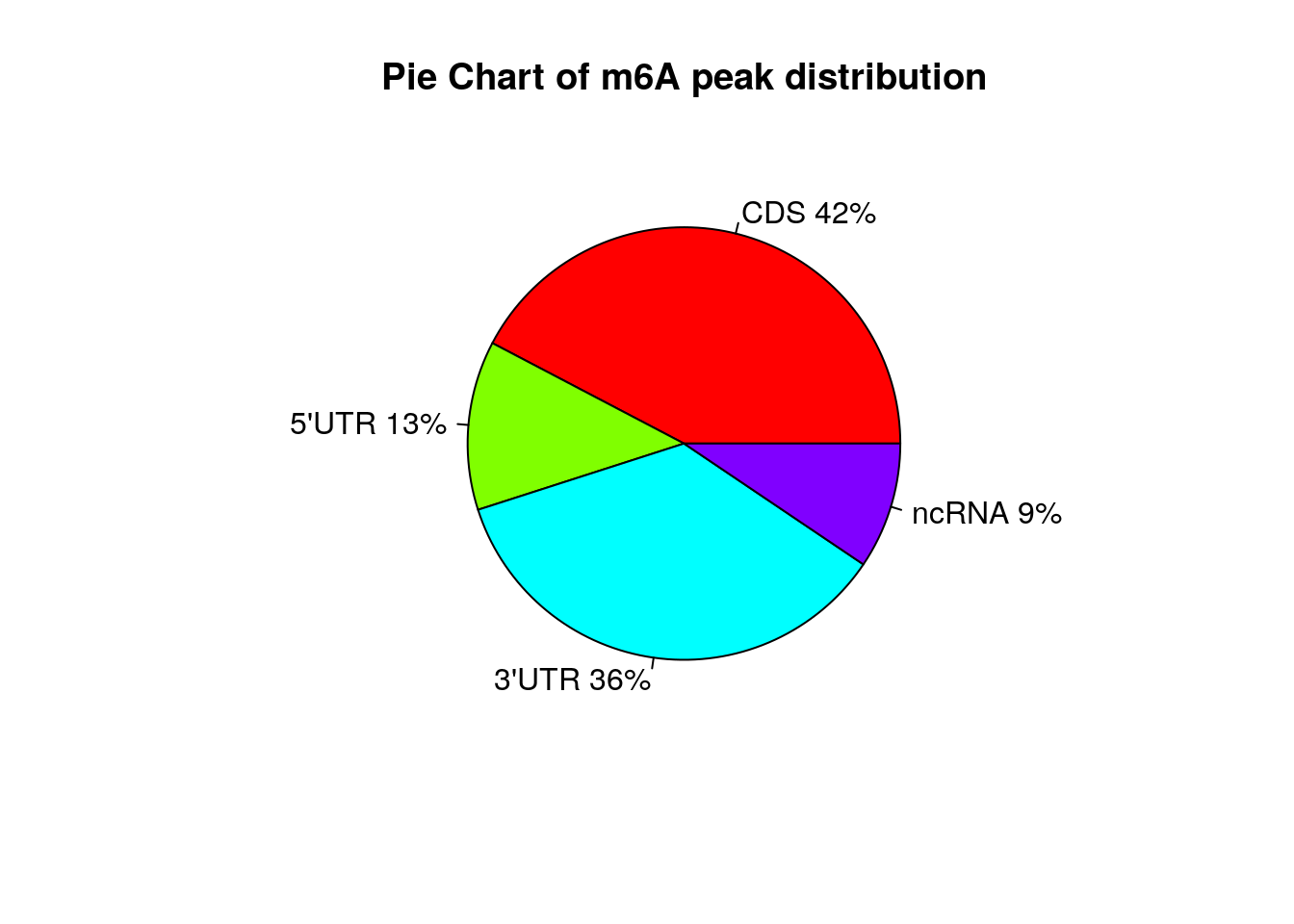
Show RSV infected peaks distribution on Pie chart
peakDistribution(peak = InfectedPeak, gtf = "~/Database/genome/hg38/hg38_UCSC.gtf" )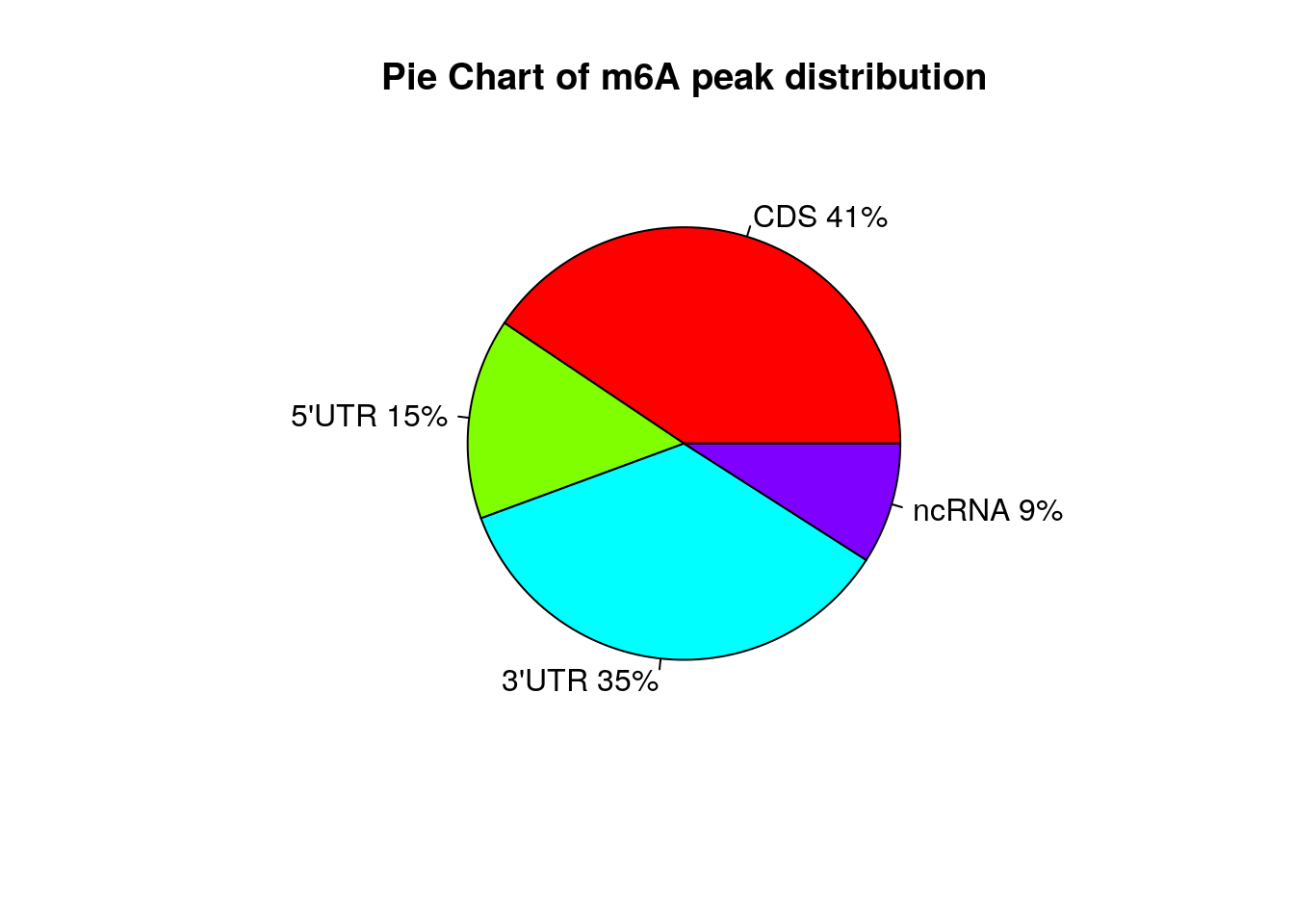
Differential methylation test with QNB
#RSV.Hela <- JointPeakCount(RSV.Hela,joint_threshold = 2)
#RSV.Hela <- normalizePeak(RSV.Hela,c("Ctl","Ctl","Infected","Infected"))
#RSV.Hela <- QNBtest(RSV.Hela)
hist(qvalue::qvalue(RSV.Hela$all.est.peak$p_value))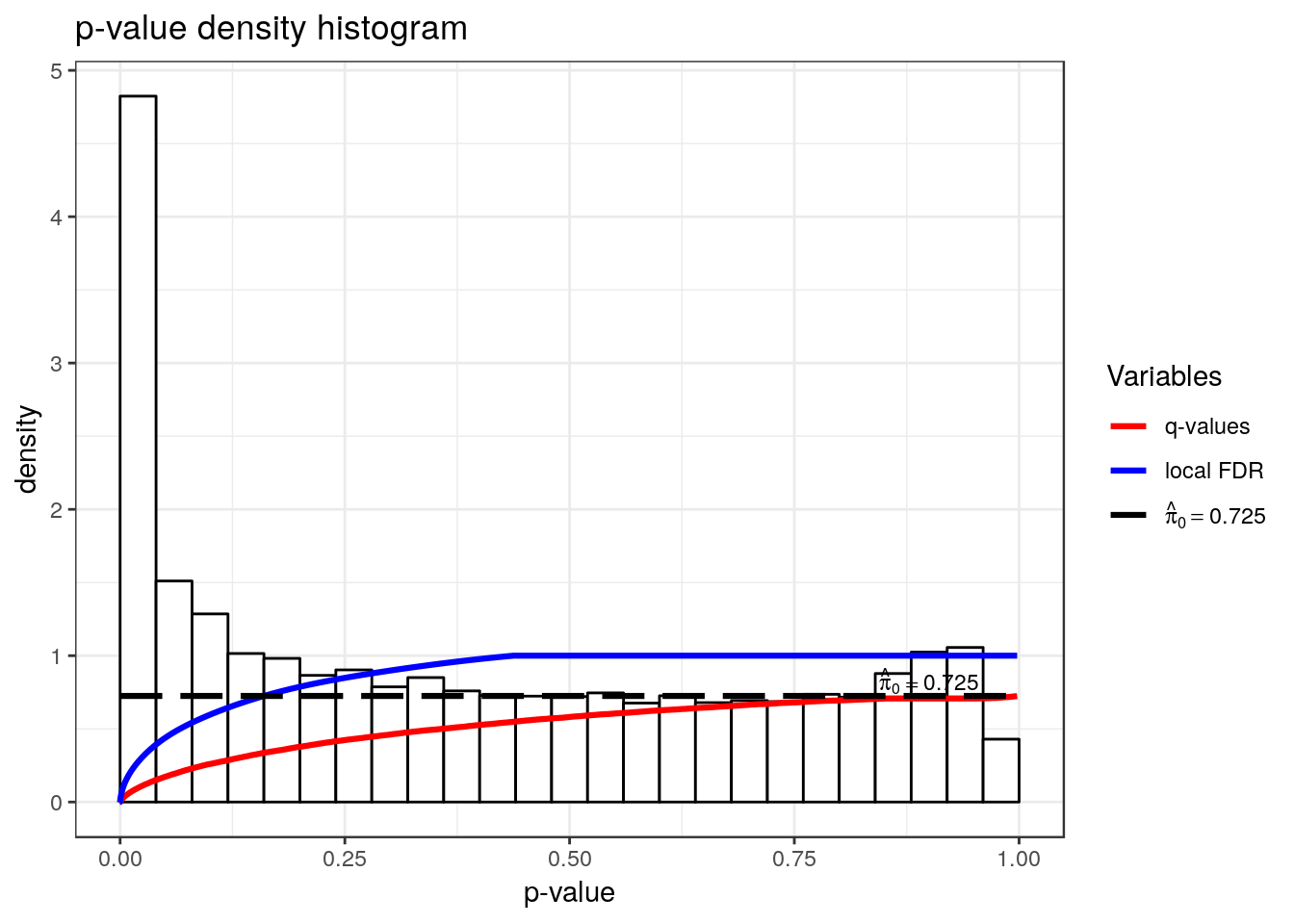
Display differential methylated peaks
jointPeak <- reportJointPeak(RSV.Hela,threads = 20)Reporting joint peak with PoissonGamma test statistics...
Reporting 18114 peaks...
Hyper-thread registered: TRUE
Using 20 thread(s) to report merged report...
Time used to report peaks: 3.46580692529678 mins... #write.table(jointPeak[which(jointPeak$fdr<0.1 & abs(jointPeak$logFC)>0.5 ),], file = "~/RSV/2017/Host_diff_meth.xls",sep = "\t", quote = F,row.names = F)
knitr::kable(jointPeak[which(jointPeak$fdr<0.1 & abs(jointPeak$logFC)>0.5 ),], format = "pandoc", row.names = F)| chr | start | end | name | score | strand | thickStart | thickEnd | itemRgb | blockCount | blockSizes | blockStarts | logFC | p_value | fdr |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chr2 | 218264179 | 218264228 | AAMP | 0 | - | 218264179 | 218264228 | 0 | 1 | 49 | 0 | 5.2144851 | 0.0000364 | 0.0011679 |
| chr20 | 36239899 | 36240195 | AAR2 | 0 | + | 36239899 | 36240195 | 0 | 1 | 296 | 0 | -1.0571236 | 0.0000921 | 0.0024380 |
| chrX | 153725611 | 153726105 | ABCD1 | 0 | + | 153725611 | 153726105 | 0 | 1 | 494 | 0 | -1.0287961 | 0.0004289 | 0.0082539 |
| chr7 | 151226365 | 151226464 | ABCF2 | 0 | - | 151226365 | 151226464 | 0 | 1 | 99 | 0 | 1.1304662 | 0.0041922 | 0.0456205 |
| chr19 | 1877025 | 1877270 | ABHD17A | 0 | - | 1877025 | 1877270 | 0 | 1 | 245 | 0 | -1.0515146 | 0.0000223 | 0.0007821 |
| chr19 | 17300911 | 17301358 | ABHD8 | 0 | - | 17300911 | 17301358 | 0 | 1 | 447 | 0 | -0.7294186 | 0.0022919 | 0.0292196 |
| chr9 | 130884104 | 130884252 | ABL1 | 0 | + | 130884104 | 130884252 | 0 | 1 | 148 | 0 | -1.3050447 | 0.0034496 | 0.0395854 |
| chr9 | 130884451 | 130884649 | ABL1 | 0 | + | 130884451 | 130884649 | 0 | 1 | 198 | 0 | -2.2016470 | 0.0000011 | 0.0000586 |
| chr9 | 130884947 | 130885145 | ABL1 | 0 | + | 130884947 | 130885145 | 0 | 1 | 198 | 0 | -2.1549088 | 0.0000001 | 0.0000070 |
| chr9 | 130885294 | 130885343 | ABL1 | 0 | + | 130885294 | 130885343 | 0 | 1 | 49 | 0 | -3.1025292 | 0.0000267 | 0.0008925 |
| chr9 | 130885394 | 130886336 | ABL1 | 0 | + | 130885394 | 130886336 | 0 | 1 | 942 | 0 | -1.0046098 | 0.0000129 | 0.0004992 |
| chr1 | 179107676 | 179109075 | ABL2 | 0 | - | 179107676 | 179109075 | 0 | 1 | 1399 | 0 | -0.5720723 | 0.0008334 | 0.0137688 |
| chr3 | 127680285 | 127680532 | ABTB1 | 0 | + | 127680285 | 127680532 | 0 | 1 | 247 | 0 | -0.9703037 | 0.0087641 | 0.0767781 |
| chr11 | 34357266 | 34358008 | ABTB2 | 0 | - | 34357266 | 34358008 | 0 | 1 | 742 | 0 | -0.6052720 | 0.0011806 | 0.0179959 |
| chr3 | 38122859 | 38123057 | ACAA1 | 0 | - | 38122859 | 38123057 | 0 | 1 | 198 | 0 | -1.8191906 | 0.0000151 | 0.0005690 |
| chr1 | 226146412 | 226146560 | ACBD3 | 0 | - | 226146412 | 226146560 | 0 | 1 | 148 | 0 | -1.8680184 | 0.0006109 | 0.0108768 |
| chr1 | 226146709 | 226152617 | ACBD3 | 0 | - | 226146709 | 226152617 | 0 | 2 | 113,283 | 0,5626 | -0.5120338 | 0.0066953 | 0.0640540 |
| chr9 | 19452060 | 19452109 | ACER2 | 0 | + | 19452060 | 19452109 | 0 | 1 | 49 | 0 | -2.9662093 | 0.0077726 | 0.0706204 |
| chr2 | 236580602 | 236580801 | ACKR3 | 0 | + | 236580602 | 236580801 | 0 | 1 | 199 | 0 | -0.5762403 | 0.0099167 | 0.0837161 |
| chr17 | 41867562 | 41867860 | ACLY | 0 | - | 41867562 | 41867860 | 0 | 1 | 298 | 0 | -0.9473264 | 0.0117882 | 0.0939985 |
| chrX | 109644027 | 109644126 | ACSL4 | 0 | - | 109644027 | 109644126 | 0 | 1 | 99 | 0 | -1.2310347 | 0.0038059 | 0.0424835 |
| chr1 | 17825986 | 17826182 | ACTL8 | 0 | + | 17825986 | 17826182 | 0 | 1 | 196 | 0 | -1.4132368 | 0.0007527 | 0.0127146 |
| chr14 | 68925639 | 68979316 | ACTN1 | 0 | - | 68925639 | 68979316 | 0 | 2 | 34,365 | 0,53313 | 0.5185801 | 0.0038096 | 0.0424835 |
| chr19 | 38647627 | 38700714 | ACTN4 | 0 | + | 38647627 | 38700714 | 0 | 2 | 281,115 | 0,52973 | 0.6023915 | 0.0069931 | 0.0658701 |
| chr19 | 38717093 | 38717142 | ACTN4 | 0 | + | 38717093 | 38717142 | 0 | 1 | 49 | 0 | -2.3759214 | 0.0057114 | 0.0573934 |
| chr19 | 38717950 | 38721610 | ACTN4 | 0 | + | 38717950 | 38721610 | 0 | 2 | 125,73 | 0,3588 | -1.1811629 | 0.0027492 | 0.0333261 |
| chr19 | 38729840 | 38730086 | ACTN4 | 0 | + | 38729840 | 38730086 | 0 | 1 | 246 | 0 | -1.4038613 | 0.0002544 | 0.0054526 |
| chr10 | 102490571 | 102502705 | ACTR1A | 0 | - | 102490571 | 102502705 | 0 | 2 | 43,106 | 0,12029 | 1.6693010 | 0.0000321 | 0.0010572 |
| chr1 | 150560241 | 150560439 | ADAMTSL4 | 0 | + | 150560241 | 150560439 | 0 | 1 | 198 | 0 | -1.5843645 | 0.0000000 | 0.0000010 |
| chr1 | 154582158 | 154582307 | ADAR | 0 | - | 154582158 | 154582307 | 0 | 1 | 149 | 0 | -1.0522790 | 0.0068151 | 0.0646932 |
| chr1 | 154601057 | 154606084 | ADAR | 0 | - | 154601057 | 154606084 | 0 | 2 | 1570,79 | 0,4949 | -1.0974519 | 0.0000000 | 0.0000001 |
| chr21 | 45175898 | 45176497 | ADARB1 | 0 | + | 45175898 | 45176497 | 0 | 1 | 599 | 0 | -0.5112422 | 0.0121087 | 0.0955815 |
| chr16 | 75612377 | 75612673 | ADAT1 | 0 | - | 75612377 | 75612673 | 0 | 1 | 296 | 0 | -1.3484275 | 0.0013358 | 0.0197490 |
| chr7 | 140673695 | 140674187 | ADCK2 | 0 | + | 140673695 | 140674187 | 0 | 1 | 492 | 0 | -0.7867378 | 0.0001093 | 0.0028073 |
| chr19 | 40691679 | 40691878 | ADCK4 | 0 | - | 40691679 | 40691878 | 0 | 1 | 199 | 0 | -1.6650473 | 0.0013733 | 0.0201353 |
| chr2 | 24820069 | 24820777 | ADCY3 | 0 | - | 24820069 | 24820777 | 0 | 2 | 46,54 | 0,655 | -3.5717915 | 0.0000014 | 0.0000730 |
| chr2 | 24918438 | 24918986 | ADCY3 | 0 | - | 24918438 | 24918986 | 0 | 1 | 548 | 0 | -1.7869018 | 0.0000000 | 0.0000015 |
| chr16 | 3965341 | 3966287 | ADCY9 | 0 | - | 3965341 | 3966287 | 0 | 1 | 946 | 0 | -0.6034758 | 0.0054939 | 0.0556707 |
| chr16 | 4114387 | 4114535 | ADCY9 | 0 | - | 4114387 | 4114535 | 0 | 1 | 148 | 0 | -2.4305062 | 0.0000526 | 0.0015500 |
| chr16 | 4114835 | 4114934 | ADCY9 | 0 | - | 4114835 | 4114934 | 0 | 1 | 99 | 0 | -2.6589168 | 0.0011075 | 0.0172053 |
| chr12 | 131139221 | 131139818 | ADGRD1 | 0 | + | 131139221 | 131139818 | 0 | 1 | 597 | 0 | 0.6288189 | 0.0006540 | 0.0114863 |
| chr19 | 14162719 | 14163365 | ADGRL1 | 0 | - | 14162719 | 14163365 | 0 | 1 | 646 | 0 | -0.7763979 | 0.0025189 | 0.0314347 |
| chr15 | 72752160 | 72752803 | ADPGK | 0 | - | 72752160 | 72752803 | 0 | 1 | 643 | 0 | -0.6322942 | 0.0015241 | 0.0215945 |
| chr1 | 36092513 | 36093733 | ADPRHL2 | 0 | + | 36092513 | 36093733 | 0 | 2 | 10,637 | 0,584 | -0.5946927 | 0.0024645 | 0.0309055 |
| chr15 | 88626405 | 88626701 | AEN | 0 | + | 88626405 | 88626701 | 0 | 1 | 296 | 0 | -0.6985635 | 0.0007188 | 0.0122790 |
| chr15 | 88629376 | 88630649 | AEN | 0 | + | 88629376 | 88630649 | 0 | 2 | 51,592 | 0,682 | -0.5935024 | 0.0037706 | 0.0422315 |
| chr4 | 87047181 | 87047579 | AFF1 | 0 | + | 87047181 | 87047579 | 0 | 1 | 398 | 0 | -1.0119256 | 0.0000520 | 0.0015358 |
| chr5 | 132896793 | 132897191 | AFF4 | 0 | - | 132896793 | 132897191 | 0 | 1 | 398 | 0 | -0.8607216 | 0.0077258 | 0.0703006 |
| chr5 | 132934375 | 132934873 | AFF4 | 0 | - | 132934375 | 132934873 | 0 | 1 | 498 | 0 | -0.8272063 | 0.0000680 | 0.0019228 |
| chr16 | 89994737 | 89995527 | AFG3L1P | 0 | + | 89994737 | 89995527 | 0 | 1 | 790 | 0 | -1.0383583 | 0.0011267 | 0.0173989 |
| chr16 | 89995577 | 89995725 | AFG3L1P | 0 | + | 89995577 | 89995725 | 0 | 1 | 148 | 0 | -1.9332277 | 0.0022683 | 0.0290069 |
| chr2 | 64552675 | 64553221 | AFTPH | 0 | + | 64552675 | 64553221 | 0 | 1 | 546 | 0 | -0.7005081 | 0.0109928 | 0.0899598 |
| chr12 | 57727813 | 57728257 | AGAP2-AS1 | 0 | + | 57727813 | 57728257 | 0 | 1 | 444 | 0 | -0.7468229 | 0.0005322 | 0.0096576 |
| chr9 | 136673193 | 136673841 | AGPAT2 | 0 | - | 136673193 | 136673841 | 0 | 1 | 648 | 0 | -1.0147999 | 0.0000559 | 0.0016316 |
| chr1 | 246851040 | 246851239 | AHCTF1 | 0 | - | 246851040 | 246851239 | 0 | 1 | 199 | 0 | -0.9114310 | 0.0104658 | 0.0869734 |
| chr11 | 62516999 | 62517048 | AHNAK | 0 | - | 62516999 | 62517048 | 0 | 1 | 49 | 0 | -1.0793613 | 0.0016137 | 0.0225463 |
| chr11 | 62517448 | 62517647 | AHNAK | 0 | - | 62517448 | 62517647 | 0 | 1 | 199 | 0 | -1.9774340 | 0.0000000 | 0.0000000 |
| chr11 | 62517898 | 62518296 | AHNAK | 0 | - | 62517898 | 62518296 | 0 | 1 | 398 | 0 | -1.1975415 | 0.0000000 | 0.0000000 |
| chr11 | 62518347 | 62518645 | AHNAK | 0 | - | 62518347 | 62518645 | 0 | 1 | 298 | 0 | -1.9573787 | 0.0000000 | 0.0000000 |
| chr11 | 62518946 | 62519045 | AHNAK | 0 | - | 62518946 | 62519045 | 0 | 1 | 99 | 0 | -1.7788671 | 0.0000002 | 0.0000130 |
| chr11 | 62519545 | 62519643 | AHNAK | 0 | - | 62519545 | 62519643 | 0 | 1 | 98 | 0 | -2.0845095 | 0.0000003 | 0.0000202 |
| chr11 | 62520493 | 62520941 | AHNAK | 0 | - | 62520493 | 62520941 | 0 | 1 | 448 | 0 | -1.1998491 | 0.0000004 | 0.0000266 |
| chr11 | 62521092 | 62521141 | AHNAK | 0 | - | 62521092 | 62521141 | 0 | 1 | 49 | 0 | -2.2530428 | 0.0000000 | 0.0000001 |
| chr11 | 62521241 | 62521340 | AHNAK | 0 | - | 62521241 | 62521340 | 0 | 1 | 99 | 0 | -1.3466811 | 0.0000686 | 0.0019357 |
| chr11 | 62522040 | 62522189 | AHNAK | 0 | - | 62522040 | 62522189 | 0 | 1 | 149 | 0 | -1.8588767 | 0.0000002 | 0.0000135 |
| chr11 | 62523188 | 62523437 | AHNAK | 0 | - | 62523188 | 62523437 | 0 | 1 | 249 | 0 | -2.0222805 | 0.0000000 | 0.0000000 |
| chr11 | 62524486 | 62524634 | AHNAK | 0 | - | 62524486 | 62524634 | 0 | 1 | 148 | 0 | -1.5184940 | 0.0001143 | 0.0029069 |
| chr11 | 62525534 | 62525682 | AHNAK | 0 | - | 62525534 | 62525682 | 0 | 1 | 148 | 0 | -1.9439177 | 0.0000000 | 0.0000000 |
| chr11 | 62526133 | 62526481 | AHNAK | 0 | - | 62526133 | 62526481 | 0 | 1 | 348 | 0 | -1.4906561 | 0.0000000 | 0.0000000 |
| chr11 | 62526632 | 62526880 | AHNAK | 0 | - | 62526632 | 62526880 | 0 | 1 | 248 | 0 | -1.4263890 | 0.0000000 | 0.0000004 |
| chr11 | 62526981 | 62527230 | AHNAK | 0 | - | 62526981 | 62527230 | 0 | 1 | 249 | 0 | -1.6009712 | 0.0000000 | 0.0000000 |
| chr11 | 62527780 | 62527829 | AHNAK | 0 | - | 62527780 | 62527829 | 0 | 1 | 49 | 0 | -1.5123549 | 0.0007677 | 0.0129194 |
| chr11 | 62528977 | 62529176 | AHNAK | 0 | - | 62528977 | 62529176 | 0 | 1 | 199 | 0 | -1.7154497 | 0.0000001 | 0.0000072 |
| chr11 | 62532721 | 62533119 | AHNAK | 0 | - | 62532721 | 62533119 | 0 | 1 | 398 | 0 | -0.8487974 | 0.0001867 | 0.0042286 |
| chr14 | 104938751 | 104938950 | AHNAK2 | 0 | - | 104938751 | 104938950 | 0 | 1 | 199 | 0 | -1.5066009 | 0.0000084 | 0.0003467 |
| chr14 | 104941596 | 104941695 | AHNAK2 | 0 | - | 104941596 | 104941695 | 0 | 1 | 99 | 0 | -1.9822887 | 0.0001046 | 0.0027106 |
| chr14 | 104941746 | 104941945 | AHNAK2 | 0 | - | 104941746 | 104941945 | 0 | 1 | 199 | 0 | -1.3765929 | 0.0021772 | 0.0280791 |
| chr14 | 104942095 | 104942194 | AHNAK2 | 0 | - | 104942095 | 104942194 | 0 | 1 | 99 | 0 | -1.7112411 | 0.0015839 | 0.0222334 |
| chr14 | 104951528 | 104951577 | AHNAK2 | 0 | - | 104951528 | 104951577 | 0 | 1 | 49 | 0 | -2.1542005 | 0.0012789 | 0.0191079 |
| chr14 | 104953924 | 104954622 | AHNAK2 | 0 | - | 104953924 | 104954622 | 0 | 1 | 698 | 0 | -0.9299827 | 0.0000012 | 0.0000646 |
| chr7 | 17339312 | 17339461 | AHR | 0 | + | 17339312 | 17339461 | 0 | 1 | 149 | 0 | -1.3913339 | 0.0070531 | 0.0662779 |
| chr17 | 57105778 | 57105877 | AKAP1 | 0 | + | 57105778 | 57105877 | 0 | 1 | 99 | 0 | -1.5563095 | 0.0051557 | 0.0532629 |
| chr17 | 57105977 | 57106423 | AKAP1 | 0 | + | 57105977 | 57106423 | 0 | 1 | 446 | 0 | -0.9211036 | 0.0003223 | 0.0065797 |
| chr17 | 19905101 | 19905249 | AKAP10 | 0 | - | 19905101 | 19905249 | 0 | 1 | 148 | 0 | -1.3753316 | 0.0088336 | 0.0771207 |
| chr15 | 85579021 | 85579619 | AKAP13 | 0 | + | 85579021 | 85579619 | 0 | 1 | 598 | 0 | -1.1406361 | 0.0000382 | 0.0012134 |
| chr15 | 85579918 | 85580117 | AKAP13 | 0 | + | 85579918 | 85580117 | 0 | 1 | 199 | 0 | -2.3881686 | 0.0000004 | 0.0000255 |
| chr15 | 85580317 | 85580466 | AKAP13 | 0 | + | 85580317 | 85580466 | 0 | 1 | 149 | 0 | -1.5704284 | 0.0122014 | 0.0961534 |
| chr15 | 85580865 | 85581014 | AKAP13 | 0 | + | 85580865 | 85581014 | 0 | 1 | 149 | 0 | -1.6865617 | 0.0029403 | 0.0350278 |
| chr15 | 85581065 | 85581114 | AKAP13 | 0 | + | 85581065 | 85581114 | 0 | 1 | 49 | 0 | -2.8385579 | 0.0071450 | 0.0666773 |
| chr15 | 85744724 | 85745371 | AKAP13 | 0 | + | 85744724 | 85745371 | 0 | 1 | 647 | 0 | -0.5781172 | 0.0126130 | 0.0983616 |
| chr9 | 110136473 | 110136622 | AKAP2 | 0 | + | 110136473 | 110136622 | 0 | 1 | 149 | 0 | -1.1231923 | 0.0036177 | 0.0409150 |
| chr9 | 110137423 | 110137522 | AKAP2 | 0 | + | 110137423 | 110137522 | 0 | 1 | 99 | 0 | -1.2514970 | 0.0013836 | 0.0202278 |
| chr9 | 110137572 | 110137821 | AKAP2 | 0 | + | 110137572 | 110137821 | 0 | 1 | 249 | 0 | -1.4526295 | 0.0000055 | 0.0002357 |
| chr6 | 87701772 | 87702267 | AKIRIN2 | 0 | - | 87701772 | 87702267 | 0 | 1 | 495 | 0 | 1.7826391 | 0.0000000 | 0.0000000 |
| chr19 | 40233167 | 40233216 | AKT2 | 0 | - | 40233167 | 40233216 | 0 | 1 | 49 | 0 | 1.4596721 | 0.0083483 | 0.0741400 |
| chr15 | 100908481 | 100915094 | ALDH1A3 | 0 | + | 100908481 | 100915094 | 0 | 2 | 2,394 | 0,6220 | -0.7510519 | 0.0108694 | 0.0893462 |
| chr9 | 38396091 | 38396289 | ALDH1B1 | 0 | + | 38396091 | 38396289 | 0 | 1 | 198 | 0 | -1.2572779 | 0.0001547 | 0.0036662 |
| chr9 | 38397081 | 38397328 | ALDH1B1 | 0 | + | 38397081 | 38397328 | 0 | 1 | 247 | 0 | -1.1192617 | 0.0000039 | 0.0001776 |
| chr17 | 19676055 | 19676154 | ALDH3A2 | 0 | + | 19676055 | 19676154 | 0 | 1 | 99 | 0 | -2.0262703 | 0.0015717 | 0.0220959 |
| chr11 | 68027940 | 68028387 | ALDH3B1 | 0 | + | 68027940 | 68028387 | 0 | 1 | 447 | 0 | -0.7676826 | 0.0032330 | 0.0377412 |
| chr11 | 68028537 | 68028884 | ALDH3B1 | 0 | + | 68028537 | 68028884 | 0 | 1 | 347 | 0 | -0.9611457 | 0.0000022 | 0.0001058 |
| chr22 | 49903503 | 49903996 | ALG12 | 0 | - | 49903503 | 49903996 | 0 | 1 | 493 | 0 | -0.6779602 | 0.0039883 | 0.0439827 |
| chr9 | 99221616 | 99221914 | ALG2 | 0 | - | 99221616 | 99221914 | 0 | 1 | 298 | 0 | 1.3305440 | 0.0000005 | 0.0000312 |
| chr1 | 63370921 | 63370970 | ALG6 | 0 | + | 63370921 | 63370970 | 0 | 1 | 49 | 0 | -2.4459001 | 0.0116240 | 0.0933974 |
| chr12 | 109092681 | 109092930 | ALKBH2 | 0 | - | 109092681 | 109092930 | 0 | 1 | 249 | 0 | -0.9223523 | 0.0059891 | 0.0592723 |
| chr17 | 18184100 | 18184697 | ALKBH5 | 0 | + | 18184100 | 18184697 | 0 | 1 | 597 | 0 | -1.9133562 | 0.0000000 | 0.0000000 |
| chr11 | 46542531 | 46542878 | AMBRA1 | 0 | - | 46542531 | 46542878 | 0 | 1 | 347 | 0 | -1.7076817 | 0.0000023 | 0.0001129 |
| chr11 | 94799421 | 94800270 | AMOTL1 | 0 | + | 94799421 | 94800270 | 0 | 1 | 849 | 0 | -1.0372071 | 0.0000000 | 0.0000028 |
| chr11 | 94871605 | 94871704 | AMOTL1 | 0 | + | 94871605 | 94871704 | 0 | 1 | 99 | 0 | -1.3501143 | 0.0021535 | 0.0278544 |
| chr5 | 140529123 | 140529371 | ANKHD1 | 0 | + | 140529123 | 140529371 | 0 | 1 | 248 | 0 | -1.3067473 | 0.0042602 | 0.0462766 |
| chr5 | 140526149 | 140526348 | ANKHD1-EIF4EBP3 | 0 | + | 140526149 | 140526348 | 0 | 1 | 199 | 0 | -1.6225288 | 0.0114682 | 0.0924901 |
| chr5 | 140529142 | 140529341 | ANKHD1-EIF4EBP3 | 0 | + | 140529142 | 140529341 | 0 | 1 | 199 | 0 | -1.2074635 | 0.0114025 | 0.0922179 |
| chr16 | 89279784 | 89279983 | ANKRD11 | 0 | - | 89279784 | 89279983 | 0 | 1 | 199 | 0 | -1.9350640 | 0.0000023 | 0.0001116 |
| chr16 | 89280033 | 89280281 | ANKRD11 | 0 | - | 89280033 | 89280281 | 0 | 1 | 248 | 0 | -2.4041705 | 0.0000000 | 0.0000023 |
| chr19 | 32597644 | 32597792 | ANKRD27 | 0 | - | 32597644 | 32597792 | 0 | 1 | 148 | 0 | -1.5085993 | 0.0094478 | 0.0807336 |
| chr5 | 10654872 | 10655121 | ANKRD33B | 0 | + | 10654872 | 10655121 | 0 | 1 | 249 | 0 | -1.1371220 | 0.0102091 | 0.0854673 |
| chr12 | 45216136 | 45216284 | ANO6 | 0 | + | 45216136 | 45216284 | 0 | 1 | 148 | 0 | 1.2957477 | 0.0015590 | 0.0219692 |
| chr15 | 60354173 | 60357172 | ANXA2 | 0 | - | 60354173 | 60357172 | 0 | 3 | 41,80,27 | 0,1746,2973 | -2.3014330 | 0.0014474 | 0.0208342 |
| chr19 | 16198003 | 16198052 | AP1M1 | 0 | + | 16198003 | 16198052 | 0 | 1 | 49 | 0 | 2.5069225 | 0.0003657 | 0.0073001 |
| chr5 | 78294556 | 78294655 | AP3B1 | 0 | - | 78294556 | 78294655 | 0 | 1 | 99 | 0 | 1.6936518 | 0.0014019 | 0.0203898 |
| chr5 | 115841922 | 115842021 | AP3S1 | 0 | + | 115841922 | 115842021 | 0 | 1 | 99 | 0 | 1.4399194 | 0.0027386 | 0.0332664 |
| chr15 | 89835213 | 89835560 | AP3S2 | 0 | - | 89835213 | 89835560 | 0 | 1 | 347 | 0 | -0.8475240 | 0.0000888 | 0.0023718 |
| chr19 | 3759706 | 3761675 | APBA3 | 0 | - | 3759706 | 3761675 | 0 | 2 | 596,140 | 0,1830 | -0.8228132 | 0.0006169 | 0.0109728 |
| chrX | 55007076 | 55007474 | APEX2 | 0 | + | 55007076 | 55007474 | 0 | 1 | 398 | 0 | -1.0588049 | 0.0000002 | 0.0000109 |
| chr20 | 24963191 | 24963338 | APMAP | 0 | - | 24963191 | 24963338 | 0 | 1 | 147 | 0 | -1.5662454 | 0.0014474 | 0.0208342 |
| chr22 | 35648427 | 35656431 | APOL6 | 0 | + | 35648427 | 35656431 | 0 | 2 | 197,53 | 0,7952 | 1.0987065 | 0.0066477 | 0.0638951 |
| chr22 | 35658671 | 35658720 | APOL6 | 0 | + | 35658671 | 35658720 | 0 | 1 | 49 | 0 | -2.0760623 | 0.0016340 | 0.0227430 |
| chr22 | 35658870 | 35659519 | APOL6 | 0 | + | 35658870 | 35659519 | 0 | 1 | 649 | 0 | -0.8516070 | 0.0000235 | 0.0008090 |
| chr21 | 26170618 | 26170766 | APP | 0 | - | 26170618 | 26170766 | 0 | 1 | 148 | 0 | 1.3217957 | 0.0003456 | 0.0069686 |
| chr9 | 33443808 | 33443857 | AQP3 | 0 | - | 33443808 | 33443857 | 0 | 1 | 49 | 0 | -3.3906416 | 0.0008076 | 0.0134411 |
| chr11 | 118600753 | 118601000 | ARCN1 | 0 | + | 118600753 | 118601000 | 0 | 1 | 247 | 0 | -1.1587415 | 0.0029318 | 0.0349502 |
| chr13 | 106543897 | 106557076 | ARGLU1 | 0 | - | 106543897 | 106557076 | 0 | 2 | 264,29 | 0,13151 | 0.8066467 | 0.0027273 | 0.0331849 |
| chr10 | 31908614 | 31908862 | ARHGAP12 | 0 | - | 31908614 | 31908862 | 0 | 1 | 248 | 0 | -1.1914926 | 0.0020169 | 0.0265225 |
| chr10 | 24585351 | 24585549 | ARHGAP21 | 0 | - | 24585351 | 24585549 | 0 | 1 | 198 | 0 | -1.4542033 | 0.0000025 | 0.0001186 |
| chr10 | 24619794 | 24619992 | ARHGAP21 | 0 | - | 24619794 | 24619992 | 0 | 1 | 198 | 0 | -0.9998310 | 0.0044045 | 0.0473052 |
| chr10 | 24620242 | 24620440 | ARHGAP21 | 0 | - | 24620242 | 24620440 | 0 | 1 | 198 | 0 | -1.2200232 | 0.0019963 | 0.0263599 |
| chr19 | 46918676 | 46919224 | ARHGAP35 | 0 | + | 46918676 | 46919224 | 0 | 1 | 548 | 0 | -1.0710746 | 0.0004256 | 0.0081991 |
| chr19 | 46921722 | 46921871 | ARHGAP35 | 0 | + | 46921722 | 46921871 | 0 | 1 | 149 | 0 | -2.3273711 | 0.0004653 | 0.0087954 |
| chr19 | 46922171 | 46922220 | ARHGAP35 | 0 | + | 46922171 | 46922220 | 0 | 1 | 49 | 0 | -2.0731602 | 0.0105502 | 0.0874739 |
| chr19 | 47000583 | 47000632 | ARHGAP35 | 0 | + | 47000583 | 47000632 | 0 | 1 | 49 | 0 | -2.7276641 | 0.0081712 | 0.0730578 |
| chr17 | 81867917 | 81868264 | ARHGDIA | 0 | - | 81867917 | 81868264 | 0 | 1 | 347 | 0 | -1.0312192 | 0.0027980 | 0.0336819 |
| chr17 | 81869761 | 81870736 | ARHGDIA | 0 | - | 81869761 | 81870736 | 0 | 2 | 197,101 | 0,875 | 0.8051223 | 0.0012142 | 0.0184160 |
| chr1 | 3454731 | 3463083 | ARHGEF16 | 0 | + | 3454731 | 3463083 | 0 | 2 | 81,18 | 0,8335 | 1.8859039 | 0.0061808 | 0.0607058 |
| chr11 | 73310061 | 73310259 | ARHGEF17 | 0 | + | 73310061 | 73310259 | 0 | 1 | 198 | 0 | -1.4552499 | 0.0008997 | 0.0146116 |
| chr11 | 73310559 | 73311155 | ARHGEF17 | 0 | + | 73310559 | 73311155 | 0 | 1 | 596 | 0 | -1.1415607 | 0.0000861 | 0.0023205 |
| chr11 | 73311255 | 73311752 | ARHGEF17 | 0 | + | 73311255 | 73311752 | 0 | 1 | 497 | 0 | -1.9007715 | 0.0000000 | 0.0000000 |
| chr1 | 155961909 | 155962153 | ARHGEF2 | 0 | - | 155961909 | 155962153 | 0 | 2 | 1,49 | 0,196 | 2.9586807 | 0.0047166 | 0.0497713 |
| chr7 | 144284174 | 144284421 | ARHGEF34P | 0 | - | 144284174 | 144284421 | 0 | 1 | 247 | 0 | -1.1661195 | 0.0025327 | 0.0315415 |
| chr7 | 144284965 | 144285261 | ARHGEF34P | 0 | - | 144284965 | 144285261 | 0 | 1 | 296 | 0 | -1.4972229 | 0.0000991 | 0.0025942 |
| chr7 | 144362801 | 144363399 | ARHGEF5 | 0 | + | 144362801 | 144363399 | 0 | 1 | 598 | 0 | -0.6542319 | 0.0086068 | 0.0758769 |
| chr7 | 144364448 | 144364646 | ARHGEF5 | 0 | + | 144364448 | 144364646 | 0 | 1 | 198 | 0 | -1.4337329 | 0.0028302 | 0.0339620 |
| chr7 | 144365196 | 144365445 | ARHGEF5 | 0 | + | 144365196 | 144365445 | 0 | 1 | 249 | 0 | -1.4291138 | 0.0001511 | 0.0036094 |
| chr1 | 26779266 | 26779664 | ARID1A | 0 | + | 26779266 | 26779664 | 0 | 1 | 398 | 0 | -1.2370356 | 0.0001487 | 0.0035672 |
| chr1 | 26779715 | 26780613 | ARID1A | 0 | + | 26779715 | 26780613 | 0 | 1 | 898 | 0 | -1.3854854 | 0.0000000 | 0.0000000 |
| chr10 | 62091075 | 62091273 | ARID5B | 0 | + | 62091075 | 62091273 | 0 | 1 | 198 | 0 | -1.2930682 | 0.0063190 | 0.0616839 |
| chr10 | 62091373 | 62091621 | ARID5B | 0 | + | 62091373 | 62091621 | 0 | 1 | 248 | 0 | -1.2811733 | 0.0020034 | 0.0264217 |
| chr10 | 62091772 | 62092966 | ARID5B | 0 | + | 62091772 | 62092966 | 0 | 1 | 1194 | 0 | -0.9019597 | 0.0000565 | 0.0016431 |
| chr10 | 62093165 | 62093563 | ARID5B | 0 | + | 62093165 | 62093563 | 0 | 1 | 398 | 0 | -1.4977537 | 0.0000000 | 0.0000029 |
| chr15 | 72474326 | 72474969 | ARIH1 | 0 | + | 72474326 | 72474969 | 0 | 1 | 643 | 0 | 0.5235716 | 0.0010044 | 0.0159119 |
| chr15 | 72583782 | 72583880 | ARIH1 | 0 | + | 72583782 | 72583880 | 0 | 1 | 98 | 0 | -1.2999458 | 0.0114491 | 0.0924595 |
| chr17 | 43399948 | 43400195 | ARL4D | 0 | + | 43399948 | 43400195 | 0 | 1 | 247 | 0 | -0.5397290 | 0.0093105 | 0.0799197 |
| chr3 | 69084939 | 69085136 | ARL6IP5 | 0 | + | 69084939 | 69085136 | 0 | 1 | 197 | 0 | 1.1512947 | 0.0002591 | 0.0055203 |
| chr17 | 75129077 | 75129424 | ARMC7 | 0 | + | 75129077 | 75129424 | 0 | 1 | 347 | 0 | -1.2784212 | 0.0027211 | 0.0331591 |
| chr7 | 99325970 | 99338200 | ARPC1A | 0 | + | 99325970 | 99338200 | 0 | 3 | 35,93,20 | 0,7355,12211 | 1.3936621 | 0.0050628 | 0.0525607 |
| chr1 | 183627484 | 183627583 | ARPC5 | 0 | - | 183627484 | 183627583 | 0 | 1 | 99 | 0 | -1.6730051 | 0.0000031 | 0.0001462 |
| chr17 | 68307073 | 68307322 | ARSG | 0 | + | 68307073 | 68307322 | 0 | 1 | 249 | 0 | -1.2667081 | 0.0106122 | 0.0876272 |
| chr2 | 9207063 | 9207212 | ASAP2 | 0 | + | 9207063 | 9207212 | 0 | 1 | 149 | 0 | 2.2139661 | 0.0002112 | 0.0046476 |
| chr9 | 129637437 | 129638181 | ASB6 | 0 | - | 129637437 | 129638181 | 0 | 1 | 744 | 0 | -1.0232431 | 0.0000000 | 0.0000000 |
| chr1 | 155438548 | 155438997 | ASH1L | 0 | - | 155438548 | 155438997 | 0 | 1 | 449 | 0 | -1.4911170 | 0.0003832 | 0.0075756 |
| chr7 | 97871862 | 97872010 | ASNS | 0 | - | 97871862 | 97872010 | 0 | 1 | 148 | 0 | 1.6262800 | 0.0108796 | 0.0893462 |
| chr16 | 29901515 | 29901761 | ASPHD1 | 0 | + | 29901515 | 29901761 | 0 | 1 | 246 | 0 | 1.4059159 | 0.0000000 | 0.0000000 |
| chr2 | 25743917 | 25744365 | ASXL2 | 0 | - | 25743917 | 25744365 | 0 | 1 | 448 | 0 | -0.8343363 | 0.0011640 | 0.0177893 |
| chr1 | 212619431 | 212619679 | ATF3 | 0 | + | 212619431 | 212619679 | 0 | 1 | 248 | 0 | -0.8191354 | 0.0007498 | 0.0126766 |
| chr22 | 39521846 | 39521895 | ATF4 | 0 | + | 39521846 | 39521895 | 0 | 1 | 49 | 0 | -1.0903081 | 0.0069139 | 0.0654112 |
| chr19 | 49930922 | 49932512 | ATF5 | 0 | + | 49930922 | 49932512 | 0 | 2 | 107,91 | 0,1500 | 0.8730225 | 0.0026400 | 0.0324988 |
| chr12 | 52070073 | 52070172 | ATG101 | 0 | + | 52070073 | 52070172 | 0 | 1 | 99 | 0 | 0.9013843 | 0.0128047 | 0.0994285 |
| chr11 | 46673928 | 46674027 | ATG13 | 0 | + | 46673928 | 46674027 | 0 | 1 | 99 | 0 | -2.1077304 | 0.0004646 | 0.0087918 |
| chr2 | 219224514 | 219224662 | ATG9A | 0 | - | 219224514 | 219224662 | 0 | 1 | 148 | 0 | -2.0931457 | 0.0000798 | 0.0021896 |
| chr2 | 38295990 | 38296188 | ATL2 | 0 | - | 38295990 | 38296188 | 0 | 1 | 198 | 0 | -1.2406976 | 0.0042328 | 0.0460070 |
| chr16 | 81043229 | 81043327 | ATMIN | 0 | + | 81043229 | 81043327 | 0 | 1 | 98 | 0 | -1.4512394 | 0.0016256 | 0.0226434 |
| chr16 | 81043874 | 81044220 | ATMIN | 0 | + | 81043874 | 81044220 | 0 | 1 | 346 | 0 | -0.8433880 | 0.0026605 | 0.0326395 |
| chr16 | 81044420 | 81045014 | ATMIN | 0 | + | 81044420 | 81045014 | 0 | 1 | 594 | 0 | -0.7289201 | 0.0000432 | 0.0013319 |
| chr12 | 6935559 | 6935906 | ATN1 | 0 | + | 6935559 | 6935906 | 0 | 1 | 347 | 0 | -0.6385576 | 0.0120276 | 0.0953156 |
| chr1 | 169130072 | 169131722 | ATP1B1 | 0 | + | 169130072 | 169131722 | 0 | 2 | 19,431 | 0,1220 | -0.6979087 | 0.0051898 | 0.0534874 |
| chr3 | 141876628 | 141876872 | ATP1B3 | 0 | + | 141876628 | 141876872 | 0 | 1 | 244 | 0 | 1.0288682 | 0.0014873 | 0.0211891 |
| chr17 | 3944718 | 3944767 | ATP2A3 | 0 | - | 3944718 | 3944767 | 0 | 1 | 49 | 0 | 3.8830973 | 0.0008196 | 0.0135657 |
| chr1 | 203682907 | 203698162 | ATP2B4 | 0 | + | 203682907 | 203698162 | 0 | 2 | 492,6 | 0,15250 | -1.0898908 | 0.0000018 | 0.0000921 |
| chr20 | 59028727 | 59028776 | ATP5E | 0 | - | 59028727 | 59028776 | 0 | 1 | 49 | 0 | 3.1496781 | 0.0018890 | 0.0253619 |
| chr7 | 99419782 | 99420081 | ATP5J2-PTCD1 | 0 | - | 99419782 | 99420081 | 0 | 1 | 299 | 0 | -0.8274498 | 0.0004439 | 0.0084694 |
| chr5 | 173034456 | 173034848 | ATP6V0E1 | 0 | + | 173034456 | 173034848 | 0 | 1 | 392 | 0 | -1.0265970 | 0.0000000 | 0.0000001 |
| chr9 | 114587763 | 114592638 | ATP6V1G1 | 0 | + | 114587763 | 114592638 | 0 | 2 | 158,87 | 0,4789 | 1.5005773 | 0.0000000 | 0.0000010 |
| chr1 | 109483989 | 109484038 | ATXN7L2 | 0 | + | 109483989 | 109484038 | 0 | 1 | 49 | 0 | 3.1437684 | 0.0060591 | 0.0598246 |
| chr17 | 8206502 | 8206551 | AURKB | 0 | - | 8206502 | 8206551 | 0 | 1 | 49 | 0 | -2.2624110 | 0.0013561 | 0.0199644 |
| chr10 | 97677573 | 97677920 | AVPI1 | 0 | - | 97677573 | 97677920 | 0 | 1 | 347 | 0 | -0.6062106 | 0.0000616 | 0.0017664 |
| chr2 | 62223133 | 62223332 | B3GNT2 | 0 | + | 62223133 | 62223332 | 0 | 1 | 199 | 0 | -1.2155449 | 0.0058507 | 0.0583712 |
| chr3 | 119229904 | 119230152 | B4GALT4 | 0 | - | 119229904 | 119230152 | 0 | 1 | 248 | 0 | -1.1357671 | 0.0050930 | 0.0527600 |
| chr21 | 29329687 | 29342839 | BACH1 | 0 | + | 29329687 | 29342839 | 0 | 2 | 7,441 | 0,12712 | -0.8685888 | 0.0004692 | 0.0088499 |
| chr9 | 33264414 | 33264711 | BAG1 | 0 | - | 33264414 | 33264711 | 0 | 1 | 297 | 0 | 0.6342497 | 0.0105659 | 0.0875388 |
| chr10 | 119676484 | 119676632 | BAG3 | 0 | + | 119676484 | 119676632 | 0 | 1 | 148 | 0 | -1.0336799 | 0.0072101 | 0.0669889 |
| chr10 | 119676780 | 119677372 | BAG3 | 0 | + | 119676780 | 119677372 | 0 | 1 | 592 | 0 | -0.9839485 | 0.0001079 | 0.0027844 |
| chr14 | 103559865 | 103560558 | BAG5 | 0 | - | 103559865 | 103560558 | 0 | 1 | 693 | 0 | -0.6981052 | 0.0012149 | 0.0184160 |
| chr14 | 103560658 | 103561922 | BAG5 | 0 | - | 103560658 | 103561922 | 0 | 2 | 535,10 | 0,1255 | -0.6284487 | 0.0095668 | 0.0814842 |
| chr17 | 81442426 | 81443222 | BAHCC1 | 0 | + | 81442426 | 81443222 | 0 | 1 | 796 | 0 | -1.0906803 | 0.0002241 | 0.0048895 |
| chr16 | 1349292 | 1349441 | BAIAP3 | 0 | + | 1349292 | 1349441 | 0 | 1 | 149 | 0 | -5.4319768 | 0.0000002 | 0.0000126 |
| chr10 | 28677495 | 28677741 | BAMBI | 0 | + | 28677495 | 28677741 | 0 | 1 | 246 | 0 | 0.5734225 | 0.0019858 | 0.0263237 |
| chr5 | 17276453 | 17276502 | BASP1 | 0 | + | 17276453 | 17276502 | 0 | 1 | 49 | 0 | -0.8268847 | 0.0116337 | 0.0934168 |
| chr7 | 73477327 | 73477426 | BAZ1B | 0 | - | 73477327 | 73477426 | 0 | 1 | 99 | 0 | -2.3719261 | 0.0000004 | 0.0000221 |
| chr7 | 73477476 | 73477575 | BAZ1B | 0 | - | 73477476 | 73477575 | 0 | 1 | 99 | 0 | -2.7032102 | 0.0000005 | 0.0000318 |
| chr7 | 73477924 | 73478370 | BAZ1B | 0 | - | 73477924 | 73478370 | 0 | 1 | 446 | 0 | -0.7343220 | 0.0015524 | 0.0219101 |
| chr12 | 56609837 | 56609886 | BAZ2A | 0 | - | 56609837 | 56609886 | 0 | 1 | 49 | 0 | -1.1792537 | 0.0019858 | 0.0263237 |
| chr16 | 75229979 | 75235651 | BCAR1 | 0 | - | 75229979 | 75235651 | 0 | 3 | 45,90,763 | 0,3867,4910 | -0.5358987 | 0.0014447 | 0.0208283 |
| chr16 | 75235702 | 75235751 | BCAR1 | 0 | - | 75235702 | 75235751 | 0 | 1 | 49 | 0 | -1.4859492 | 0.0069576 | 0.0656874 |
| chr1 | 93674882 | 93681195 | BCAR3 | 0 | - | 93674882 | 93681195 | 0 | 2 | 60,89 | 0,6225 | 0.9946768 | 0.0037867 | 0.0423530 |
| chr20 | 31665885 | 31722705 | BCL2L1 | 0 | - | 31665885 | 31722705 | 0 | 3 | 202,694,88 | 0,55770,56733 | -0.8872553 | 0.0000000 | 0.0000001 |
| chr22 | 17706783 | 17727776 | BCL2L13 | 0 | + | 17706783 | 17727776 | 0 | 2 | 43,1100 | 0,19894 | -0.6407924 | 0.0019929 | 0.0263467 |
| chr3 | 187729290 | 187729488 | BCL6 | 0 | - | 187729290 | 187729488 | 0 | 1 | 198 | 0 | -1.6095774 | 0.0000000 | 0.0000001 |
| chr1 | 147624372 | 147625170 | BCL9 | 0 | + | 147624372 | 147625170 | 0 | 1 | 798 | 0 | -0.9787583 | 0.0000395 | 0.0012395 |
| chr11 | 118901013 | 118902660 | BCL9L | 0 | - | 118901013 | 118902660 | 0 | 1 | 1647 | 0 | -1.2790555 | 0.0000000 | 0.0000000 |
| chr6 | 136276136 | 136276285 | BCLAF1 | 0 | - | 136276136 | 136276285 | 0 | 1 | 149 | 0 | -1.4579336 | 0.0023734 | 0.0300538 |
| chrX | 40072613 | 40072812 | BCOR | 0 | - | 40072613 | 40072812 | 0 | 1 | 199 | 0 | -1.5491581 | 0.0007214 | 0.0123073 |
| chrX | 130015379 | 130015878 | BCORL1 | 0 | + | 130015379 | 130015878 | 0 | 1 | 499 | 0 | -1.7186003 | 0.0000917 | 0.0024374 |
| chr22 | 23181810 | 23182059 | BCR | 0 | + | 23181810 | 23182059 | 0 | 1 | 249 | 0 | -1.1930059 | 0.0012167 | 0.0184212 |
| chr6 | 107068610 | 107068759 | BEND3 | 0 | - | 107068610 | 107068759 | 0 | 1 | 149 | 0 | -2.0522814 | 0.0069829 | 0.0658575 |
| chr6 | 107069207 | 107069256 | BEND3 | 0 | - | 107069207 | 107069256 | 0 | 1 | 49 | 0 | -3.4063277 | 0.0078403 | 0.0709503 |
| chr11 | 203023 | 203170 | BET1L | 0 | - | 203023 | 203170 | 0 | 1 | 147 | 0 | -1.3297360 | 0.0054255 | 0.0551938 |
| chr3 | 4982943 | 4983787 | BHLHE40 | 0 | + | 4982943 | 4983787 | 0 | 1 | 844 | 0 | -0.6748723 | 0.0000001 | 0.0000056 |
| chr9 | 92719160 | 92719507 | BICD2 | 0 | - | 92719160 | 92719507 | 0 | 1 | 347 | 0 | -1.9170438 | 0.0000001 | 0.0000056 |
| chr2 | 32414803 | 32415101 | BIRC6 | 0 | + | 32414803 | 32415101 | 0 | 1 | 298 | 0 | -1.4228520 | 0.0028232 | 0.0339013 |
| chr13 | 102862312 | 102862761 | BIVM-ERCC5 | 0 | + | 102862312 | 102862761 | 0 | 1 | 449 | 0 | -0.8491252 | 0.0040888 | 0.0447642 |
| chr20 | 37518811 | 37518960 | BLCAP | 0 | - | 37518811 | 37518960 | 0 | 1 | 149 | 0 | 1.8783529 | 0.0000074 | 0.0003116 |
| chr6 | 7882630 | 7882925 | BLOC1S5-TXNDC5 | 0 | - | 7882630 | 7882925 | 0 | 1 | 295 | 0 | -1.0577011 | 0.0015059 | 0.0213876 |
| chr20 | 6768098 | 6768347 | BMP2 | 0 | + | 6768098 | 6768347 | 0 | 1 | 249 | 0 | -1.1361278 | 0.0000132 | 0.0005089 |
| chr6 | 7726778 | 7727172 | BMP6 | 0 | + | 7726778 | 7727172 | 0 | 1 | 394 | 0 | -0.7513169 | 0.0000097 | 0.0003939 |
| chr6 | 7880250 | 7880595 | BMP6 | 0 | + | 7880250 | 7880595 | 0 | 1 | 345 | 0 | -0.5164488 | 0.0044775 | 0.0478055 |
| chr6 | 7881038 | 7881087 | BMP6 | 0 | + | 7881038 | 7881087 | 0 | 1 | 49 | 0 | -1.7488817 | 0.0000000 | 0.0000040 |
| chr10 | 131973900 | 131981914 | BNIP3 | 0 | - | 131973900 | 131981914 | 0 | 2 | 44,154 | 0,7861 | 1.3001171 | 0.0000007 | 0.0000378 |
| chr5 | 173607839 | 173608235 | BOD1 | 0 | - | 173607839 | 173608235 | 0 | 1 | 396 | 0 | -0.7161935 | 0.0015302 | 0.0216644 |
| chr2 | 241572741 | 241572939 | BOK | 0 | + | 241572741 | 241572939 | 0 | 1 | 198 | 0 | -2.1952801 | 0.0000000 | 0.0000014 |
| chr2 | 241572990 | 241573289 | BOK | 0 | + | 241572990 | 241573289 | 0 | 1 | 299 | 0 | -1.7216406 | 0.0000006 | 0.0000329 |
| chr17 | 67854265 | 67854464 | BPTF | 0 | + | 67854265 | 67854464 | 0 | 1 | 199 | 0 | -2.0527651 | 0.0005554 | 0.0100166 |
| chr12 | 111644039 | 111644288 | BRAP | 0 | - | 111644039 | 111644288 | 0 | 1 | 249 | 0 | -0.7727937 | 0.0082282 | 0.0734056 |
| chr22 | 49823116 | 49823364 | BRD1 | 0 | - | 49823116 | 49823364 | 0 | 1 | 248 | 0 | -1.7955859 | 0.0000246 | 0.0008364 |
| chr22 | 49823662 | 49823959 | BRD1 | 0 | - | 49823662 | 49823959 | 0 | 1 | 297 | 0 | -1.4830247 | 0.0000165 | 0.0006075 |
| chr5 | 138164385 | 138165132 | BRD8 | 0 | - | 138164385 | 138165132 | 0 | 2 | 29,419 | 0,329 | -0.6048867 | 0.0007609 | 0.0128296 |
| chr5 | 864082 | 864428 | BRD9 | 0 | - | 864082 | 864428 | 0 | 1 | 346 | 0 | -0.9708834 | 0.0000010 | 0.0000534 |
| chr7 | 98281667 | 98281861 | BRI3 | 0 | + | 98281667 | 98281861 | 0 | 1 | 194 | 0 | 1.1050101 | 0.0004809 | 0.0089582 |
| chr3 | 9734439 | 9739543 | BRPF1 | 0 | + | 9734439 | 9739543 | 0 | 2 | 301,545 | 0,4560 | -0.9793415 | 0.0000118 | 0.0004588 |
| chr3 | 9747369 | 9747716 | BRPF1 | 0 | + | 9747369 | 9747716 | 0 | 1 | 347 | 0 | -1.3990763 | 0.0001097 | 0.0028128 |
| chr6 | 36201194 | 36201591 | BRPF3 | 0 | + | 36201194 | 36201591 | 0 | 1 | 397 | 0 | -1.9804241 | 0.0000049 | 0.0002125 |
| chr1 | 32376320 | 32376568 | BSDC1 | 0 | - | 32376320 | 32376568 | 0 | 1 | 248 | 0 | -1.1157549 | 0.0000382 | 0.0012134 |
| chr14 | 105249902 | 105250149 | BTBD6 | 0 | + | 105249902 | 105250149 | 0 | 1 | 247 | 0 | -1.0924676 | 0.0018209 | 0.0246429 |
| chr6 | 26468112 | 26468360 | BTN2A1 | 0 | + | 26468112 | 26468360 | 0 | 1 | 248 | 0 | -1.6474502 | 0.0000214 | 0.0007629 |
| chr2 | 110660021 | 110661615 | BUB1 | 0 | - | 110660021 | 110661615 | 0 | 2 | 16,34 | 0,1561 | -2.3438193 | 0.0111132 | 0.0905658 |
| chr10 | 100988526 | 100988575 | C10orf2 | 0 | + | 100988526 | 100988575 | 0 | 1 | 49 | 0 | -2.0906232 | 0.0093978 | 0.0803852 |
| chr10 | 100989071 | 100989170 | C10orf2 | 0 | + | 100989071 | 100989170 | 0 | 1 | 99 | 0 | -2.5143898 | 0.0000001 | 0.0000070 |
| chr12 | 121003310 | 121003556 | C12orf43 | 0 | - | 121003310 | 121003556 | 0 | 1 | 246 | 0 | -0.9509215 | 0.0067398 | 0.0643012 |
| chr12 | 104986320 | 104988211 | C12orf45 | 0 | + | 104986320 | 104988211 | 0 | 2 | 148,45 | 0,1847 | 0.9916386 | 0.0070119 | 0.0659882 |
| chr14 | 75651089 | 75651187 | C14orf1 | 0 | - | 75651089 | 75651187 | 0 | 1 | 98 | 0 | -1.5148136 | 0.0014290 | 0.0206678 |
| chr14 | 73492414 | 73492858 | C14orf169 | 0 | + | 73492414 | 73492858 | 0 | 1 | 444 | 0 | -1.0844548 | 0.0000045 | 0.0001999 |
| chr14 | 73493105 | 73493204 | C14orf169 | 0 | + | 73493105 | 73493204 | 0 | 1 | 99 | 0 | -2.4762048 | 0.0096560 | 0.0820123 |
| chr15 | 89835216 | 89835563 | C15orf38-AP3S2 | 0 | - | 89835216 | 89835563 | 0 | 1 | 347 | 0 | -0.8117778 | 0.0001965 | 0.0043927 |
| chr17 | 21534182 | 21534380 | C17orf51 | 0 | - | 21534182 | 21534380 | 0 | 1 | 198 | 0 | -1.1112207 | 0.0071196 | 0.0665740 |
| chr17 | 73236812 | 73242258 | C17orf80 | 0 | + | 73236812 | 73242258 | 0 | 2 | 184,15 | 0,5432 | -1.6936887 | 0.0002273 | 0.0049285 |
| chr18 | 46215900 | 46216147 | C18orf25 | 0 | + | 46215900 | 46216147 | 0 | 1 | 247 | 0 | -1.4121543 | 0.0029881 | 0.0355038 |
| chr18 | 46216297 | 46216545 | C18orf25 | 0 | + | 46216297 | 46216545 | 0 | 1 | 248 | 0 | -1.4944639 | 0.0000020 | 0.0001005 |
| chr22 | 37180763 | 37181011 | C1QTNF6 | 0 | - | 37180763 | 37181011 | 0 | 1 | 248 | 0 | -0.8737502 | 0.0000576 | 0.0016704 |
| chr20 | 36605782 | 36606024 | C20orf24 | 0 | + | 36605782 | 36606024 | 0 | 1 | 242 | 0 | 0.9527833 | 0.0021323 | 0.0276386 |
| chr20 | 36612027 | 36612316 | C20orf24 | 0 | + | 36612027 | 36612316 | 0 | 1 | 289 | 0 | -0.9328254 | 0.0000009 | 0.0000508 |
| chr2 | 24037929 | 24037978 | C2orf44 | 0 | - | 24037929 | 24037978 | 0 | 1 | 49 | 0 | -2.3773267 | 0.0038378 | 0.0427131 |
| chr5 | 41917062 | 41917409 | C5orf51 | 0 | + | 41917062 | 41917409 | 0 | 1 | 347 | 0 | -0.8868844 | 0.0042278 | 0.0459807 |
| chr6 | 34590315 | 34606866 | C6orf106 | 0 | - | 34590315 | 34606866 | 0 | 2 | 286,312 | 0,16240 | -0.9041437 | 0.0000022 | 0.0001075 |
| chr9 | 83956551 | 83956649 | C9orf64 | 0 | - | 83956551 | 83956649 | 0 | 1 | 98 | 0 | 3.9621403 | 0.0007423 | 0.0126098 |
| chr9 | 136115025 | 136115419 | C9orf69 | 0 | - | 136115025 | 136115419 | 0 | 1 | 394 | 0 | -1.0868320 | 0.0000267 | 0.0008925 |
| chr9 | 129827678 | 129827923 | C9orf78 | 0 | - | 129827678 | 129827923 | 0 | 1 | 245 | 0 | -0.6848788 | 0.0005282 | 0.0096425 |
| chr12 | 48828297 | 48828644 | CACNB3 | 0 | + | 48828297 | 48828644 | 0 | 1 | 347 | 0 | -1.7415515 | 0.0000040 | 0.0001803 |
| chr19 | 3611274 | 3611820 | CACTIN | 0 | - | 3611274 | 3611820 | 0 | 1 | 546 | 0 | -0.7127856 | 0.0057654 | 0.0577647 |
| chr10 | 118754498 | 118754946 | CACUL1 | 0 | - | 118754498 | 118754946 | 0 | 1 | 448 | 0 | 1.0911622 | 0.0000097 | 0.0003939 |
| chr12 | 53711917 | 53713144 | CALCOCO1 | 0 | - | 53711917 | 53713144 | 0 | 2 | 205,45 | 0,1183 | 0.9133752 | 0.0008365 | 0.0138081 |
| chr14 | 90405356 | 90405504 | CALM1 | 0 | + | 90405356 | 90405504 | 0 | 1 | 148 | 0 | -0.8917810 | 0.0055533 | 0.0561185 |
| chr12 | 67305329 | 67306127 | CAND1 | 0 | + | 67305329 | 67306127 | 0 | 1 | 798 | 0 | -0.5985397 | 0.0126707 | 0.0986416 |
| chr17 | 78997115 | 78997412 | CANT1 | 0 | - | 78997115 | 78997412 | 0 | 1 | 297 | 0 | -0.5176794 | 0.0070150 | 0.0659882 |
| chr1 | 40071659 | 40071857 | CAP1 | 0 | + | 40071659 | 40071857 | 0 | 1 | 198 | 0 | -0.8843822 | 0.0101349 | 0.0849640 |
| chr1 | 223712576 | 223712823 | CAPN2 | 0 | + | 223712576 | 223712823 | 0 | 1 | 247 | 0 | 1.0418173 | 0.0045717 | 0.0485822 |
| chr1 | 223775472 | 223775620 | CAPN2 | 0 | + | 223775472 | 223775620 | 0 | 1 | 148 | 0 | 1.2744363 | 0.0002834 | 0.0059262 |
| chr11 | 34051733 | 34052530 | CAPRIN1 | 0 | + | 34051733 | 34052530 | 0 | 2 | 139,110 | 0,688 | 1.1972660 | 0.0011972 | 0.0182018 |
| chr11 | 34099628 | 34099677 | CAPRIN1 | 0 | + | 34099628 | 34099677 | 0 | 1 | 49 | 0 | -2.7814597 | 0.0000365 | 0.0011689 |
| chr11 | 34099727 | 34099776 | CAPRIN1 | 0 | + | 34099727 | 34099776 | 0 | 1 | 49 | 0 | -2.9628347 | 0.0014762 | 0.0211306 |
| chr13 | 110705886 | 110706082 | CARS2 | 0 | - | 110705886 | 110706082 | 0 | 1 | 196 | 0 | 1.2960111 | 0.0000618 | 0.0017704 |
| chr17 | 40163496 | 40166822 | CASC3 | 0 | + | 40163496 | 40166822 | 0 | 2 | 671,26 | 0,3301 | -1.0092656 | 0.0000861 | 0.0023205 |
| chr11 | 104954936 | 104956628 | CASP4 | 0 | - | 104954936 | 104956628 | 0 | 2 | 66,32 | 0,1661 | 1.2154717 | 0.0020079 | 0.0264241 |
| chr5 | 96662335 | 96695857 | CAST | 0 | + | 96662335 | 96695857 | 0 | 3 | 163,63,22 | 0,13204,33501 | 0.8520498 | 0.0050709 | 0.0525615 |
| chr5 | 96695907 | 96729204 | CAST | 0 | + | 96695907 | 96729204 | 0 | 6 | 1,127,60,66,42,52 | 0,6882,26732,30887,31582,33246 | 0.8650331 | 0.0018687 | 0.0251393 |
| chr7 | 116526656 | 116559060 | CAV1 | 0 | + | 116526656 | 116559060 | 0 | 2 | 34,115 | 0,32290 | -1.7853622 | 0.0000028 | 0.0001327 |
| chr7 | 116559505 | 116559702 | CAV1 | 0 | + | 116559505 | 116559702 | 0 | 1 | 197 | 0 | -0.9795389 | 0.0088402 | 0.0771207 |
| chr7 | 116560148 | 116560246 | CAV1 | 0 | + | 116560148 | 116560246 | 0 | 1 | 98 | 0 | -1.6926924 | 0.0011365 | 0.0175189 |
| chr7 | 116560444 | 116560839 | CAV1 | 0 | + | 116560444 | 116560839 | 0 | 1 | 395 | 0 | -1.1866819 | 0.0003862 | 0.0076041 |
| chr7 | 107758747 | 107759192 | CBLL1 | 0 | + | 107758747 | 107759192 | 0 | 1 | 445 | 0 | -0.7655066 | 0.0059319 | 0.0589146 |
| chr17 | 79784154 | 79784453 | CBX2 | 0 | + | 79784154 | 79784453 | 0 | 1 | 299 | 0 | -1.2675967 | 0.0000001 | 0.0000069 |
| chr22 | 38866249 | 38866696 | CBX6 | 0 | - | 38866249 | 38866696 | 0 | 1 | 447 | 0 | 0.7908135 | 0.0127213 | 0.0989504 |
| chr5 | 205054 | 205494 | CCDC127 | 0 | - | 205054 | 205494 | 0 | 1 | 440 | 0 | -0.7201677 | 0.0000035 | 0.0001628 |
| chr5 | 205739 | 205836 | CCDC127 | 0 | - | 205739 | 205836 | 0 | 1 | 97 | 0 | -0.9792171 | 0.0003488 | 0.0070261 |
| chr19 | 13762798 | 13763243 | CCDC130 | 0 | + | 13762798 | 13763243 | 0 | 1 | 445 | 0 | -0.6674826 | 0.0123697 | 0.0968820 |
| chr13 | 36168758 | 36168907 | CCDC169-SOHLH2 | 0 | - | 36168758 | 36168907 | 0 | 1 | 149 | 0 | -1.3538102 | 0.0016689 | 0.0231217 |
| chr17 | 44678730 | 44679027 | CCDC43 | 0 | - | 44678730 | 44679027 | 0 | 1 | 297 | 0 | -0.7458200 | 0.0048770 | 0.0510775 |
| chr3 | 49163024 | 49163122 | CCDC71 | 0 | - | 49163024 | 49163122 | 0 | 1 | 98 | 0 | -1.4251060 | 0.0009898 | 0.0157913 |
| chr3 | 112638452 | 112638501 | CCDC80 | 0 | - | 112638452 | 112638501 | 0 | 1 | 49 | 0 | -2.4037225 | 0.0073767 | 0.0680472 |
| chr11 | 65891389 | 65891586 | CCDC85B | 0 | + | 65891389 | 65891586 | 0 | 1 | 197 | 0 | -0.5548414 | 0.0002652 | 0.0056160 |
| chr19 | 41323842 | 41324239 | CCDC97 | 0 | + | 41323842 | 41324239 | 0 | 1 | 397 | 0 | -1.0406453 | 0.0010771 | 0.0168579 |
| chr11 | 69641155 | 69641503 | CCND1 | 0 | + | 69641155 | 69641503 | 0 | 1 | 348 | 0 | 0.8827720 | 0.0000216 | 0.0007653 |
| chr5 | 163443733 | 163443832 | CCNG1 | 0 | + | 163443733 | 163443832 | 0 | 1 | 99 | 0 | -1.2065812 | 0.0004972 | 0.0091775 |
| chr12 | 48689106 | 48689155 | CCNT1 | 0 | - | 48689106 | 48689155 | 0 | 1 | 49 | 0 | -1.6176179 | 0.0013457 | 0.0198757 |
| chr12 | 48692695 | 48693891 | CCNT1 | 0 | - | 48692695 | 48693891 | 0 | 1 | 1196 | 0 | -0.6048699 | 0.0006883 | 0.0118822 |
| chr15 | 55360553 | 55360652 | CCPG1 | 0 | - | 55360553 | 55360652 | 0 | 1 | 99 | 0 | -0.9313251 | 0.0110460 | 0.0901661 |
| chr2 | 61885064 | 61888671 | CCT4 | 0 | - | 61885064 | 61888671 | 0 | 2 | 9,291 | 0,3317 | 0.7669481 | 0.0112115 | 0.0909985 |
| chr11 | 838386 | 838535 | CD151 | 0 | + | 838386 | 838535 | 0 | 1 | 149 | 0 | -1.2512745 | 0.0092517 | 0.0796622 |
| chr6 | 109382214 | 109382460 | CD164 | 0 | - | 109382214 | 109382460 | 0 | 1 | 246 | 0 | 1.0785150 | 0.0001852 | 0.0042135 |
| chr19 | 8302175 | 8302464 | CD320 | 0 | - | 8302175 | 8302464 | 0 | 1 | 289 | 0 | -0.6286256 | 0.0000757 | 0.0020993 |
| chr11 | 35139169 | 35139368 | CD44 | 0 | + | 35139169 | 35139368 | 0 | 1 | 199 | 0 | 1.3803241 | 0.0000000 | 0.0000000 |
| chr11 | 35229313 | 35229412 | CD44 | 0 | + | 35229313 | 35229412 | 0 | 1 | 99 | 0 | -0.9513434 | 0.0011159 | 0.0172859 |
| chr1 | 207360125 | 207360174 | CD55 | 0 | + | 207360125 | 207360174 | 0 | 1 | 49 | 0 | -1.4358492 | 0.0023412 | 0.0297086 |
| chr11 | 33705703 | 33705752 | CD59 | 0 | - | 33705703 | 33705752 | 0 | 1 | 49 | 0 | -2.2978124 | 0.0106891 | 0.0880214 |
| chr17 | 47120682 | 47122456 | CDC27 | 0 | - | 47120682 | 47122456 | 0 | 2 | 336,13 | 0,1762 | -0.8889875 | 0.0052573 | 0.0539292 |
| chr22 | 37568814 | 37569108 | CDC42EP1 | 0 | + | 37568814 | 37569108 | 0 | 1 | 294 | 0 | -0.8463789 | 0.0000102 | 0.0004072 |
| chr17 | 73285100 | 73285986 | CDC42EP4 | 0 | - | 73285100 | 73285986 | 0 | 1 | 886 | 0 | -0.5541861 | 0.0000799 | 0.0021896 |
| chr17 | 73286331 | 73286429 | CDC42EP4 | 0 | - | 73286331 | 73286429 | 0 | 1 | 98 | 0 | -1.2852970 | 0.0001072 | 0.0027694 |
| chr1 | 151055953 | 151056758 | CDC42SE1 | 0 | - | 151055953 | 151056758 | 0 | 2 | 41,108 | 0,698 | 2.1639090 | 0.0000002 | 0.0000116 |
| chr17 | 39471018 | 39471566 | CDK12 | 0 | + | 39471018 | 39471566 | 0 | 1 | 548 | 0 | -0.9384996 | 0.0010606 | 0.0166421 |
| chr7 | 39987650 | 39988098 | CDK13 | 0 | + | 39987650 | 39988098 | 0 | 1 | 448 | 0 | -1.0745333 | 0.0012445 | 0.0187319 |
| chr1 | 205530668 | 205531503 | CDK18 | 0 | + | 205530668 | 205531503 | 0 | 2 | 38,160 | 0,676 | 1.5035701 | 0.0005069 | 0.0093279 |
| chr9 | 120458597 | 120460595 | CDK5RAP2 | 0 | - | 120458597 | 120460595 | 0 | 2 | 26,24 | 0,1975 | -Inf | 0.0000000 | 0.0000000 |
| chr17 | 47981638 | 47981687 | CDK5RAP3 | 0 | + | 47981638 | 47981687 | 0 | 1 | 49 | 0 | -1.4977873 | 0.0104750 | 0.0870094 |
| chr12 | 12717814 | 12718867 | CDKN1B | 0 | + | 12717814 | 12718867 | 0 | 2 | 501,43 | 0,1011 | -0.9844096 | 0.0000047 | 0.0002073 |
| chr4 | 183446611 | 183446759 | CDKN2AIP | 0 | + | 183446611 | 183446759 | 0 | 1 | 148 | 0 | -1.6553781 | 0.0006404 | 0.0112810 |
| chr17 | 75003360 | 75004105 | CDR2L | 0 | + | 75003360 | 75004105 | 0 | 1 | 745 | 0 | -0.8248054 | 0.0000021 | 0.0001048 |
| chr6 | 4891781 | 4935561 | CDYL | 0 | + | 4891781 | 4935561 | 0 | 2 | 599,47 | 0,43734 | -0.7248100 | 0.0001379 | 0.0033561 |
| chr20 | 50192005 | 50192201 | CEBPB | 0 | + | 50192005 | 50192201 | 0 | 1 | 196 | 0 | -0.5931532 | 0.0000761 | 0.0021081 |
| chr11 | 47466785 | 47466934 | CELF1 | 0 | - | 47466785 | 47466934 | 0 | 1 | 149 | 0 | 1.1726507 | 0.0092875 | 0.0798565 |
| chr11 | 47471627 | 47471925 | CELF1 | 0 | - | 47471627 | 47471925 | 0 | 1 | 298 | 0 | 0.9523519 | 0.0014982 | 0.0213084 |
| chr11 | 47472026 | 47472313 | CELF1 | 0 | - | 47472026 | 47472313 | 0 | 2 | 77,123 | 0,165 | -1.1523825 | 0.0010982 | 0.0171087 |
| chr22 | 46533874 | 46534523 | CELSR1 | 0 | - | 46533874 | 46534523 | 0 | 1 | 649 | 0 | -1.7636180 | 0.0000000 | 0.0000031 |
| chr22 | 46534573 | 46534772 | CELSR1 | 0 | - | 46534573 | 46534772 | 0 | 1 | 199 | 0 | -1.7558995 | 0.0009769 | 0.0156415 |
| chr22 | 46535173 | 46535472 | CELSR1 | 0 | - | 46535173 | 46535472 | 0 | 1 | 299 | 0 | -1.0083886 | 0.0064747 | 0.0627642 |
| chr22 | 46535622 | 46536021 | CELSR1 | 0 | - | 46535622 | 46536021 | 0 | 1 | 399 | 0 | -1.5615246 | 0.0000474 | 0.0014347 |
| chr22 | 46536072 | 46536371 | CELSR1 | 0 | - | 46536072 | 46536371 | 0 | 1 | 299 | 0 | -2.4944764 | 0.0000000 | 0.0000002 |
| chr1 | 109250568 | 109251017 | CELSR2 | 0 | + | 109250568 | 109251017 | 0 | 1 | 449 | 0 | -1.3690295 | 0.0000001 | 0.0000051 |
| chr1 | 109251118 | 109253365 | CELSR2 | 0 | + | 109251118 | 109253365 | 0 | 1 | 2247 | 0 | -1.0738119 | 0.0000000 | 0.0000002 |
| chr20 | 3785545 | 3786192 | CENPB | 0 | - | 3785545 | 3786192 | 0 | 1 | 647 | 0 | -1.1784406 | 0.0000000 | 0.0000000 |
| chr16 | 89970515 | 89970613 | CENPBD1 | 0 | - | 89970515 | 89970613 | 0 | 1 | 98 | 0 | -1.8331493 | 0.0055445 | 0.0560888 |
| chr1 | 214603239 | 214603288 | CENPF | 0 | + | 214603239 | 214603288 | 0 | 1 | 49 | 0 | -1.8490255 | 0.0120208 | 0.0953031 |
| chr1 | 214647094 | 214647293 | CENPF | 0 | + | 214647094 | 214647293 | 0 | 1 | 199 | 0 | -1.1795357 | 0.0116612 | 0.0935165 |
| chr14 | 104886398 | 104887938 | CEP170B | 0 | + | 104886398 | 104887938 | 0 | 1 | 1540 | 0 | -0.9973339 | 0.0000000 | 0.0000001 |
| chr18 | 13049204 | 13049353 | CEP192 | 0 | + | 13049204 | 13049353 | 0 | 1 | 149 | 0 | -2.2350750 | 0.0119291 | 0.0947418 |
| chr20 | 35503006 | 35503105 | CEP250 | 0 | + | 35503006 | 35503105 | 0 | 1 | 99 | 0 | -2.8345449 | 0.0072083 | 0.0669889 |
| chr2 | 65072075 | 65072571 | CEP68 | 0 | + | 65072075 | 65072571 | 0 | 1 | 496 | 0 | -0.9104365 | 0.0000443 | 0.0013593 |
| chr9 | 128420854 | 128423158 | CERCAM | 0 | + | 128420854 | 128423158 | 0 | 3 | 221,111,13 | 0,2014,2292 | 0.8056902 | 0.0078229 | 0.0708333 |
| chr1 | 150966021 | 150966269 | CERS2 | 0 | - | 150966021 | 150966269 | 0 | 1 | 248 | 0 | -0.8867142 | 0.0087499 | 0.0767280 |
| chr16 | 66934741 | 66935976 | CES2 | 0 | + | 66934741 | 66935976 | 0 | 1 | 1235 | 0 | -1.4765124 | 0.0000000 | 0.0000000 |
| chr16 | 66973859 | 66974007 | CES3 | 0 | + | 66973859 | 66974007 | 0 | 1 | 148 | 0 | -2.0623521 | 0.0014541 | 0.0208977 |
| chr14 | 34713402 | 34713451 | CFL2 | 0 | - | 34713402 | 34713451 | 0 | 1 | 49 | 0 | -2.0581576 | 0.0112050 | 0.0909868 |
| chr3 | 88055664 | 88055911 | CGGBP1 | 0 | - | 88055664 | 88055911 | 0 | 1 | 247 | 0 | -0.8911133 | 0.0059344 | 0.0589146 |
| chr1 | 151519069 | 151519217 | CGN | 0 | + | 151519069 | 151519217 | 0 | 1 | 148 | 0 | -1.9069257 | 0.0039642 | 0.0437703 |
| chr14 | 54538077 | 54538371 | CGRRF1 | 0 | + | 54538077 | 54538371 | 0 | 1 | 294 | 0 | -1.1823988 | 0.0040420 | 0.0444670 |
| chr19 | 4408949 | 4409697 | CHAF1A | 0 | + | 4408949 | 4409697 | 0 | 1 | 748 | 0 | -0.8306926 | 0.0000035 | 0.0001640 |
| chr19 | 4433162 | 4433211 | CHAF1A | 0 | + | 4433162 | 4433211 | 0 | 1 | 49 | 0 | 2.2544955 | 0.0015233 | 0.0215945 |
| chr13 | 114325593 | 114325791 | CHAMP1 | 0 | + | 114325593 | 114325791 | 0 | 1 | 198 | 0 | -1.5030047 | 0.0093293 | 0.0799884 |
| chr7 | 133081920 | 133082068 | CHCHD3 | 0 | - | 133081920 | 133082068 | 0 | 1 | 148 | 0 | 1.4143710 | 0.0009810 | 0.0156793 |
| chr2 | 86529354 | 86563447 | CHMP3 | 0 | - | 86529354 | 86563447 | 0 | 3 | 44,61,144 | 0,12898,33950 | 0.8764836 | 0.0080448 | 0.0722953 |
| chr14 | 24210743 | 24211535 | CHMP4A | 0 | - | 24210743 | 24211535 | 0 | 2 | 26,121 | 0,672 | -1.6826464 | 0.0017462 | 0.0238456 |
| chr20 | 33811304 | 33811603 | CHMP4B | 0 | + | 33811304 | 33811603 | 0 | 1 | 299 | 0 | 0.8156585 | 0.0030900 | 0.0363338 |
| chr20 | 33853769 | 33853818 | CHMP4B | 0 | + | 33853769 | 33853818 | 0 | 1 | 49 | 0 | -2.1050439 | 0.0036457 | 0.0411058 |
| chr2 | 219538997 | 219539095 | CHPF | 0 | - | 219538997 | 219539095 | 0 | 1 | 98 | 0 | -1.3294118 | 0.0009860 | 0.0157448 |
| chr2 | 219539691 | 219539988 | CHPF | 0 | - | 219539691 | 219539988 | 0 | 1 | 297 | 0 | -1.1521410 | 0.0000340 | 0.0011041 |
| chr7 | 151235206 | 151235604 | CHPF2 | 0 | + | 151235206 | 151235604 | 0 | 1 | 398 | 0 | -0.9139047 | 0.0003280 | 0.0066704 |
| chr8 | 140515156 | 140515404 | CHRAC1 | 0 | + | 140515156 | 140515404 | 0 | 1 | 248 | 0 | -1.0062532 | 0.0004768 | 0.0089010 |
| chr4 | 40354129 | 40354475 | CHRNA9 | 0 | + | 40354129 | 40354475 | 0 | 1 | 346 | 0 | -0.8965705 | 0.0035550 | 0.0405124 |
| chr12 | 104757391 | 104757738 | CHST11 | 0 | + | 104757391 | 104757738 | 0 | 1 | 347 | 0 | -1.2181229 | 0.0001775 | 0.0040686 |
| chr10 | 72007887 | 72008285 | CHST3 | 0 | + | 72007887 | 72008285 | 0 | 1 | 398 | 0 | -1.8515184 | 0.0000002 | 0.0000135 |
| chr10 | 72009182 | 72009430 | CHST3 | 0 | + | 72009182 | 72009430 | 0 | 1 | 248 | 0 | -0.9330887 | 0.0076454 | 0.0698504 |
| chr2 | 96266295 | 96266344 | CIAO1 | 0 | + | 96266295 | 96266344 | 0 | 1 | 49 | 0 | 1.7400578 | 0.0027036 | 0.0329898 |
| chr2 | 96272917 | 96273066 | CIAO1 | 0 | + | 96272917 | 96273066 | 0 | 1 | 149 | 0 | -1.7427636 | 0.0119137 | 0.0947031 |
| chr19 | 42272015 | 42272064 | CIC | 0 | + | 42272015 | 42272064 | 0 | 1 | 49 | 0 | -4.7876298 | 0.0000924 | 0.0024380 |
| chr19 | 42272215 | 42272264 | CIC | 0 | + | 42272215 | 42272264 | 0 | 1 | 49 | 0 | -2.2747855 | 0.0126265 | 0.0984247 |
| chr19 | 42272315 | 42272364 | CIC | 0 | + | 42272315 | 42272364 | 0 | 1 | 49 | 0 | -4.5901926 | 0.0002864 | 0.0059565 |
| chr19 | 42273363 | 42273711 | CIC | 0 | + | 42273363 | 42273711 | 0 | 1 | 348 | 0 | -2.0791346 | 0.0000011 | 0.0000573 |
| chr19 | 42274211 | 42274260 | CIC | 0 | + | 42274211 | 42274260 | 0 | 1 | 49 | 0 | -Inf | 0.0102726 | 0.0858039 |
| chr19 | 42290758 | 42291056 | CIC | 0 | + | 42290758 | 42291056 | 0 | 1 | 298 | 0 | -1.2965338 | 0.0028998 | 0.0345912 |
| chr19 | 42295048 | 42295297 | CIC | 0 | + | 42295048 | 42295297 | 0 | 1 | 249 | 0 | -1.1760014 | 0.0074112 | 0.0683308 |
| chr6 | 139373682 | 139373830 | CITED2 | 0 | - | 139373682 | 139373830 | 0 | 1 | 148 | 0 | -0.5556149 | 0.0019507 | 0.0259732 |
| chr13 | 52461363 | 52461860 | CKAP2 | 0 | + | 52461363 | 52461860 | 0 | 1 | 497 | 0 | -1.1702693 | 0.0000227 | 0.0007861 |
| chr12 | 106238976 | 106240269 | CKAP4 | 0 | - | 106238976 | 106240269 | 0 | 1 | 1293 | 0 | -0.6179122 | 0.0000033 | 0.0001545 |
| chr9 | 89311195 | 89315251 | CKS2 | 0 | + | 89311195 | 89315251 | 0 | 2 | 157,82 | 0,3975 | 0.7567018 | 0.0004161 | 0.0080662 |
| chr1 | 24745418 | 24745617 | CLIC4 | 0 | + | 24745418 | 24745617 | 0 | 1 | 199 | 0 | 0.7012671 | 0.0025286 | 0.0315126 |
| chr5 | 157787389 | 157787438 | CLINT1 | 0 | - | 157787389 | 157787438 | 0 | 1 | 49 | 0 | -2.2113062 | 0.0065553 | 0.0632448 |
| chr12 | 122340974 | 122341669 | CLIP1 | 0 | - | 122340974 | 122341669 | 0 | 1 | 695 | 0 | -0.6891431 | 0.0002539 | 0.0054474 |
| chr12 | 122377654 | 122377952 | CLIP1 | 0 | - | 122377654 | 122377952 | 0 | 1 | 298 | 0 | -1.8121597 | 0.0000006 | 0.0000356 |
| chr5 | 176416226 | 176416569 | CLTB | 0 | - | 176416226 | 176416569 | 0 | 1 | 343 | 0 | 0.7327736 | 0.0106470 | 0.0877944 |
| chr16 | 81445219 | 81445516 | CMIP | 0 | + | 81445219 | 81445516 | 0 | 1 | 297 | 0 | 1.1936393 | 0.0012473 | 0.0187594 |
| chr3 | 99817884 | 99818033 | CMSS1 | 0 | + | 99817884 | 99818033 | 0 | 1 | 149 | 0 | 0.9223190 | 0.0014699 | 0.0210913 |
| chr2 | 96761952 | 96762301 | CNNM4 | 0 | + | 96761952 | 96762301 | 0 | 1 | 349 | 0 | -1.1559418 | 0.0016937 | 0.0233831 |
| chr3 | 32685244 | 32695550 | CNOT10 | 0 | + | 32685244 | 32695550 | 0 | 2 | 239,10 | 0,10297 | 2.0311186 | 0.0000005 | 0.0000301 |
| chr1 | 246591705 | 246591903 | CNST | 0 | + | 246591705 | 246591903 | 0 | 1 | 198 | 0 | -1.4025343 | 0.0085536 | 0.0755184 |
| chr7 | 51016711 | 51016860 | COBL | 0 | - | 51016711 | 51016860 | 0 | 1 | 149 | 0 | -1.5428508 | 0.0043641 | 0.0471791 |
| chr7 | 51026573 | 51027967 | COBL | 0 | - | 51026573 | 51027967 | 0 | 2 | 93,256 | 0,1139 | -1.0702034 | 0.0003532 | 0.0071070 |
| chr7 | 51028018 | 51028117 | COBL | 0 | - | 51028018 | 51028117 | 0 | 1 | 99 | 0 | -1.5177920 | 0.0015509 | 0.0219060 |
| chr7 | 51028268 | 51028366 | COBL | 0 | - | 51028268 | 51028366 | 0 | 1 | 98 | 0 | -2.0359883 | 0.0005323 | 0.0096576 |
| chr7 | 51028867 | 51029265 | COBL | 0 | - | 51028867 | 51029265 | 0 | 1 | 398 | 0 | -1.1054997 | 0.0000000 | 0.0000027 |
| chr16 | 69334658 | 69335256 | COG8 | 0 | - | 69334658 | 69335256 | 0 | 1 | 598 | 0 | -0.6521463 | 0.0003771 | 0.0074787 |
| chr17 | 56949981 | 56950477 | COIL | 0 | - | 56949981 | 56950477 | 0 | 1 | 496 | 0 | -1.0343971 | 0.0006237 | 0.0110617 |
| chr13 | 110149206 | 110149305 | COL4A1 | 0 | - | 110149206 | 110149305 | 0 | 1 | 99 | 0 | -1.8463545 | 0.0010019 | 0.0158865 |
| chr19 | 17582508 | 17582707 | COLGALT1 | 0 | + | 17582508 | 17582707 | 0 | 1 | 199 | 0 | -1.2945844 | 0.0068166 | 0.0646932 |
| chr19 | 18907099 | 18919390 | COPE | 0 | - | 18907099 | 18919390 | 0 | 4 | 14,101,63,168 | 0,3872,5885,12124 | 0.7482197 | 0.0116436 | 0.0934168 |
| chr4 | 83284567 | 83284665 | COQ2 | 0 | - | 83284567 | 83284665 | 0 | 1 | 98 | 0 | 1.6159668 | 0.0009935 | 0.0158370 |
| chr11 | 67440216 | 67441406 | CORO1B | 0 | - | 67440216 | 67441406 | 0 | 4 | 48,105,120,74 | 0,119,909,1117 | -1.5728461 | 0.0000001 | 0.0000091 |
| chr12 | 108647096 | 108647492 | CORO1C | 0 | - | 108647096 | 108647492 | 0 | 1 | 396 | 0 | -0.6417140 | 0.0072548 | 0.0672460 |
| chr16 | 84617549 | 84618096 | COTL1 | 0 | - | 84617549 | 84618096 | 0 | 2 | 35,259 | 0,289 | 0.9573147 | 0.0057165 | 0.0574126 |
| chr17 | 14076846 | 14076993 | COX10 | 0 | + | 14076846 | 14076993 | 0 | 1 | 147 | 0 | -1.4911271 | 0.0067945 | 0.0645605 |
| chr2 | 42350844 | 42350989 | COX7A2L | 0 | - | 42350844 | 42350989 | 0 | 1 | 145 | 0 | -1.0728747 | 0.0009970 | 0.0158596 |
| chr12 | 68855510 | 68855559 | CPM | 0 | - | 68855510 | 68855559 | 0 | 1 | 49 | 0 | -2.1301784 | 0.0017889 | 0.0243193 |
| chr12 | 68855610 | 68856506 | CPM | 0 | - | 68855610 | 68856506 | 0 | 1 | 896 | 0 | -0.5831160 | 0.0018892 | 0.0253619 |
| chr12 | 38874496 | 38905618 | CPNE8 | 0 | - | 38874496 | 38905618 | 0 | 2 | 16,182 | 0,30941 | 1.0961710 | 0.0039391 | 0.0436189 |
| chr1 | 53210907 | 53211005 | CPT2 | 0 | + | 53210907 | 53211005 | 0 | 1 | 98 | 0 | -1.6580375 | 0.0008579 | 0.0140584 |
| chr1 | 53213898 | 53213947 | CPT2 | 0 | + | 53213898 | 53213947 | 0 | 1 | 49 | 0 | -2.9408755 | 0.0010200 | 0.0161307 |
| chr1 | 1327701 | 1327899 | CPTP | 0 | + | 1327701 | 1327899 | 0 | 1 | 198 | 0 | -1.2212012 | 0.0098303 | 0.0832197 |
| chr16 | 3728254 | 3728603 | CREBBP | 0 | - | 3728254 | 3728603 | 0 | 1 | 349 | 0 | -1.0816296 | 0.0022584 | 0.0289213 |
| chr16 | 3728704 | 3728853 | CREBBP | 0 | - | 3728704 | 3728853 | 0 | 1 | 149 | 0 | -2.3381584 | 0.0001313 | 0.0032168 |
| chr2 | 36356227 | 36356524 | CRIM1 | 0 | + | 36356227 | 36356524 | 0 | 1 | 297 | 0 | 1.0334409 | 0.0057675 | 0.0577647 |
| chr22 | 20918023 | 20934003 | CRKL | 0 | + | 20918023 | 20934003 | 0 | 2 | 223,225 | 0,15756 | -1.0689039 | 0.0000496 | 0.0014884 |
| chr15 | 90641196 | 90643166 | CRTC3 | 0 | + | 90641196 | 90643166 | 0 | 2 | 4,1235 | 0,736 | -0.9254357 | 0.0000000 | 0.0000000 |
| chr15 | 90644454 | 90644602 | CRTC3 | 0 | + | 90644454 | 90644602 | 0 | 1 | 148 | 0 | -2.9815074 | 0.0000000 | 0.0000011 |
| chr12 | 56274846 | 56275117 | CS | 0 | - | 56274846 | 56275117 | 0 | 2 | 33,116 | 0,156 | 3.9920560 | 0.0000000 | 0.0000000 |
| chr12 | 56282553 | 56282902 | CS | 0 | - | 56282553 | 56282902 | 0 | 2 | 56,43 | 0,307 | 3.9550149 | 0.0000000 | 0.0000000 |
| chr12 | 56282953 | 56283802 | CS | 0 | - | 56282953 | 56283802 | 0 | 2 | 39,11 | 0,839 | 2.1500883 | 0.0038858 | 0.0431415 |
| chr12 | 56283852 | 56286008 | CS | 0 | - | 56283852 | 56286008 | 0 | 2 | 6,93 | 0,2064 | 3.0500682 | 0.0000000 | 0.0000019 |
| chr1 | 114739708 | 114741542 | CSDE1 | 0 | - | 114739708 | 114741542 | 0 | 2 | 183,16 | 0,1819 | -1.3839923 | 0.0017460 | 0.0238456 |
| chr19 | 1941310 | 1978334 | CSNK1G2 | 0 | + | 1941310 | 1978334 | 0 | 3 | 109,452,30 | 0,28198,36995 | -0.6642486 | 0.0011506 | 0.0176416 |
| chr15 | 75675171 | 75675520 | CSPG4 | 0 | - | 75675171 | 75675520 | 0 | 1 | 349 | 0 | -0.8778874 | 0.0029668 | 0.0352974 |
| chr15 | 75689710 | 75690008 | CSPG4 | 0 | - | 75689710 | 75690008 | 0 | 1 | 298 | 0 | -1.8113121 | 0.0000038 | 0.0001761 |
| chr3 | 39143495 | 39143892 | CSRNP1 | 0 | - | 39143495 | 39143892 | 0 | 1 | 397 | 0 | -1.0252510 | 0.0003152 | 0.0064563 |
| chr21 | 43774110 | 43774258 | CSTB | 0 | - | 43774110 | 43774258 | 0 | 1 | 148 | 0 | -1.3091785 | 0.0000552 | 0.0016157 |
| chr10 | 51698011 | 51698902 | CSTF2T | 0 | - | 51698011 | 51698902 | 0 | 1 | 891 | 0 | -0.8794092 | 0.0000005 | 0.0000300 |
| chr4 | 1241476 | 1249021 | CTBP1 | 0 | - | 1241476 | 1249021 | 0 | 2 | 44,106 | 0,7440 | 2.4450692 | 0.0000001 | 0.0000083 |
| chr16 | 67610888 | 67611486 | CTCF | 0 | + | 67610888 | 67611486 | 0 | 1 | 598 | 0 | -0.7491454 | 0.0037462 | 0.0420035 |
| chr12 | 57822660 | 57823703 | CTDSP2 | 0 | - | 57822660 | 57823703 | 0 | 1 | 1043 | 0 | -0.6590810 | 0.0000749 | 0.0020871 |
| chr18 | 48758070 | 48761395 | CTIF | 0 | + | 48758070 | 48761395 | 0 | 2 | 342,6 | 0,3320 | -1.0219470 | 0.0072075 | 0.0669889 |
| chr1 | 40979695 | 40983340 | CTPS1 | 0 | + | 40979695 | 40983340 | 0 | 2 | 135,63 | 0,3583 | 0.9711030 | 0.0098847 | 0.0835198 |
| chr8 | 11844170 | 11844269 | CTSB | 0 | - | 11844170 | 11844269 | 0 | 1 | 99 | 0 | -0.6002540 | 0.0047912 | 0.0504702 |
| chr11 | 1752999 | 1753048 | CTSD | 0 | - | 1752999 | 1753048 | 0 | 1 | 49 | 0 | -1.5388800 | 0.0000002 | 0.0000152 |
| chr9 | 87726108 | 87728138 | CTSL | 0 | + | 87726108 | 87728138 | 0 | 3 | 291,136,112 | 0,1486,1919 | 0.7182987 | 0.0000039 | 0.0001776 |
| chr20 | 58996666 | 58996765 | CTSZ | 0 | - | 58996666 | 58996765 | 0 | 1 | 99 | 0 | -0.8555667 | 0.0024019 | 0.0303297 |
| chr11 | 70435441 | 70435888 | CTTN | 0 | + | 70435441 | 70435888 | 0 | 1 | 447 | 0 | 1.5651436 | 0.0000003 | 0.0000177 |
| chr1 | 112456115 | 112456314 | CTTNBP2NL | 0 | + | 112456115 | 112456314 | 0 | 1 | 199 | 0 | -1.6310273 | 0.0001259 | 0.0031155 |
| chr1 | 112456365 | 112456614 | CTTNBP2NL | 0 | + | 112456365 | 112456614 | 0 | 1 | 249 | 0 | -2.0450772 | 0.0000046 | 0.0002036 |
| chr1 | 112457965 | 112458114 | CTTNBP2NL | 0 | + | 112457965 | 112458114 | 0 | 1 | 149 | 0 | -2.2114091 | 0.0077645 | 0.0705936 |
| chr17 | 57862137 | 57862634 | CUEDC1 | 0 | - | 57862137 | 57862634 | 0 | 1 | 497 | 0 | -0.8609800 | 0.0071394 | 0.0666773 |
| chr13 | 113209633 | 113210029 | CUL4A | 0 | + | 113209633 | 113210029 | 0 | 2 | 143,57 | 0,340 | 0.9378000 | 0.0041866 | 0.0455872 |
| chr13 | 113260704 | 113263780 | CUL4A | 0 | + | 113260704 | 113263780 | 0 | 2 | 56,294 | 0,2783 | -0.6271602 | 0.0002499 | 0.0053688 |
| chr2 | 136115522 | 136115767 | CXCR4 | 0 | - | 136115522 | 136115767 | 0 | 1 | 245 | 0 | -0.8195768 | 0.0000048 | 0.0002088 |
| chr22 | 42619128 | 42619523 | CYB5R3 | 0 | - | 42619128 | 42619523 | 0 | 1 | 395 | 0 | -0.7961391 | 0.0092152 | 0.0794994 |
| chr2 | 38070774 | 38074750 | CYP1B1 | 0 | - | 38070774 | 38074750 | 0 | 2 | 537,405 | 0,3572 | -0.5990999 | 0.0002163 | 0.0047534 |
| chr5 | 140175259 | 140194564 | CYSTM1 | 0 | + | 140175259 | 140194564 | 0 | 2 | 27,119 | 0,19187 | 1.1439387 | 0.0000475 | 0.0014347 |
| chr19 | 48480779 | 48481422 | CYTH2 | 0 | + | 48480779 | 48481422 | 0 | 1 | 643 | 0 | 0.6573759 | 0.0024474 | 0.0307546 |
| chr5 | 39381559 | 39383005 | DAB2 | 0 | - | 39381559 | 39383005 | 0 | 2 | 58,388 | 0,1059 | -0.5628749 | 0.0026545 | 0.0326395 |
| chr9 | 121760177 | 121760276 | DAB2IP | 0 | + | 121760177 | 121760276 | 0 | 1 | 99 | 0 | -1.6353265 | 0.0024008 | 0.0303297 |
| chr9 | 121784982 | 121785031 | DAB2IP | 0 | + | 121784982 | 121785031 | 0 | 1 | 49 | 0 | -1.7103785 | 0.0077112 | 0.0702034 |
| chr3 | 49531024 | 49531373 | DAG1 | 0 | + | 49531024 | 49531373 | 0 | 1 | 349 | 0 | -1.0849091 | 0.0000000 | 0.0000001 |
| chr3 | 49531973 | 49532222 | DAG1 | 0 | + | 49531973 | 49532222 | 0 | 1 | 249 | 0 | -2.9298240 | 0.0000000 | 0.0000000 |
| chr3 | 49532273 | 49532721 | DAG1 | 0 | + | 49532273 | 49532721 | 0 | 1 | 448 | 0 | -2.0935773 | 0.0000000 | 0.0000000 |
| chr3 | 49535319 | 49535418 | DAG1 | 0 | + | 49535319 | 49535418 | 0 | 1 | 99 | 0 | -1.3290023 | 0.0001631 | 0.0038005 |
| chr7 | 6409165 | 6409214 | DAGLB | 0 | - | 6409165 | 6409214 | 0 | 1 | 49 | 0 | -3.3725898 | 0.0003620 | 0.0072439 |
| chr5 | 10679579 | 10679628 | DAP | 0 | - | 10679579 | 10679628 | 0 | 1 | 49 | 0 | -1.5925907 | 0.0007125 | 0.0121830 |
| chr1 | 155689091 | 155689239 | DAP3 | 0 | + | 155689091 | 155689239 | 0 | 1 | 148 | 0 | 1.5918903 | 0.0000000 | 0.0000036 |
| chr1 | 173824709 | 173825258 | DARS2 | 0 | + | 173824709 | 173825258 | 0 | 1 | 549 | 0 | -1.4221967 | 0.0000000 | 0.0000000 |
| chr14 | 69053910 | 69054607 | DCAF5 | 0 | - | 69053910 | 69054607 | 0 | 1 | 697 | 0 | -0.7242427 | 0.0004621 | 0.0087620 |
| chr1 | 113911445 | 113911893 | DCLRE1B | 0 | + | 113911445 | 113911893 | 0 | 1 | 448 | 0 | -0.6315203 | 0.0063349 | 0.0617678 |
| chr1 | 113912093 | 113912342 | DCLRE1B | 0 | + | 113912093 | 113912342 | 0 | 1 | 249 | 0 | -1.0698474 | 0.0006265 | 0.0111009 |
| chr16 | 23669027 | 23669275 | DCTN5 | 0 | + | 23669027 | 23669275 | 0 | 1 | 248 | 0 | -0.9759172 | 0.0062575 | 0.0612823 |
| chr1 | 85464757 | 85465154 | DDAH1 | 0 | - | 85464757 | 85465154 | 0 | 1 | 397 | 0 | 1.2047511 | 0.0001119 | 0.0028550 |
| chr11 | 108723100 | 108723346 | DDX10 | 0 | + | 108723100 | 108723346 | 0 | 1 | 246 | 0 | -1.0775305 | 0.0118882 | 0.0945699 |
| chr22 | 38485886 | 38492106 | DDX17 | 0 | - | 38485886 | 38492106 | 0 | 3 | 555,243,51 | 0,1987,6170 | 0.7559572 | 0.0018482 | 0.0249035 |
| chr1 | 111765847 | 111767182 | DDX20 | 0 | + | 111765847 | 111767182 | 0 | 1 | 1335 | 0 | -0.5394621 | 0.0000236 | 0.0008109 |
| chr10 | 68969065 | 68970242 | DDX21 | 0 | + | 68969065 | 68970242 | 0 | 2 | 57,42 | 0,1136 | -2.0920251 | 0.0010666 | 0.0167073 |
| chr12 | 48836658 | 48836757 | DDX23 | 0 | - | 48836658 | 48836757 | 0 | 1 | 99 | 0 | -1.4981730 | 0.0018420 | 0.0248550 |
| chr16 | 68021723 | 68022221 | DDX28 | 0 | - | 68021723 | 68022221 | 0 | 1 | 498 | 0 | -0.9155818 | 0.0000077 | 0.0003228 |
| chrX | 41347412 | 41347999 | DDX3X | 0 | + | 41347412 | 41347999 | 0 | 2 | 40,360 | 0,228 | -0.8360109 | 0.0000688 | 0.0019376 |
| chr12 | 113185330 | 113185479 | DDX54 | 0 | - | 113185330 | 113185479 | 0 | 1 | 149 | 0 | 1.0965422 | 0.0044463 | 0.0476471 |
| chr19 | 42198692 | 42198940 | DEDD2 | 0 | - | 42198692 | 42198940 | 0 | 1 | 248 | 0 | -1.1403991 | 0.0000340 | 0.0011041 |
| chr1 | 224192409 | 224192654 | DEGS1 | 0 | + | 224192409 | 224192654 | 0 | 1 | 245 | 0 | -0.6625878 | 0.0002185 | 0.0047892 |
| chr11 | 9203724 | 9204269 | DENND5A | 0 | - | 9203724 | 9204269 | 0 | 1 | 545 | 0 | -1.0058208 | 0.0009190 | 0.0148720 |
| chr22 | 19122255 | 19122354 | DGCR2 | 0 | - | 19122255 | 19122354 | 0 | 1 | 99 | 0 | 1.7411680 | 0.0021750 | 0.0280791 |
| chr11 | 71434658 | 71434953 | DHCR7 | 0 | - | 71434658 | 71434953 | 0 | 1 | 295 | 0 | -1.2728401 | 0.0055067 | 0.0557689 |
| chr5 | 55281513 | 55283746 | DHX29 | 0 | - | 55281513 | 55283746 | 0 | 2 | 3,544 | 0,1690 | -0.8329389 | 0.0063826 | 0.0620040 |
| chr2 | 38861563 | 38861712 | DHX57 | 0 | - | 38861563 | 38861712 | 0 | 1 | 149 | 0 | -2.1940706 | 0.0061810 | 0.0607058 |
| chr17 | 43484016 | 43484065 | DHX8 | 0 | + | 43484016 | 43484065 | 0 | 1 | 49 | 0 | 1.8134969 | 0.0081341 | 0.0728453 |
| chr20 | 62879186 | 62879933 | DIDO1 | 0 | - | 62879186 | 62879933 | 0 | 1 | 747 | 0 | -0.7512068 | 0.0000668 | 0.0018947 |
| chr20 | 62880084 | 62880183 | DIDO1 | 0 | - | 62880084 | 62880183 | 0 | 1 | 99 | 0 | -2.0919160 | 0.0040986 | 0.0448442 |
| chr20 | 62880384 | 62880532 | DIDO1 | 0 | - | 62880384 | 62880532 | 0 | 1 | 148 | 0 | -3.2071076 | 0.0000016 | 0.0000830 |
| chr19 | 14518262 | 14518460 | DNAJB1 | 0 | - | 14518262 | 14518460 | 0 | 1 | 198 | 0 | 0.8512802 | 0.0066650 | 0.0639926 |
| chr3 | 186570676 | 186570924 | DNAJB11 | 0 | + | 186570676 | 186570924 | 0 | 1 | 248 | 0 | 1.2226234 | 0.0001403 | 0.0034020 |
| chr2 | 219284867 | 219285015 | DNAJB2 | 0 | + | 219284867 | 219285015 | 0 | 1 | 148 | 0 | -2.0642138 | 0.0070702 | 0.0663830 |
| chr9 | 111631401 | 111631696 | DNAJC25 | 0 | + | 111631401 | 111631696 | 0 | 1 | 295 | 0 | 1.2437187 | 0.0007936 | 0.0132930 |
| chr9 | 111631402 | 111666822 | DNAJC25-GNG10 | 0 | + | 111631402 | 111666822 | 0 | 2 | 342,8 | 0,35413 | 1.2568052 | 0.0002248 | 0.0048932 |
| chr13 | 95791496 | 95791694 | DNAJC3 | 0 | + | 95791496 | 95791694 | 0 | 1 | 198 | 0 | -1.3303917 | 0.0053777 | 0.0548615 |
| chr1 | 28205314 | 28208355 | DNAJC8 | 0 | - | 28205314 | 28208355 | 0 | 2 | 36,14 | 0,3028 | -2.2511563 | 0.0022744 | 0.0290445 |
| chr19 | 12875407 | 12875554 | DNASE2 | 0 | - | 12875407 | 12875554 | 0 | 1 | 147 | 0 | -1.2363783 | 0.0022979 | 0.0292616 |
| chr19 | 12875652 | 12875945 | DNASE2 | 0 | - | 12875652 | 12875945 | 0 | 1 | 293 | 0 | -0.5490406 | 0.0074406 | 0.0684970 |
| chr19 | 12876190 | 12876239 | DNASE2 | 0 | - | 12876190 | 12876239 | 0 | 1 | 49 | 0 | -1.2727282 | 0.0092477 | 0.0796622 |
| chr10 | 99955316 | 99955565 | DNMBP | 0 | - | 99955316 | 99955565 | 0 | 1 | 249 | 0 | -2.1813783 | 0.0000414 | 0.0012867 |
| chr10 | 99956116 | 99956215 | DNMBP | 0 | - | 99956116 | 99956215 | 0 | 1 | 99 | 0 | -1.8516673 | 0.0124750 | 0.0975798 |
| chr10 | 99956266 | 99956565 | DNMBP | 0 | - | 99956266 | 99956565 | 0 | 1 | 299 | 0 | -1.8307877 | 0.0000208 | 0.0007436 |
| chr1 | 93876508 | 93876557 | DNTTIP2 | 0 | - | 93876508 | 93876557 | 0 | 1 | 49 | 0 | -2.1195484 | 0.0054324 | 0.0552329 |
| chr1 | 93876656 | 93877398 | DNTTIP2 | 0 | - | 93876656 | 93877398 | 0 | 1 | 742 | 0 | -0.6329973 | 0.0015630 | 0.0220085 |
| chr8 | 25184772 | 25184920 | DOCK5 | 0 | + | 25184772 | 25184920 | 0 | 1 | 148 | 0 | 2.2592105 | 0.0000005 | 0.0000301 |
| chr17 | 2042983 | 2043381 | DPH1 | 0 | + | 2042983 | 2043381 | 0 | 1 | 398 | 0 | -0.5050042 | 0.0009976 | 0.0158596 |
| chr15 | 65512479 | 65515769 | DPP8 | 0 | - | 65512479 | 65515769 | 0 | 2 | 86,114 | 0,3177 | 1.0815234 | 0.0020055 | 0.0264241 |
| chr15 | 65517548 | 65517647 | DPP8 | 0 | - | 65517548 | 65517647 | 0 | 1 | 99 | 0 | 1.7213979 | 0.0008717 | 0.0142330 |
| chr1 | 93345921 | 93346809 | DR1 | 0 | + | 93345921 | 93346809 | 0 | 1 | 888 | 0 | -0.7677203 | 0.0003977 | 0.0077846 |
| chr1 | 111118126 | 111118175 | DRAM2 | 0 | - | 111118126 | 111118175 | 0 | 1 | 49 | 0 | -1.5522638 | 0.0045257 | 0.0481857 |
| chr11 | 65919400 | 65919971 | DRAP1 | 0 | + | 65919400 | 65919971 | 0 | 3 | 144,73,24 | 0,380,548 | -0.6662892 | 0.0065410 | 0.0631949 |
| chr18 | 31545728 | 31545975 | DSG2 | 0 | + | 31545728 | 31545975 | 0 | 1 | 247 | 0 | -1.3361354 | 0.0007664 | 0.0129096 |
| chr20 | 36771134 | 36773728 | DSN1 | 0 | - | 36771134 | 36773728 | 0 | 3 | 60,49,335 | 0,291,2260 | 1.4446076 | 0.0001211 | 0.0030588 |
| chr6 | 7579405 | 7579603 | DSP | 0 | + | 7579405 | 7579603 | 0 | 1 | 198 | 0 | -0.9984773 | 0.0105206 | 0.0873333 |
| chr6 | 7581251 | 7581500 | DSP | 0 | + | 7581251 | 7581500 | 0 | 1 | 249 | 0 | -0.9264604 | 0.0006357 | 0.0112198 |
| chr6 | 7582872 | 7583220 | DSP | 0 | + | 7582872 | 7583220 | 0 | 1 | 348 | 0 | -0.9500689 | 0.0002182 | 0.0047892 |
| chr6 | 7583371 | 7584168 | DSP | 0 | + | 7583371 | 7584168 | 0 | 1 | 797 | 0 | -0.9088827 | 0.0000216 | 0.0007653 |
| chr6 | 7584319 | 7584468 | DSP | 0 | + | 7584319 | 7584468 | 0 | 1 | 149 | 0 | -1.8995413 | 0.0000114 | 0.0004477 |
| chr1 | 205169307 | 205187737 | DSTYK | 0 | - | 205169307 | 205187737 | 0 | 2 | 526,320 | 0,18111 | -0.8990787 | 0.0002708 | 0.0057151 |
| chr1 | 212100951 | 212101050 | DTL | 0 | + | 212100951 | 212101050 | 0 | 1 | 99 | 0 | -1.6270604 | 0.0067575 | 0.0644116 |
| chr3 | 122564487 | 122565881 | DTX3L | 0 | + | 122564487 | 122565881 | 0 | 2 | 127,23 | 0,1372 | 1.5328391 | 0.0000888 | 0.0023718 |
| chr3 | 122568697 | 122568796 | DTX3L | 0 | + | 122568697 | 122568796 | 0 | 1 | 99 | 0 | -1.3384410 | 0.0027903 | 0.0336402 |
| chr3 | 122569492 | 122570618 | DTX3L | 0 | + | 122569492 | 122570618 | 0 | 2 | 533,164 | 0,963 | -0.8950355 | 0.0001622 | 0.0037902 |
| chr2 | 241686678 | 241686775 | DTYMK | 0 | - | 241686678 | 241686775 | 0 | 1 | 97 | 0 | 1.0661536 | 0.0000667 | 0.0018947 |
| chr12 | 12476363 | 12476561 | DUSP16 | 0 | - | 12476363 | 12476561 | 0 | 1 | 198 | 0 | -0.8969708 | 0.0075891 | 0.0694504 |
| chr12 | 12476761 | 12480259 | DUSP16 | 0 | - | 12476761 | 12480259 | 0 | 2 | 1255,37 | 0,3462 | -1.4426268 | 0.0000000 | 0.0000000 |
| chr12 | 12520967 | 12521364 | DUSP16 | 0 | - | 12520967 | 12521364 | 0 | 1 | 397 | 0 | -1.0183481 | 0.0000007 | 0.0000373 |
| chr17 | 43766220 | 43766667 | DUSP3 | 0 | - | 43766220 | 43766667 | 0 | 1 | 447 | 0 | 1.2594885 | 0.0000002 | 0.0000158 |
| chr17 | 43768356 | 43768703 | DUSP3 | 0 | - | 43768356 | 43768703 | 0 | 1 | 347 | 0 | -0.7735465 | 0.0012994 | 0.0193173 |
| chr17 | 43769498 | 43769646 | DUSP3 | 0 | - | 43769498 | 43769646 | 0 | 1 | 148 | 0 | -1.1446269 | 0.0045264 | 0.0481857 |
| chr14 | 101964628 | 101964827 | DYNC1H1 | 0 | + | 101964628 | 101964827 | 0 | 1 | 199 | 0 | 1.3735330 | 0.0052348 | 0.0537712 |
| chr14 | 101985761 | 101986060 | DYNC1H1 | 0 | + | 101985761 | 101986060 | 0 | 1 | 299 | 0 | -1.5575013 | 0.0003236 | 0.0065918 |
| chr14 | 101986210 | 101986509 | DYNC1H1 | 0 | + | 101986210 | 101986509 | 0 | 1 | 299 | 0 | -1.9170562 | 0.0000000 | 0.0000002 |
| chr14 | 101986560 | 101986709 | DYNC1H1 | 0 | + | 101986560 | 101986709 | 0 | 1 | 149 | 0 | -1.5964490 | 0.0077991 | 0.0707776 |
| chr12 | 67649942 | 67657548 | DYRK2 | 0 | + | 67649942 | 67657548 | 0 | 2 | 4,443 | 0,7164 | -0.7290503 | 0.0022613 | 0.0289387 |
| chr15 | 55360347 | 55360845 | DYX1C1-CCPG1 | 0 | - | 55360347 | 55360845 | 0 | 1 | 498 | 0 | -0.9268988 | 0.0000026 | 0.0001222 |
| chr6 | 20488222 | 20490736 | E2F3 | 0 | + | 20488222 | 20490736 | 0 | 2 | 27,569 | 0,1946 | -0.8672912 | 0.0020268 | 0.0266139 |
| chr3 | 15427707 | 15427756 | EAF1 | 0 | + | 15427707 | 15427756 | 0 | 1 | 49 | 0 | 1.9627808 | 0.0049079 | 0.0513383 |
| chrX | 48528571 | 48528668 | EBP | 0 | + | 48528571 | 48528668 | 0 | 1 | 97 | 0 | -0.9252834 | 0.0118077 | 0.0941082 |
| chr10 | 133370719 | 133373355 | ECHS1 | 0 | - | 133370719 | 133373355 | 0 | 2 | 39,110 | 0,2527 | 1.0445840 | 0.0118917 | 0.0945699 |
| chr16 | 2240063 | 2243059 | ECI1 | 0 | - | 2240063 | 2243059 | 0 | 2 | 83,14 | 0,2983 | -2.1971748 | 0.0000001 | 0.0000066 |
| chr9 | 136862311 | 136862409 | EDF1 | 0 | - | 136862311 | 136862409 | 0 | 1 | 98 | 0 | -0.9052476 | 0.0004369 | 0.0083716 |
| chr11 | 86244781 | 86245078 | EED | 0 | + | 86244781 | 86245078 | 0 | 1 | 297 | 0 | -0.8827518 | 0.0068845 | 0.0652015 |
| chr3 | 128341386 | 128341832 | EEFSEC | 0 | + | 128341386 | 128341832 | 0 | 1 | 446 | 0 | -0.9657788 | 0.0018045 | 0.0244761 |
| chr7 | 55019082 | 55019181 | EGFR | 0 | + | 55019082 | 55019181 | 0 | 1 | 99 | 0 | 1.5111539 | 0.0001768 | 0.0040616 |
| chr7 | 55019281 | 55019330 | EGFR | 0 | + | 55019281 | 55019330 | 0 | 1 | 49 | 0 | 3.1180822 | 0.0001219 | 0.0030654 |
| chr1 | 231421047 | 231421296 | EGLN1 | 0 | - | 231421047 | 231421296 | 0 | 1 | 249 | 0 | -0.8090457 | 0.0020191 | 0.0265323 |
| chr19 | 47740945 | 47741641 | EHD2 | 0 | + | 47740945 | 47741641 | 0 | 1 | 696 | 0 | -0.6217438 | 0.0004666 | 0.0088100 |
| chr11 | 125584394 | 125584443 | EI24 | 0 | + | 125584394 | 125584443 | 0 | 1 | 49 | 0 | -4.1459439 | 0.0015880 | 0.0222733 |
| chr3 | 150571976 | 150572224 | EIF2A | 0 | + | 150571976 | 150572224 | 0 | 1 | 248 | 0 | -0.5738604 | 0.0058834 | 0.0586007 |
| chr20 | 34105481 | 34112229 | EIF2S2 | 0 | - | 34105481 | 34112229 | 0 | 2 | 65,134 | 0,6615 | 1.0917971 | 0.0002303 | 0.0049823 |
| chr10 | 119038294 | 119049828 | EIF3A | 0 | - | 119038294 | 119049828 | 0 | 4 | 146,779,89,28 | 0,3700,5760,11507 | -1.1439321 | 0.0000000 | 0.0000000 |
| chr1 | 32231409 | 32231555 | EIF3I | 0 | + | 32231409 | 32231555 | 0 | 1 | 146 | 0 | -1.2906491 | 0.0001868 | 0.0042286 |
| chr22 | 37849422 | 37851318 | EIF3L | 0 | + | 37849422 | 37851318 | 0 | 3 | 61,49,39 | 0,593,1858 | 1.0305375 | 0.0011267 | 0.0173989 |
| chr22 | 37876002 | 37878012 | EIF3L | 0 | + | 37876002 | 37878012 | 0 | 2 | 10,339 | 0,1672 | -0.7327411 | 0.0022020 | 0.0283194 |
| chr3 | 186783572 | 186784598 | EIF4A2 | 0 | + | 186783572 | 186784598 | 0 | 3 | 68,46,35 | 0,860,992 | 2.4724965 | 0.0000000 | 0.0000000 |
| chr17 | 80135313 | 80135362 | EIF4A3 | 0 | - | 80135313 | 80135362 | 0 | 1 | 49 | 0 | 1.5574832 | 0.0117833 | 0.0939985 |
| chr17 | 80144239 | 80147133 | EIF4A3 | 0 | - | 80144239 | 80147133 | 0 | 2 | 6,341 | 0,2554 | 0.5763238 | 0.0023133 | 0.0294025 |
| chr3 | 184321340 | 184322089 | EIF4G1 | 0 | + | 184321340 | 184322089 | 0 | 1 | 749 | 0 | -0.5828407 | 0.0004730 | 0.0088666 |
| chr14 | 103334201 | 103334527 | EIF5 | 0 | + | 103334201 | 103334527 | 0 | 2 | 60,139 | 0,188 | 1.0221440 | 0.0032469 | 0.0378107 |
| chr13 | 40933464 | 40933763 | ELF1 | 0 | - | 40933464 | 40933763 | 0 | 1 | 299 | 0 | -1.1124925 | 0.0092295 | 0.0795848 |
| chrX | 130066062 | 130066111 | ELF4 | 0 | - | 130066062 | 130066111 | 0 | 1 | 49 | 0 | -2.1511733 | 0.0000009 | 0.0000478 |
| chrX | 130066509 | 130066608 | ELF4 | 0 | - | 130066509 | 130066608 | 0 | 1 | 99 | 0 | -1.3949643 | 0.0042900 | 0.0465173 |
| chr22 | 37372775 | 37373074 | ELFN2 | 0 | - | 37372775 | 37373074 | 0 | 1 | 299 | 0 | -1.0147202 | 0.0093535 | 0.0801195 |
| chrX | 47650011 | 47650555 | ELK1 | 0 | - | 47650011 | 47650555 | 0 | 2 | 16,133 | 0,412 | 1.0414465 | 0.0105907 | 0.0876272 |
| chr1 | 205620446 | 205620695 | ELK4 | 0 | - | 205620446 | 205620695 | 0 | 1 | 249 | 0 | -1.3759114 | 0.0000414 | 0.0012874 |
| chr20 | 46366598 | 46367543 | ELMO2 | 0 | - | 46366598 | 46367543 | 0 | 1 | 945 | 0 | -0.8580491 | 0.0005836 | 0.0104520 |
| chr14 | 73739556 | 73739754 | ELMSAN1 | 0 | - | 73739556 | 73739754 | 0 | 1 | 198 | 0 | -1.5204408 | 0.0048087 | 0.0506200 |
| chr19 | 11553285 | 11553431 | ELOF1 | 0 | - | 11553285 | 11553431 | 0 | 1 | 146 | 0 | 1.1980796 | 0.0004106 | 0.0079773 |
| chr3 | 9986544 | 9986838 | EMC3 | 0 | - | 9986544 | 9986838 | 0 | 1 | 294 | 0 | 0.9106337 | 0.0001022 | 0.0026585 |
| chr15 | 34084453 | 34088068 | EMC7 | 0 | - | 34084453 | 34088068 | 0 | 2 | 34,16 | 0,3600 | -2.3000798 | 0.0006860 | 0.0118528 |
| chr2 | 42329768 | 42330264 | EML4 | 0 | + | 42329768 | 42330264 | 0 | 1 | 496 | 0 | -0.7518137 | 0.0010988 | 0.0171087 |
| chr16 | 10532445 | 10532792 | EMP2 | 0 | - | 10532445 | 10532792 | 0 | 1 | 347 | 0 | -0.6143309 | 0.0106068 | 0.0876272 |
| chr11 | 95128408 | 95129106 | ENDOD1 | 0 | + | 95128408 | 95129106 | 0 | 1 | 698 | 0 | -0.5426711 | 0.0051281 | 0.0530929 |
| chr11 | 95129357 | 95129556 | ENDOD1 | 0 | + | 95129357 | 95129556 | 0 | 1 | 199 | 0 | -1.3433164 | 0.0004745 | 0.0088765 |
| chr22 | 41117345 | 41117743 | EP300 | 0 | + | 41117345 | 41117743 | 0 | 1 | 398 | 0 | -0.9294055 | 0.0004718 | 0.0088575 |
| chr2 | 46386157 | 46386256 | EPAS1 | 0 | + | 46386157 | 46386256 | 0 | 1 | 99 | 0 | -1.9342104 | 0.0003848 | 0.0075985 |
| chr20 | 36229869 | 36230017 | EPB41L1 | 0 | + | 36229869 | 36230017 | 0 | 1 | 148 | 0 | -1.9329644 | 0.0000585 | 0.0016940 |
| chr17 | 19282971 | 19283618 | EPN2 | 0 | + | 19282971 | 19283618 | 0 | 1 | 647 | 0 | -0.8250760 | 0.0003067 | 0.0063110 |
| chr8 | 143858017 | 143858116 | EPPK1 | 0 | - | 143858017 | 143858116 | 0 | 1 | 99 | 0 | -1.3565415 | 0.0022107 | 0.0283905 |
| chr8 | 143870633 | 143870682 | EPPK1 | 0 | - | 143870633 | 143870682 | 0 | 1 | 49 | 0 | -1.8786637 | 0.0088712 | 0.0772281 |
| chr1 | 51356656 | 51356705 | EPS15 | 0 | - | 51356656 | 51356705 | 0 | 1 | 49 | 0 | 4.7018015 | 0.0000131 | 0.0005072 |
| chr11 | 727379 | 727677 | EPS8L2 | 0 | + | 727379 | 727677 | 0 | 1 | 298 | 0 | -0.9466460 | 0.0035668 | 0.0405446 |
| chr17 | 28859079 | 28861005 | ERAL1 | 0 | + | 28859079 | 28861005 | 0 | 3 | 35,81,575 | 0,124,1352 | -0.7752922 | 0.0013545 | 0.0199567 |
| chr5 | 66078429 | 66078528 | ERBB2IP | 0 | + | 66078429 | 66078528 | 0 | 1 | 99 | 0 | -1.1473902 | 0.0124962 | 0.0977037 |
| chr12 | 1027967 | 1028265 | ERC1 | 0 | + | 1027967 | 1028265 | 0 | 1 | 298 | 0 | -1.1980251 | 0.0035736 | 0.0405707 |
| chr12 | 1028565 | 1083504 | ERC1 | 0 | + | 1028565 | 1083504 | 0 | 2 | 8,341 | 0,54599 | -2.2528555 | 0.0000000 | 0.0000000 |
| chr19 | 45409359 | 45409655 | ERCC1 | 0 | - | 45409359 | 45409655 | 0 | 1 | 296 | 0 | -2.3224317 | 0.0000216 | 0.0007653 |
| chr13 | 102862218 | 102862766 | ERCC5 | 0 | + | 102862218 | 102862766 | 0 | 1 | 548 | 0 | -0.9321523 | 0.0007725 | 0.0129641 |
| chr10 | 100151279 | 100151527 | ERLIN1 | 0 | - | 100151279 | 100151527 | 0 | 1 | 248 | 0 | -1.0331007 | 0.0057476 | 0.0576928 |
| chr12 | 112022386 | 112022583 | ERP29 | 0 | + | 112022386 | 112022583 | 0 | 1 | 197 | 0 | -0.8404496 | 0.0004511 | 0.0085886 |
| chr1 | 157124866 | 157125213 | ETV3 | 0 | - | 157124866 | 157125213 | 0 | 1 | 347 | 0 | -0.7571230 | 0.0045407 | 0.0483095 |
| chr1 | 157125710 | 157125957 | ETV3 | 0 | - | 157125710 | 157125957 | 0 | 1 | 247 | 0 | -1.3129765 | 0.0002905 | 0.0060263 |
| chr3 | 186048244 | 186048441 | ETV5 | 0 | - | 186048244 | 186048441 | 0 | 1 | 197 | 0 | -1.3387914 | 0.0006532 | 0.0114835 |
| chr4 | 5811322 | 5811570 | EVC | 0 | + | 5811322 | 5811570 | 0 | 1 | 248 | 0 | -1.9442901 | 0.0055536 | 0.0561185 |
| chr4 | 5813857 | 5814155 | EVC | 0 | + | 5813857 | 5814155 | 0 | 1 | 298 | 0 | -1.8023982 | 0.0001365 | 0.0033256 |
| chr17 | 76081092 | 76081488 | EXOC7 | 0 | - | 76081092 | 76081488 | 0 | 1 | 396 | 0 | -0.7065112 | 0.0006319 | 0.0111634 |
| chr17 | 76082727 | 76082925 | EXOC7 | 0 | - | 76082727 | 76082925 | 0 | 1 | 198 | 0 | -1.2029929 | 0.0069952 | 0.0658701 |
| chr1 | 231335626 | 231336275 | EXOC8 | 0 | - | 231335626 | 231336275 | 0 | 1 | 649 | 0 | -0.9161369 | 0.0002432 | 0.0052424 |
| chr1 | 231336575 | 231336973 | EXOC8 | 0 | - | 231336575 | 231336973 | 0 | 1 | 398 | 0 | -0.9067867 | 0.0021787 | 0.0280791 |
| chr1 | 231337074 | 231337323 | EXOC8 | 0 | - | 231337074 | 231337323 | 0 | 1 | 249 | 0 | -1.1160524 | 0.0030752 | 0.0362145 |
| chr9 | 130703152 | 130703959 | EXOSC2 | 0 | + | 130703152 | 130703959 | 0 | 2 | 30,266 | 0,542 | -0.6366390 | 0.0041266 | 0.0450957 |
| chr8 | 118110090 | 118110682 | EXT1 | 0 | - | 118110090 | 118110682 | 0 | 1 | 592 | 0 | -1.0742366 | 0.0000000 | 0.0000024 |
| chr8 | 118110979 | 118111226 | EXT1 | 0 | - | 118110979 | 118111226 | 0 | 1 | 247 | 0 | -1.5170906 | 0.0003863 | 0.0076041 |
| chr11 | 44244196 | 44244788 | EXT2 | 0 | + | 44244196 | 44244788 | 0 | 1 | 592 | 0 | -0.8519939 | 0.0002491 | 0.0053626 |
| chr8 | 28717550 | 28718198 | EXTL3 | 0 | + | 28717550 | 28718198 | 0 | 1 | 648 | 0 | -0.7493388 | 0.0006650 | 0.0116120 |
| chr7 | 148883198 | 148883247 | EZH2 | 0 | - | 148883198 | 148883247 | 0 | 1 | 49 | 0 | 1.8735782 | 0.0001364 | 0.0033256 |
| chr6 | 158765988 | 158766086 | EZR | 0 | - | 158765988 | 158766086 | 0 | 1 | 98 | 0 | -1.8346337 | 0.0005316 | 0.0096576 |
| chr21 | 44976680 | 44976973 | FAM207A | 0 | + | 44976680 | 44976973 | 0 | 1 | 293 | 0 | -0.6693941 | 0.0011111 | 0.0172411 |
| chr10 | 5748880 | 5749428 | FAM208B | 0 | + | 5748880 | 5749428 | 0 | 1 | 548 | 0 | -0.8412883 | 0.0025074 | 0.0313502 |
| chr6 | 81749107 | 81749206 | FAM46A | 0 | - | 81749107 | 81749206 | 0 | 1 | 99 | 0 | -1.6596107 | 0.0108914 | 0.0893893 |
| chr10 | 124622538 | 124622587 | FAM53B | 0 | - | 124622538 | 124622587 | 0 | 1 | 49 | 0 | -2.3123696 | 0.0111953 | 0.0909652 |
| chr20 | 38947937 | 38947986 | FAM83D | 0 | + | 38947937 | 38947986 | 0 | 1 | 49 | 0 | Inf | 0.0009378 | 0.0150947 |
| chr20 | 38951723 | 38952466 | FAM83D | 0 | + | 38951723 | 38952466 | 0 | 1 | 743 | 0 | -0.6884877 | 0.0000503 | 0.0015002 |
| chr6 | 5368837 | 5404593 | FARS2 | 0 | + | 5368837 | 5404593 | 0 | 2 | 346,52 | 0,35705 | -0.7878674 | 0.0059330 | 0.0589146 |
| chr17 | 82078736 | 82079034 | FASN | 0 | - | 82078736 | 82079034 | 0 | 1 | 298 | 0 | -1.3608941 | 0.0006806 | 0.0117716 |
| chr4 | 186596710 | 186596958 | FAT1 | 0 | - | 186596710 | 186596958 | 0 | 1 | 248 | 0 | -1.0262028 | 0.0079848 | 0.0718985 |
| chr4 | 186603242 | 186603391 | FAT1 | 0 | - | 186603242 | 186603391 | 0 | 1 | 149 | 0 | -1.8128570 | 0.0048431 | 0.0508419 |
| chr4 | 186617796 | 186617945 | FAT1 | 0 | - | 186617796 | 186617945 | 0 | 1 | 149 | 0 | -1.4914914 | 0.0011641 | 0.0177893 |
| chr19 | 39846317 | 39846366 | FBL | 0 | - | 39846317 | 39846366 | 0 | 1 | 49 | 0 | 1.3028651 | 0.0078528 | 0.0709938 |
| chr3 | 13570932 | 13570981 | FBLN2 | 0 | + | 13570932 | 13570981 | 0 | 1 | 49 | 0 | -1.4719672 | 0.0075241 | 0.0690801 |
| chr3 | 13571081 | 13571626 | FBLN2 | 0 | + | 13571081 | 13571626 | 0 | 1 | 545 | 0 | -0.7955024 | 0.0004992 | 0.0091960 |
| chr7 | 5500518 | 5500866 | FBXL18 | 0 | - | 5500518 | 5500866 | 0 | 1 | 348 | 0 | -1.3661483 | 0.0014006 | 0.0203878 |
| chr7 | 5501116 | 5501265 | FBXL18 | 0 | - | 5501116 | 5501265 | 0 | 1 | 149 | 0 | -1.9729083 | 0.0010015 | 0.0158865 |
| chr16 | 30947136 | 30947434 | FBXL19 | 0 | + | 30947136 | 30947434 | 0 | 1 | 298 | 0 | 1.0402000 | 0.0019259 | 0.0257082 |
| chr4 | 15612397 | 15625633 | FBXL5 | 0 | - | 15612397 | 15625633 | 0 | 2 | 18,382 | 0,12855 | -0.9304070 | 0.0016620 | 0.0230617 |
| chr19 | 39025251 | 39025497 | FBXO27 | 0 | - | 39025251 | 39025497 | 0 | 1 | 246 | 0 | -1.0892968 | 0.0067394 | 0.0643012 |
| chr11 | 33774426 | 33774525 | FBXO3 | 0 | - | 33774426 | 33774525 | 0 | 1 | 99 | 0 | 1.6668020 | 0.0061869 | 0.0607058 |
| chr16 | 87329916 | 87330015 | FBXO31 | 0 | - | 87329916 | 87330015 | 0 | 1 | 99 | 0 | -1.3989932 | 0.0071501 | 0.0666773 |
| chr16 | 87330562 | 87331256 | FBXO31 | 0 | - | 87330562 | 87331256 | 0 | 1 | 694 | 0 | -1.1637067 | 0.0000001 | 0.0000044 |
| chr5 | 148383985 | 148394808 | FBXO38 | 0 | + | 148383985 | 148394808 | 0 | 2 | 55,95 | 0,10729 | 1.2240019 | 0.0093154 | 0.0799197 |
| chr2 | 73257822 | 73257921 | FBXO41 | 0 | - | 73257822 | 73257921 | 0 | 1 | 99 | 0 | -3.7069243 | 0.0001291 | 0.0031771 |
| chr1 | 155320467 | 155320616 | FDPS | 0 | + | 155320467 | 155320616 | 0 | 1 | 149 | 0 | -0.9962956 | 0.0035769 | 0.0405829 |
| chr19 | 4793463 | 4793662 | FEM1A | 0 | + | 4793463 | 4793662 | 0 | 1 | 199 | 0 | -1.8819838 | 0.0000000 | 0.0000033 |
| chr15 | 68278650 | 68290337 | FEM1B | 0 | + | 68278650 | 68290337 | 0 | 2 | 16,731 | 0,10957 | -0.8336687 | 0.0000220 | 0.0007759 |
| chr15 | 68290487 | 68290686 | FEM1B | 0 | + | 68290487 | 68290686 | 0 | 1 | 199 | 0 | -0.9865553 | 0.0082533 | 0.0735391 |
| chr5 | 115525233 | 115525531 | FEM1C | 0 | - | 115525233 | 115525531 | 0 | 1 | 298 | 0 | -1.0709265 | 0.0053860 | 0.0549152 |
| chr11 | 61795747 | 61796245 | FEN1 | 0 | + | 61795747 | 61796245 | 0 | 1 | 498 | 0 | -0.6620234 | 0.0000002 | 0.0000159 |
| chr11 | 61796345 | 61796444 | FEN1 | 0 | + | 61796345 | 61796444 | 0 | 1 | 99 | 0 | -0.7084655 | 0.0092043 | 0.0794429 |
| chr2 | 36597891 | 36598139 | FEZ2 | 0 | - | 36597891 | 36598139 | 0 | 1 | 248 | 0 | 1.0800339 | 0.0054460 | 0.0553214 |
| chr3 | 14945217 | 14947094 | FGD5-AS1 | 0 | - | 14945217 | 14947094 | 0 | 2 | 938,8 | 0,1870 | -0.7622827 | 0.0000000 | 0.0000003 |
| chr9 | 113165620 | 113166117 | FKBP15 | 0 | - | 113165620 | 113166117 | 0 | 1 | 497 | 0 | -1.0383854 | 0.0032202 | 0.0376204 |
| chr9 | 113207235 | 113221211 | FKBP15 | 0 | - | 113207235 | 113221211 | 0 | 3 | 62,116,21 | 0,4242,13956 | 1.5721702 | 0.0002281 | 0.0049402 |
| chr1 | 154988517 | 154988862 | FLAD1 | 0 | + | 154988517 | 154988862 | 0 | 1 | 345 | 0 | -0.8206889 | 0.0000000 | 0.0000026 |
| chrX | 154364305 | 154364606 | FLNA | 0 | - | 154364305 | 154364606 | 0 | 2 | 68,81 | 0,221 | -2.2051708 | 0.0058749 | 0.0585488 |
| chr3 | 58170862 | 58171110 | FLNB | 0 | + | 58170862 | 58171110 | 0 | 1 | 248 | 0 | -1.3446579 | 0.0006790 | 0.0117655 |
| chr13 | 48976012 | 48976111 | FNDC3A | 0 | + | 48976012 | 48976111 | 0 | 1 | 99 | 0 | 1.9750486 | 0.0054098 | 0.0550649 |
| chr3 | 172397260 | 172397359 | FNDC3B | 0 | + | 172397260 | 172397359 | 0 | 1 | 99 | 0 | -1.9703195 | 0.0004720 | 0.0088575 |
| chr8 | 43056299 | 43059142 | FNTA | 0 | + | 43056299 | 43059142 | 0 | 2 | 248,51 | 0,2793 | 0.6830971 | 0.0108365 | 0.0891939 |
| chr11 | 72195625 | 72195674 | FOLR1 | 0 | + | 72195625 | 72195674 | 0 | 1 | 49 | 0 | -1.5242812 | 0.0000000 | 0.0000001 |
| chr14 | 37591242 | 37591688 | FOXA1 | 0 | - | 37591242 | 37591688 | 0 | 1 | 446 | 0 | -1.0348422 | 0.0002784 | 0.0058416 |
| chr14 | 37591739 | 37592583 | FOXA1 | 0 | - | 37591739 | 37592583 | 0 | 1 | 844 | 0 | -0.6239806 | 0.0083678 | 0.0742036 |
| chr6 | 1610846 | 1611095 | FOXC1 | 0 | + | 1610846 | 1611095 | 0 | 1 | 249 | 0 | -1.5779779 | 0.0000000 | 0.0000000 |
| chr16 | 86567745 | 86567893 | FOXC2 | 0 | + | 86567745 | 86567893 | 0 | 1 | 148 | 0 | -1.2922105 | 0.0071508 | 0.0666773 |
| chr7 | 4762351 | 4763150 | FOXK1 | 0 | + | 4762351 | 4763150 | 0 | 1 | 799 | 0 | -0.9545546 | 0.0000009 | 0.0000509 |
| chr7 | 4763201 | 4763699 | FOXK1 | 0 | + | 4763201 | 4763699 | 0 | 1 | 498 | 0 | -1.6762335 | 0.0000023 | 0.0001134 |
| chr7 | 4766548 | 4766747 | FOXK1 | 0 | + | 4766548 | 4766747 | 0 | 1 | 199 | 0 | 1.5930555 | 0.0005910 | 0.0105646 |
| chr7 | 4766798 | 4767546 | FOXK1 | 0 | + | 4766798 | 4767546 | 0 | 1 | 748 | 0 | 0.5116606 | 0.0093169 | 0.0799197 |
| chr17 | 82602572 | 82602621 | FOXK2 | 0 | + | 82602572 | 82602621 | 0 | 1 | 49 | 0 | -2.6407338 | 0.0033645 | 0.0387557 |
| chr12 | 2872233 | 2874261 | FOXM1 | 0 | - | 2872233 | 2874261 | 0 | 2 | 15,285 | 0,1744 | -0.7571049 | 0.0104408 | 0.0868454 |
| chr6 | 41600335 | 41600384 | FOXP4 | 0 | + | 41600335 | 41600384 | 0 | 1 | 49 | 0 | -3.3015555 | 0.0003188 | 0.0065163 |
| chr11 | 65411452 | 65412241 | FRMD8 | 0 | + | 65411452 | 65412241 | 0 | 1 | 789 | 0 | 0.5929356 | 0.0023138 | 0.0294025 |
| chr12 | 69574180 | 69574924 | FRS2 | 0 | + | 69574180 | 69574924 | 0 | 1 | 744 | 0 | -0.8051976 | 0.0005321 | 0.0096576 |
| chr7 | 5593055 | 5593203 | FSCN1 | 0 | + | 5593055 | 5593203 | 0 | 1 | 148 | 0 | -1.5172252 | 0.0003854 | 0.0076030 |
| chr7 | 5593404 | 5603291 | FSCN1 | 0 | + | 5593404 | 5603291 | 0 | 2 | 365,35 | 0,9853 | -1.2505434 | 0.0001469 | 0.0035275 |
| chr11 | 61964874 | 61965018 | FTH1 | 0 | - | 61964874 | 61965018 | 0 | 2 | 18,32 | 0,113 | -3.3449883 | 0.0000097 | 0.0003939 |
| chr11 | 61965068 | 61965422 | FTH1 | 0 | - | 61965068 | 61965422 | 0 | 2 | 45,54 | 0,301 | -3.1287083 | 0.0000000 | 0.0000001 |
| chr16 | 53826018 | 53826117 | FTO | 0 | + | 53826018 | 53826117 | 0 | 1 | 99 | 0 | -2.1766481 | 0.0019933 | 0.0263467 |
| chr16 | 53826168 | 53844179 | FTO | 0 | + | 53826168 | 53844179 | 0 | 2 | 324,25 | 0,17987 | -2.0707443 | 0.0000007 | 0.0000376 |
| chrX | 48485916 | 48486064 | FTSJ1 | 0 | + | 48485916 | 48486064 | 0 | 1 | 148 | 0 | -1.5843629 | 0.0084575 | 0.0748408 |
| chrX | 44524077 | 44524272 | FUNDC1 | 0 | - | 44524077 | 44524272 | 0 | 1 | 195 | 0 | -0.8052428 | 0.0112208 | 0.0910330 |
| chr3 | 45968104 | 45968452 | FYCO1 | 0 | - | 45968104 | 45968452 | 0 | 1 | 348 | 0 | -1.5761358 | 0.0000006 | 0.0000336 |
| chr19 | 3506347 | 3523026 | FZR1 | 0 | + | 3506347 | 3523026 | 0 | 2 | 128,71 | 0,16609 | 1.1792583 | 0.0093611 | 0.0801469 |
| chr4 | 75644649 | 75645390 | G3BP2 | 0 | - | 75644649 | 75645390 | 0 | 1 | 741 | 0 | -0.9211237 | 0.0000020 | 0.0001003 |
| chr4 | 75645440 | 75645687 | G3BP2 | 0 | - | 75645440 | 75645687 | 0 | 1 | 247 | 0 | -1.0650818 | 0.0003339 | 0.0067559 |
| chr17 | 80119732 | 80119830 | GAA | 0 | + | 80119732 | 80119830 | 0 | 1 | 98 | 0 | -0.9629202 | 0.0017001 | 0.0234426 |
| chr11 | 78218533 | 78218881 | GAB2 | 0 | - | 78218533 | 78218881 | 0 | 1 | 348 | 0 | -1.2485427 | 0.0017223 | 0.0236085 |
| chr1 | 67685374 | 67685522 | GADD45A | 0 | + | 67685374 | 67685522 | 0 | 1 | 148 | 0 | 0.8252765 | 0.0027288 | 0.0331849 |
| chr1 | 67687698 | 67687993 | GADD45A | 0 | + | 67687698 | 67687993 | 0 | 1 | 295 | 0 | -0.6933705 | 0.0001497 | 0.0035868 |
| chr12 | 51353092 | 51353141 | GALNT6 | 0 | - | 51353092 | 51353141 | 0 | 1 | 49 | 0 | -1.1908094 | 0.0078943 | 0.0712965 |
| chr12 | 51353986 | 51354432 | GALNT6 | 0 | - | 51353986 | 51354432 | 0 | 1 | 446 | 0 | -0.5540325 | 0.0091388 | 0.0790661 |
| chr12 | 51379358 | 51379705 | GALNT6 | 0 | - | 51379358 | 51379705 | 0 | 1 | 347 | 0 | -0.6946015 | 0.0004910 | 0.0090915 |
| chr4 | 173322384 | 173322433 | GALNT7 | 0 | + | 173322384 | 173322433 | 0 | 1 | 49 | 0 | -2.6385201 | 0.0108757 | 0.0893462 |
| chr9 | 86946247 | 86946395 | GAS1 | 0 | - | 86946247 | 86946395 | 0 | 1 | 148 | 0 | -1.0665295 | 0.0035263 | 0.0402567 |
| chr9 | 35737064 | 35737360 | GBA2 | 0 | - | 35737064 | 35737360 | 0 | 1 | 296 | 0 | -0.7340138 | 0.0025223 | 0.0314557 |
| chr9 | 35744699 | 35748931 | GBA2 | 0 | - | 35744699 | 35748931 | 0 | 2 | 8,586 | 0,3647 | -0.5137028 | 0.0071961 | 0.0669612 |
| chr7 | 127582022 | 127582320 | GCC1 | 0 | - | 127582022 | 127582320 | 0 | 1 | 298 | 0 | -1.0121298 | 0.0013542 | 0.0199567 |
| chr7 | 127582420 | 127582768 | GCC1 | 0 | - | 127582420 | 127582768 | 0 | 1 | 348 | 0 | -1.0582030 | 0.0008663 | 0.0141705 |
| chr16 | 19503136 | 19503235 | GDE1 | 0 | - | 19503136 | 19503235 | 0 | 1 | 99 | 0 | -1.8908984 | 0.0000817 | 0.0022204 |
| chr16 | 19503336 | 19507690 | GDE1 | 0 | - | 19503336 | 19507690 | 0 | 3 | 282,212,4 | 0,1545,4351 | -1.2831171 | 0.0000000 | 0.0000023 |
| chr19 | 18386208 | 18386257 | GDF15 | 0 | + | 18386208 | 18386257 | 0 | 1 | 49 | 0 | 2.1722301 | 0.0001242 | 0.0031018 |
| chr17 | 745419 | 745617 | GEMIN4 | 0 | - | 745419 | 745617 | 0 | 1 | 198 | 0 | -1.0078284 | 0.0115042 | 0.0926272 |
| chr17 | 746765 | 746814 | GEMIN4 | 0 | - | 746765 | 746814 | 0 | 1 | 49 | 0 | -1.2447229 | 0.0102391 | 0.0856395 |
| chr17 | 747164 | 747363 | GEMIN4 | 0 | - | 747164 | 747363 | 0 | 1 | 199 | 0 | -1.9903074 | 0.0000000 | 0.0000000 |
| chr5 | 180353217 | 180353315 | GFPT2 | 0 | - | 180353217 | 180353315 | 0 | 1 | 98 | 0 | 1.6245303 | 0.0011415 | 0.0175189 |
| chr17 | 75237336 | 75237781 | GGA3 | 0 | - | 75237336 | 75237781 | 0 | 1 | 445 | 0 | -0.7522739 | 0.0098732 | 0.0835051 |
| chr2 | 232857982 | 232858131 | GIGYF2 | 0 | + | 232857982 | 232858131 | 0 | 1 | 149 | 0 | -2.2207495 | 0.0027391 | 0.0332664 |
| chr1 | 34785484 | 34785876 | GJB3 | 0 | + | 34785484 | 34785876 | 0 | 1 | 392 | 0 | -1.1703320 | 0.0001272 | 0.0031385 |
| chr17 | 44804423 | 44805369 | GJC1 | 0 | - | 44804423 | 44805369 | 0 | 1 | 946 | 0 | -1.0313964 | 0.0000014 | 0.0000711 |
| chrX | 101397889 | 101397938 | GLA | 0 | - | 101397889 | 101397938 | 0 | 1 | 49 | 0 | -1.9839885 | 0.0025086 | 0.0313502 |
| chr15 | 69255978 | 69256177 | GLCE | 0 | + | 69255978 | 69256177 | 0 | 1 | 199 | 0 | -1.5675079 | 0.0012257 | 0.0185269 |
| chr14 | 95535376 | 95544279 | GLRX5 | 0 | + | 95535376 | 95544279 | 0 | 2 | 9,333 | 0,8571 | -0.9419387 | 0.0000472 | 0.0014329 |
| chr10 | 87094473 | 87094920 | GLUD1 | 0 | - | 87094473 | 87094920 | 0 | 1 | 447 | 0 | 0.9261826 | 0.0073339 | 0.0677732 |
| chr6 | 24774931 | 24775174 | GMNN | 0 | + | 24774931 | 24775174 | 0 | 1 | 243 | 0 | 1.2594977 | 0.0002895 | 0.0060125 |
| chr7 | 2730304 | 2730403 | GNA12 | 0 | - | 2730304 | 2730403 | 0 | 1 | 99 | 0 | -1.5525603 | 0.0056322 | 0.0567859 |
| chr7 | 2730803 | 2731352 | GNA12 | 0 | - | 2730803 | 2731352 | 0 | 1 | 549 | 0 | -0.7596988 | 0.0020678 | 0.0269966 |
| chr9 | 111661619 | 111666815 | GNG10 | 0 | + | 111661619 | 111666815 | 0 | 2 | 97,1 | 0,5196 | 1.8790538 | 0.0012335 | 0.0185819 |
| chr3 | 52686169 | 52687351 | GNL3 | 0 | + | 52686169 | 52687351 | 0 | 3 | 34,59,106 | 0,600,1077 | 0.5524149 | 0.0127980 | 0.0994189 |
| chr12 | 64759297 | 64759396 | GNS | 0 | - | 64759297 | 64759396 | 0 | 1 | 99 | 0 | 1.2269761 | 0.0117697 | 0.0939985 |
| chr12 | 132771405 | 132771654 | GOLGA3 | 0 | - | 132771405 | 132771654 | 0 | 1 | 249 | 0 | 0.9929478 | 0.0022872 | 0.0291866 |
| chr12 | 132772853 | 132774164 | GOLGA3 | 0 | - | 132772853 | 132774164 | 0 | 2 | 442,8 | 0,1304 | 0.8396155 | 0.0083943 | 0.0744024 |
| chr12 | 21515826 | 21515974 | GOLT1B | 0 | + | 21515826 | 21515974 | 0 | 1 | 148 | 0 | 1.2368109 | 0.0002313 | 0.0049972 |
| chr1 | 155752473 | 155753235 | GON4L | 0 | - | 155752473 | 155753235 | 0 | 2 | 118,32 | 0,731 | -1.4982012 | 0.0088415 | 0.0771207 |
| chr10 | 99397216 | 99397611 | GOT1 | 0 | - | 99397216 | 99397611 | 0 | 1 | 395 | 0 | -0.6205694 | 0.0063430 | 0.0617678 |
| chr8 | 41598350 | 41598846 | GPAT4 | 0 | + | 41598350 | 41598846 | 0 | 1 | 496 | 0 | -0.9765604 | 0.0000221 | 0.0007771 |
| chr1 | 156594651 | 156594895 | GPATCH4 | 0 | - | 156594651 | 156594895 | 0 | 1 | 244 | 0 | -0.9364403 | 0.0072955 | 0.0675045 |
| chr17 | 44398269 | 44398966 | GPATCH8 | 0 | - | 44398269 | 44398966 | 0 | 1 | 697 | 0 | -1.3354048 | 0.0000010 | 0.0000540 |
| chr17 | 44400213 | 44400511 | GPATCH8 | 0 | - | 44400213 | 44400511 | 0 | 1 | 298 | 0 | -1.4912643 | 0.0016229 | 0.0226228 |
| chr2 | 240466086 | 240466534 | GPC1 | 0 | + | 240466086 | 240466534 | 0 | 1 | 448 | 0 | -0.5328530 | 0.0028580 | 0.0342507 |
| chr2 | 240466983 | 240467032 | GPC1 | 0 | + | 240466983 | 240467032 | 0 | 1 | 49 | 0 | -1.2970602 | 0.0116874 | 0.0936438 |
| chr3 | 32106660 | 32106758 | GPD1L | 0 | + | 32106660 | 32106758 | 0 | 1 | 98 | 0 | 1.4885404 | 0.0114821 | 0.0924901 |
| chr7 | 1092871 | 1092970 | GPER1 | 0 | + | 1092871 | 1092970 | 0 | 1 | 99 | 0 | -2.5765476 | 0.0012999 | 0.0193173 |
| chr9 | 130137073 | 130137522 | GPR107 | 0 | + | 130137073 | 130137522 | 0 | 1 | 449 | 0 | 0.8156474 | 0.0000650 | 0.0018507 |
| chr19 | 6730260 | 6731010 | GPR108 | 0 | - | 6730260 | 6731010 | 0 | 2 | 125,24 | 0,727 | 1.1775512 | 0.0091149 | 0.0789911 |
| chr12 | 12891563 | 12909074 | GPRC5A | 0 | + | 12891563 | 12909074 | 0 | 2 | 102,832 | 0,16680 | -0.6786570 | 0.0000013 | 0.0000693 |
| chr12 | 12912084 | 12912732 | GPRC5A | 0 | + | 12912084 | 12912732 | 0 | 2 | 59,286 | 0,363 | -0.6241284 | 0.0000390 | 0.0012306 |
| chr5 | 176598129 | 176598178 | GPRIN1 | 0 | - | 176598129 | 176598178 | 0 | 1 | 49 | 0 | -2.1482049 | 0.0100483 | 0.0845125 |
| chr19 | 35023521 | 35026320 | GRAMD1A | 0 | + | 35023521 | 35026320 | 0 | 2 | 27,272 | 0,2528 | -0.9955097 | 0.0001176 | 0.0029780 |
| chr7 | 50592651 | 50592999 | GRB10 | 0 | - | 50592651 | 50592999 | 0 | 1 | 348 | 0 | -0.6998678 | 0.0001272 | 0.0031385 |
| chr8 | 143990107 | 143990205 | GRINA | 0 | + | 143990107 | 143990205 | 0 | 1 | 98 | 0 | 1.4925810 | 0.0000972 | 0.0025499 |
| chr8 | 143992825 | 143993120 | GRINA | 0 | + | 143992825 | 143993120 | 0 | 1 | 295 | 0 | -1.1094058 | 0.0000077 | 0.0003231 |
| chr17 | 44352519 | 44352906 | GRN | 0 | + | 44352519 | 44352906 | 0 | 2 | 53,246 | 0,142 | -1.0154162 | 0.0004886 | 0.0090647 |
| chr17 | 3725800 | 3726349 | GSG2 | 0 | + | 3725800 | 3726349 | 0 | 1 | 549 | 0 | -1.4673154 | 0.0000203 | 0.0007296 |
| chr1 | 88853087 | 88853335 | GTF2B | 0 | - | 88853087 | 88853335 | 0 | 1 | 248 | 0 | -1.1699131 | 0.0019870 | 0.0263237 |
| chr3 | 120742761 | 120750985 | GTF2E1 | 0 | + | 120742761 | 120750985 | 0 | 2 | 34,463 | 0,7762 | -0.6388387 | 0.0059430 | 0.0589671 |
| chr19 | 6380157 | 6380354 | GTF2F1 | 0 | - | 6380157 | 6380354 | 0 | 1 | 197 | 0 | -0.9811301 | 0.0043879 | 0.0472911 |
| chr19 | 6392986 | 6393182 | GTF2F1 | 0 | - | 6392986 | 6393182 | 0 | 1 | 196 | 0 | 0.9638003 | 0.0054503 | 0.0553214 |
| chr16 | 27549732 | 27549831 | GTF3C1 | 0 | - | 27549732 | 27549831 | 0 | 1 | 99 | 0 | 2.3233378 | 0.0004131 | 0.0080178 |
| chr9 | 132678997 | 132679096 | GTF3C4 | 0 | + | 132678997 | 132679096 | 0 | 1 | 99 | 0 | -1.0134399 | 0.0005197 | 0.0095258 |
| chr9 | 132679344 | 132679492 | GTF3C4 | 0 | + | 132679344 | 132679492 | 0 | 1 | 148 | 0 | -1.0452580 | 0.0098863 | 0.0835198 |
| chr22 | 38716049 | 38716812 | GTPBP1 | 0 | + | 38716049 | 38716812 | 0 | 2 | 39,161 | 0,603 | -0.8399188 | 0.0019131 | 0.0256104 |
| chr6 | 43621072 | 43621760 | GTPBP2 | 0 | - | 43621072 | 43621760 | 0 | 1 | 688 | 0 | -0.8792759 | 0.0005419 | 0.0098129 |
| chr22 | 24540523 | 24540821 | GUCD1 | 0 | - | 24540523 | 24540821 | 0 | 1 | 298 | 0 | -0.6506218 | 0.0060507 | 0.0597742 |
| chr22 | 24542320 | 24543908 | GUCD1 | 0 | - | 24542320 | 24543908 | 0 | 2 | 781,67 | 0,1522 | -0.5010880 | 0.0033764 | 0.0388433 |
| chr1 | 228140250 | 228146026 | GUK1 | 0 | + | 228140250 | 228146026 | 0 | 5 | 114,165,50,114,1 | 0,874,4453,5261,5776 | 0.8741266 | 0.0066532 | 0.0639127 |
| chr1 | 228148807 | 228148905 | GUK1 | 0 | + | 228148807 | 228148905 | 0 | 1 | 98 | 0 | -1.0197282 | 0.0070768 | 0.0663968 |
| chr3 | 129315257 | 129315597 | H1FX | 0 | - | 129315257 | 129315597 | 0 | 1 | 340 | 0 | -1.1063020 | 0.0001028 | 0.0026707 |
| chr3 | 129315645 | 129315694 | H1FX | 0 | - | 129315645 | 129315694 | 0 | 1 | 49 | 0 | -1.4046142 | 0.0027563 | 0.0333423 |
| chr5 | 135388953 | 135399789 | H2AFY | 0 | - | 135388953 | 135399789 | 0 | 3 | 174,177,41 | 0,10109,10796 | 0.7836054 | 0.0031626 | 0.0370672 |
| chr1 | 226064465 | 226065690 | H3F3A | 0 | + | 226064465 | 226065690 | 0 | 2 | 15,35 | 0,1191 | 1.6618932 | 0.0000014 | 0.0000716 |
| chr17 | 75779069 | 75779787 | H3F3B | 0 | - | 75779069 | 75779787 | 0 | 2 | 116,131 | 0,588 | 0.6554451 | 0.0106057 | 0.0876272 |
| chr5 | 140698533 | 140699222 | HARS2 | 0 | + | 140698533 | 140699222 | 0 | 1 | 689 | 0 | -0.7089795 | 0.0004304 | 0.0082647 |
| chrX | 153447714 | 153447862 | HAUS7 | 0 | - | 153447714 | 153447862 | 0 | 1 | 148 | 0 | -1.1659119 | 0.0034852 | 0.0398419 |
| chrX | 153955038 | 153955436 | HCFC1 | 0 | - | 153955038 | 153955436 | 0 | 1 | 398 | 0 | -1.1310724 | 0.0005115 | 0.0094027 |
| chr16 | 3022669 | 3022814 | HCFC1R1 | 0 | - | 3022669 | 3022814 | 0 | 1 | 145 | 0 | -0.7668496 | 0.0095072 | 0.0810914 |
| chrX | 48822956 | 48823405 | HDAC6 | 0 | + | 48822956 | 48823405 | 0 | 1 | 449 | 0 | -1.4204728 | 0.0000111 | 0.0004356 |
| chr12 | 47783255 | 47784138 | HDAC7 | 0 | - | 47783255 | 47784138 | 0 | 2 | 632,60 | 0,824 | 0.6330726 | 0.0000039 | 0.0001776 |
| chr2 | 241248027 | 241248076 | HDLBP | 0 | - | 241248027 | 241248076 | 0 | 1 | 49 | 0 | -5.2610742 | 0.0001557 | 0.0036812 |
| chr2 | 241266906 | 241267676 | HDLBP | 0 | - | 241266906 | 241267676 | 0 | 2 | 1,99 | 0,672 | -1.2525501 | 0.0110090 | 0.0899598 |
| chr6 | 139166474 | 139166871 | HECA | 0 | + | 139166474 | 139166871 | 0 | 1 | 397 | 0 | -0.9907455 | 0.0033143 | 0.0382991 |
| chr3 | 124970039 | 124973763 | HEG1 | 0 | - | 124970039 | 124973763 | 0 | 2 | 763,33 | 0,3692 | -0.5636633 | 0.0016775 | 0.0232052 |
| chr3 | 125013557 | 125019275 | HEG1 | 0 | - | 125013557 | 125019275 | 0 | 2 | 434,14 | 0,5705 | -0.9155363 | 0.0024231 | 0.0305125 |
| chr5 | 180830541 | 180830737 | HEIH | 0 | - | 180830541 | 180830737 | 0 | 1 | 196 | 0 | -1.0428080 | 0.0026579 | 0.0326395 |
| chr20 | 63564253 | 63564351 | HELZ2 | 0 | - | 63564253 | 63564351 | 0 | 1 | 98 | 0 | -2.4164596 | 0.0001898 | 0.0042759 |
| chr20 | 63564402 | 63564501 | HELZ2 | 0 | - | 63564402 | 63564501 | 0 | 1 | 99 | 0 | -4.0145514 | 0.0000000 | 0.0000028 |
| chr20 | 63564801 | 63564900 | HELZ2 | 0 | - | 63564801 | 63564900 | 0 | 1 | 99 | 0 | -1.8119355 | 0.0011761 | 0.0179569 |
| chr20 | 63565151 | 63565499 | HELZ2 | 0 | - | 63565151 | 63565499 | 0 | 1 | 348 | 0 | -1.2378349 | 0.0025139 | 0.0313949 |
| chr20 | 63565550 | 63565599 | HELZ2 | 0 | - | 63565550 | 63565599 | 0 | 1 | 49 | 0 | -2.4177147 | 0.0020649 | 0.0269776 |
| chr15 | 63774972 | 63833898 | HERC1 | 0 | - | 63774972 | 63833898 | 0 | 2 | 678,72 | 0,58855 | -0.9891693 | 0.0011383 | 0.0175189 |
| chr7 | 35694200 | 35694595 | HERPUD2 | 0 | - | 35694200 | 35694595 | 0 | 1 | 395 | 0 | -0.7008630 | 0.0075268 | 0.0690801 |
| chr5 | 74685205 | 74685502 | HEXB | 0 | + | 74685205 | 74685502 | 0 | 1 | 297 | 0 | 0.8663635 | 0.0078234 | 0.0708333 |
| chr17 | 45148463 | 45149360 | HEXIM1 | 0 | + | 45148463 | 45149360 | 0 | 1 | 897 | 0 | -0.5846766 | 0.0000921 | 0.0024380 |
| chr17 | 45169696 | 45169844 | HEXIM2 | 0 | + | 45169696 | 45169844 | 0 | 1 | 148 | 0 | -2.1619729 | 0.0000538 | 0.0015803 |
| chr8 | 144139405 | 144140198 | HGH1 | 0 | + | 144139405 | 144140198 | 0 | 1 | 793 | 0 | -0.5272204 | 0.0001421 | 0.0034212 |
| chr7 | 27526089 | 27531257 | HIBADH | 0 | - | 27526089 | 27531257 | 0 | 2 | 284,66 | 0,5103 | -1.0935316 | 0.0013854 | 0.0202278 |
| chr14 | 61695501 | 61695800 | HIF1A | 0 | + | 61695501 | 61695800 | 0 | 1 | 299 | 0 | 0.8467906 | 0.0001253 | 0.0031081 |
| chr9 | 35813006 | 35813102 | HINT2 | 0 | - | 35813006 | 35813102 | 0 | 1 | 96 | 0 | -1.7488987 | 0.0013360 | 0.0197490 |
| chr6 | 125956814 | 125957112 | HINT3 | 0 | + | 125956814 | 125957112 | 0 | 1 | 298 | 0 | 1.4735046 | 0.0000865 | 0.0023240 |
| chr1 | 113929523 | 113941316 | HIPK1 | 0 | + | 113929523 | 113941316 | 0 | 2 | 10,935 | 0,10859 | -0.8306022 | 0.0000227 | 0.0007861 |
| chr11 | 33286449 | 33286697 | HIPK3 | 0 | + | 33286449 | 33286697 | 0 | 1 | 248 | 0 | -1.9080102 | 0.0004709 | 0.0088575 |
| chr6 | 27133436 | 27133485 | HIST1H2AG | 0 | + | 27133436 | 27133485 | 0 | 1 | 49 | 0 | 2.5839739 | 0.0093154 | 0.0799197 |
| chr6 | 27824125 | 27824174 | HIST1H4J | 0 | + | 27824125 | 27824174 | 0 | 1 | 49 | 0 | 2.6807516 | 0.0000460 | 0.0013996 |
| chr6 | 27831478 | 27831527 | HIST1H4K | 0 | - | 27831478 | 27831527 | 0 | 1 | 49 | 0 | 2.0643950 | 0.0002030 | 0.0044955 |
| chr6 | 142760099 | 142760148 | HIVEP2 | 0 | - | 142760099 | 142760148 | 0 | 1 | 49 | 0 | 5.4661403 | 0.0008689 | 0.0142003 |
| chr19 | 3575616 | 3576265 | HMG20B | 0 | + | 3575616 | 3576265 | 0 | 2 | 45,5 | 0,645 | -2.4040375 | 0.0001562 | 0.0036887 |
| chr5 | 43291170 | 43291219 | HMGCS1 | 0 | - | 43291170 | 43291219 | 0 | 1 | 49 | 0 | -1.0002687 | 0.0084603 | 0.0748408 |
| chr5 | 43298592 | 43298891 | HMGCS1 | 0 | - | 43298592 | 43298891 | 0 | 1 | 299 | 0 | -0.6385745 | 0.0006087 | 0.0108479 |
| chr21 | 39345266 | 39349073 | HMGN1 | 0 | - | 39345266 | 39349073 | 0 | 5 | 9,48,30,33,171 | 0,3026,3156,3279,3637 | 0.9024156 | 0.0000794 | 0.0021811 |
| chr1 | 26472411 | 26472558 | HMGN2 | 0 | + | 26472411 | 26472558 | 0 | 1 | 147 | 0 | 1.5039683 | 0.0000017 | 0.0000847 |
| chr6 | 79201539 | 79201686 | HMGN3 | 0 | - | 79201539 | 79201686 | 0 | 1 | 147 | 0 | 2.1026132 | 0.0006730 | 0.0117059 |
| chr6 | 26545658 | 26545952 | HMGN4 | 0 | + | 26545658 | 26545952 | 0 | 1 | 294 | 0 | -1.2063014 | 0.0000224 | 0.0007821 |
| chr5 | 150052896 | 150053044 | HMGXB3 | 0 | + | 150052896 | 150053044 | 0 | 1 | 148 | 0 | -1.3401082 | 0.0085188 | 0.0753014 |
| chr22 | 35264093 | 35265236 | HMGXB4 | 0 | + | 35264093 | 35265236 | 0 | 2 | 4,589 | 0,555 | -1.3229705 | 0.0000033 | 0.0001513 |
| chr5 | 137753757 | 137754152 | HNRNPA0 | 0 | - | 137753757 | 137754152 | 0 | 1 | 395 | 0 | -1.0955557 | 0.0000000 | 0.0000000 |
| chr19 | 41262575 | 41265132 | HNRNPUL1 | 0 | + | 41262575 | 41265132 | 0 | 3 | 8,477,11 | 0,1747,2547 | 0.9751899 | 0.0000180 | 0.0006521 |
| chr3 | 149172202 | 149172251 | HPS3 | 0 | + | 149172202 | 149172251 | 0 | 1 | 49 | 0 | 3.0418098 | 0.0096732 | 0.0820944 |
| chr22 | 26464045 | 26464343 | HPS4 | 0 | - | 26464045 | 26464343 | 0 | 1 | 298 | 0 | -0.9894770 | 0.0111452 | 0.0907102 |
| chr10 | 102067692 | 102067890 | HPS6 | 0 | + | 102067692 | 102067890 | 0 | 1 | 198 | 0 | -0.9002838 | 0.0039408 | 0.0436189 |
| chr8 | 22125666 | 22127218 | HR | 0 | - | 22125666 | 22127218 | 0 | 2 | 67,182 | 0,1371 | -1.5711867 | 0.0009186 | 0.0148720 |
| chr22 | 28742154 | 28744636 | HSCB | 0 | + | 28742154 | 28744636 | 0 | 3 | 178,97,22 | 0,1728,2461 | 0.7919312 | 0.0057778 | 0.0578328 |
| chr14 | 102082219 | 102083056 | HSP90AA1 | 0 | - | 102082219 | 102083056 | 0 | 2 | 226,23 | 0,815 | -0.8808202 | 0.0058064 | 0.0580157 |
| chr6 | 44252253 | 44253229 | HSP90AB1 | 0 | + | 44252253 | 44253229 | 0 | 2 | 15,185 | 0,792 | -1.4175109 | 0.0061919 | 0.0607058 |
| chr9 | 125236287 | 125238166 | HSPA5 | 0 | - | 125236287 | 125238166 | 0 | 2 | 868,26 | 0,1854 | -0.6174887 | 0.0049465 | 0.0515966 |
| chr9 | 125239007 | 125239205 | HSPA5 | 0 | - | 125239007 | 125239205 | 0 | 1 | 198 | 0 | -0.9802954 | 0.0110985 | 0.0904872 |
| chr11 | 123057541 | 123057836 | HSPA8 | 0 | - | 123057541 | 123057836 | 0 | 1 | 295 | 0 | -0.6161333 | 0.0031947 | 0.0373702 |
| chr11 | 123059541 | 123059935 | HSPA8 | 0 | - | 123059541 | 123059935 | 0 | 1 | 394 | 0 | -1.0102978 | 0.0000015 | 0.0000783 |
| chr1 | 16015075 | 16015224 | HSPB7 | 0 | - | 16015075 | 16015224 | 0 | 1 | 149 | 0 | -1.4267509 | 0.0036280 | 0.0409317 |
| chr11 | 20383139 | 20383188 | HTATIP2 | 0 | + | 20383139 | 20383188 | 0 | 1 | 49 | 0 | -1.8399766 | 0.0045866 | 0.0486838 |
| chr2 | 74533007 | 74533106 | HTRA2 | 0 | + | 74533007 | 74533106 | 0 | 1 | 99 | 0 | -1.2319774 | 0.0104625 | 0.0869734 |
| chrX | 53547869 | 53549002 | HUWE1 | 0 | - | 53547869 | 53549002 | 0 | 2 | 405,44 | 0,1090 | -1.2002635 | 0.0000001 | 0.0000070 |
| chr3 | 50300089 | 50300488 | HYAL1 | 0 | - | 50300089 | 50300488 | 0 | 1 | 399 | 0 | -1.4806591 | 0.0000000 | 0.0000000 |
| chr3 | 50302205 | 50302904 | HYAL1 | 0 | - | 50302205 | 50302904 | 0 | 1 | 699 | 0 | -0.5474226 | 0.0000196 | 0.0007064 |
| chr3 | 50319763 | 50321479 | HYAL2 | 0 | - | 50319763 | 50321479 | 0 | 2 | 773,114 | 0,1603 | -0.6387761 | 0.0000274 | 0.0009133 |
| chr3 | 50294924 | 50295371 | HYAL3 | 0 | - | 50294924 | 50295371 | 0 | 1 | 447 | 0 | -1.3069395 | 0.0000400 | 0.0012497 |
| chr19 | 10274828 | 10275027 | ICAM1 | 0 | + | 10274828 | 10275027 | 0 | 1 | 199 | 0 | -1.6352835 | 0.0000000 | 0.0000000 |
| chr19 | 10284882 | 10284931 | ICAM1 | 0 | + | 10284882 | 10284931 | 0 | 1 | 49 | 0 | -1.9715840 | 0.0000999 | 0.0026098 |
| chr19 | 10294511 | 10294609 | ICAM5 | 0 | + | 10294511 | 10294609 | 0 | 1 | 98 | 0 | -1.7642845 | 0.0013530 | 0.0199567 |
| chr5 | 5462791 | 5462940 | ICE1 | 0 | + | 5462791 | 5462940 | 0 | 1 | 149 | 0 | -1.1287272 | 0.0062056 | 0.0608069 |
| chr5 | 5464591 | 5464840 | ICE1 | 0 | + | 5464591 | 5464840 | 0 | 1 | 249 | 0 | -1.1343442 | 0.0011271 | 0.0173989 |
| chr1 | 23558128 | 23558375 | ID3 | 0 | - | 23558128 | 23558375 | 0 | 1 | 247 | 0 | 0.8306837 | 0.0018894 | 0.0253619 |
| chr9 | 129178113 | 129178261 | IER5L | 0 | - | 129178113 | 129178261 | 0 | 1 | 148 | 0 | -0.8821129 | 0.0008997 | 0.0146116 |
| chr10 | 89306281 | 89306480 | IFIT2 | 0 | + | 89306281 | 89306480 | 0 | 1 | 199 | 0 | -0.9893659 | 0.0021346 | 0.0276491 |
| chr10 | 89307230 | 89307578 | IFIT2 | 0 | + | 89307230 | 89307578 | 0 | 1 | 348 | 0 | 1.3976603 | 0.0000000 | 0.0000001 |
| chr10 | 89340072 | 89340221 | IFIT3 | 0 | + | 89340072 | 89340221 | 0 | 1 | 149 | 0 | -0.9186566 | 0.0019855 | 0.0263237 |
| chr11 | 313991 | 314326 | IFITM1 | 0 | + | 313991 | 314326 | 0 | 1 | 335 | 0 | 1.1384233 | 0.0000000 | 0.0000000 |
| chr11 | 308156 | 308399 | IFITM2 | 0 | + | 308156 | 308399 | 0 | 1 | 243 | 0 | 0.7077328 | 0.0038491 | 0.0427608 |
| chr21 | 33230043 | 33230092 | IFNAR2 | 0 | + | 33230043 | 33230092 | 0 | 1 | 49 | 0 | 2.2770874 | 0.0106731 | 0.0879290 |
| chrX | 70134525 | 70146634 | IGBP1 | 0 | + | 70134525 | 70146634 | 0 | 2 | 292,2 | 0,12108 | -0.9467410 | 0.0040763 | 0.0446544 |
| chr15 | 98899579 | 98908791 | IGF1R | 0 | + | 98899579 | 98908791 | 0 | 2 | 43,107 | 0,9106 | 1.5095236 | 0.0014130 | 0.0205137 |
| chr15 | 98963682 | 98963731 | IGF1R | 0 | + | 98963682 | 98963731 | 0 | 1 | 49 | 0 | -2.2987971 | 0.0011641 | 0.0177893 |
| chr17 | 48997761 | 48997909 | IGF2BP1 | 0 | + | 48997761 | 48997909 | 0 | 1 | 148 | 0 | 1.1900990 | 0.0100922 | 0.0848421 |
| chr4 | 57040857 | 57110385 | IGFBP7 | 0 | - | 57040857 | 57110385 | 0 | 2 | 77,509 | 0,69020 | 0.5628022 | 0.0108727 | 0.0893462 |
| chr1 | 116614069 | 116616171 | IGSF3 | 0 | - | 116614069 | 116616171 | 0 | 2 | 107,92 | 0,2011 | -1.5284623 | 0.0001297 | 0.0031833 |
| chr1 | 160093102 | 160093250 | IGSF8 | 0 | - | 160093102 | 160093250 | 0 | 1 | 148 | 0 | -0.9898002 | 0.0000081 | 0.0003380 |
| chr22 | 17108828 | 17109375 | IL17RA | 0 | + | 17108828 | 17109375 | 0 | 1 | 547 | 0 | -1.1041440 | 0.0046491 | 0.0492314 |
| chr11 | 112143550 | 112143799 | IL18 | 0 | - | 112143550 | 112143799 | 0 | 1 | 249 | 0 | -0.9849331 | 0.0103542 | 0.0862436 |
| chr19 | 14052216 | 14052905 | IL27RA | 0 | + | 14052216 | 14052905 | 0 | 1 | 689 | 0 | -1.1683033 | 0.0000000 | 0.0000011 |
| chr19 | 10653431 | 10653529 | ILF3-AS1 | 0 | - | 10653431 | 10653529 | 0 | 1 | 98 | 0 | 1.5940650 | 0.0123033 | 0.0966959 |
| chr11 | 6610527 | 6610725 | ILK | 0 | + | 6610527 | 6610725 | 0 | 1 | 198 | 0 | -0.9770656 | 0.0080104 | 0.0720934 |
| chr2 | 86144227 | 86144620 | IMMT | 0 | - | 86144227 | 86144620 | 0 | 1 | 393 | 0 | -0.8696038 | 0.0093008 | 0.0799197 |
| chr18 | 12028992 | 12030539 | IMPA2 | 0 | + | 12028992 | 12030539 | 0 | 2 | 2,197 | 0,1351 | 1.0828217 | 0.0009415 | 0.0151368 |
| chr14 | 104714338 | 104714834 | INF2 | 0 | + | 104714338 | 104714834 | 0 | 1 | 496 | 0 | 1.0863634 | 0.0000095 | 0.0003894 |
| chr7 | 155308348 | 155308645 | INSIG1 | 0 | + | 155308348 | 155308645 | 0 | 1 | 297 | 0 | -1.3608218 | 0.0000001 | 0.0000099 |
| chr8 | 19817457 | 19818312 | INTS10 | 0 | + | 19817457 | 19818312 | 0 | 2 | 210,38 | 0,818 | 1.1942227 | 0.0028647 | 0.0342850 |
| chr3 | 48688250 | 48688595 | IP6K2 | 0 | - | 48688250 | 48688595 | 0 | 1 | 345 | 0 | -0.8894043 | 0.0086726 | 0.0762565 |
| chr13 | 97976711 | 97982575 | IPO5 | 0 | + | 97976711 | 97982575 | 0 | 2 | 76,73 | 0,5792 | 1.9456359 | 0.0000000 | 0.0000000 |
| chr11 | 9445132 | 9445530 | IPO7 | 0 | + | 9445132 | 9445530 | 0 | 1 | 398 | 0 | -0.8266468 | 0.0008416 | 0.0138796 |
| chr3 | 12935881 | 12936080 | IQSEC1 | 0 | - | 12935881 | 12936080 | 0 | 1 | 199 | 0 | -1.6174557 | 0.0125497 | 0.0979564 |
| chrX | 154011448 | 154011745 | IRAK1 | 0 | - | 154011448 | 154011745 | 0 | 1 | 297 | 0 | -1.4215306 | 0.0032463 | 0.0378107 |
| chr19 | 45885431 | 45885973 | IRF2BP1 | 0 | - | 45885431 | 45885973 | 0 | 1 | 542 | 0 | -0.5857098 | 0.0025826 | 0.0320529 |
| chr1 | 234609079 | 234609277 | IRF2BP2 | 0 | - | 234609079 | 234609277 | 0 | 1 | 198 | 0 | -1.3825452 | 0.0007489 | 0.0126745 |
| chr14 | 77025681 | 77026621 | IRF2BPL | 0 | - | 77025681 | 77026621 | 0 | 1 | 940 | 0 | -1.1257234 | 0.0000000 | 0.0000000 |
| chr14 | 77027710 | 77027907 | IRF2BPL | 0 | - | 77027710 | 77027907 | 0 | 1 | 197 | 0 | -1.4090616 | 0.0003541 | 0.0071168 |
| chr19 | 49664748 | 49665826 | IRF3 | 0 | - | 49664748 | 49665826 | 0 | 2 | 99,343 | 0,736 | 0.7296782 | 0.0090556 | 0.0786092 |
| chr13 | 109784269 | 109784468 | IRS2 | 0 | - | 109784269 | 109784468 | 0 | 1 | 199 | 0 | -2.0934765 | 0.0000000 | 0.0000015 |
| chr13 | 109785368 | 109785417 | IRS2 | 0 | - | 109785368 | 109785417 | 0 | 1 | 49 | 0 | -2.2945370 | 0.0042691 | 0.0463457 |
| chr13 | 109785618 | 109785767 | IRS2 | 0 | - | 109785618 | 109785767 | 0 | 1 | 149 | 0 | -2.1931318 | 0.0000029 | 0.0001369 |
| chr16 | 54284627 | 54285314 | IRX3 | 0 | - | 54284627 | 54285314 | 0 | 1 | 687 | 0 | -0.9563151 | 0.0000001 | 0.0000046 |
| chr1 | 1014064 | 1014160 | ISG15 | 0 | + | 1014064 | 1014160 | 0 | 1 | 96 | 0 | -1.1555525 | 0.0000354 | 0.0011432 |
| chr1 | 156726915 | 156727657 | ISG20L2 | 0 | - | 156726915 | 156727657 | 0 | 1 | 742 | 0 | -0.7752300 | 0.0000001 | 0.0000043 |
| chr16 | 265689 | 266035 | ITFG3 | 0 | + | 265689 | 266035 | 0 | 1 | 346 | 0 | -0.9087511 | 0.0122844 | 0.0965896 |
| chr13 | 48233188 | 48253866 | ITM2B | 0 | + | 48233188 | 48253866 | 0 | 2 | 290,59 | 0,20620 | 0.6850714 | 0.0000267 | 0.0008925 |
| chr6 | 33691095 | 33691830 | ITPR3 | 0 | + | 33691095 | 33691830 | 0 | 3 | 15,105,30 | 0,520,706 | -2.1791493 | 0.0000000 | 0.0000040 |
| chr10 | 104313338 | 104314285 | ITPRIP | 0 | - | 104313338 | 104314285 | 0 | 1 | 947 | 0 | -0.9400250 | 0.0000000 | 0.0000003 |
| chr10 | 104314885 | 104315034 | ITPRIP | 0 | - | 104314885 | 104315034 | 0 | 1 | 149 | 0 | -1.7148470 | 0.0000497 | 0.0014884 |
| chr10 | 104315434 | 104315583 | ITPRIP | 0 | - | 104315434 | 104315583 | 0 | 1 | 149 | 0 | -1.2505175 | 0.0000007 | 0.0000415 |
| chr2 | 127504706 | 127523678 | IWS1 | 0 | - | 127504706 | 127523678 | 0 | 2 | 1047,3 | 0,18970 | -0.9066844 | 0.0000179 | 0.0006480 |
| chr3 | 9892982 | 9893080 | JAGN1 | 0 | + | 9892982 | 9893080 | 0 | 1 | 98 | 0 | -1.5791465 | 0.0000000 | 0.0000022 |
| chr3 | 9893275 | 9893421 | JAGN1 | 0 | + | 9893275 | 9893421 | 0 | 1 | 146 | 0 | -0.7580569 | 0.0003815 | 0.0075495 |
| chr1 | 64833520 | 64833619 | JAK1 | 0 | - | 64833520 | 64833619 | 0 | 1 | 99 | 0 | -1.6427408 | 0.0020642 | 0.0269776 |
| chr1 | 64834263 | 64834461 | JAK1 | 0 | - | 64834263 | 64834461 | 0 | 1 | 198 | 0 | -1.3212084 | 0.0006711 | 0.0116967 |
| chr6 | 15496140 | 15496834 | JARID2 | 0 | + | 15496140 | 15496834 | 0 | 1 | 694 | 0 | -0.7995372 | 0.0038192 | 0.0425583 |
| chr14 | 75469521 | 75469968 | JDP2 | 0 | + | 75469521 | 75469968 | 0 | 1 | 447 | 0 | -0.5234209 | 0.0097248 | 0.0824422 |
| chr1 | 227732237 | 227732286 | JMJD4 | 0 | - | 227732237 | 227732286 | 0 | 1 | 49 | 0 | -2.3175438 | 0.0075356 | 0.0690905 |
| chr5 | 79236651 | 79236700 | JMY | 0 | + | 79236651 | 79236700 | 0 | 1 | 49 | 0 | -2.7410023 | 0.0040470 | 0.0444951 |
| chr22 | 38685891 | 38686040 | JOSD1 | 0 | - | 38685891 | 38686040 | 0 | 1 | 149 | 0 | -1.3204608 | 0.0014869 | 0.0211891 |
| chr8 | 74244884 | 74245082 | JPH1 | 0 | - | 74244884 | 74245082 | 0 | 1 | 198 | 0 | -1.4972273 | 0.0035731 | 0.0405707 |
| chr8 | 74259440 | 74315045 | JPH1 | 0 | - | 74259440 | 74315045 | 0 | 2 | 64,185 | 0,55421 | -1.1716642 | 0.0025796 | 0.0320375 |
| chr8 | 142665238 | 142665287 | JRK | 0 | - | 142665238 | 142665287 | 0 | 1 | 49 | 0 | -3.9354036 | 0.0110082 | 0.0899598 |
| chr1 | 58783122 | 58783220 | JUN | 0 | - | 58783122 | 58783220 | 0 | 1 | 98 | 0 | -1.1860543 | 0.0036239 | 0.0409150 |
| chr19 | 12791937 | 12792281 | JUNB | 0 | + | 12791937 | 12792281 | 0 | 1 | 344 | 0 | -1.1062719 | 0.0000000 | 0.0000000 |
| chr17 | 41755101 | 41755150 | JUP | 0 | - | 41755101 | 41755150 | 0 | 1 | 49 | 0 | -1.3922259 | 0.0127918 | 0.0994133 |
| chr17 | 41755200 | 41755249 | JUP | 0 | - | 41755200 | 41755249 | 0 | 1 | 49 | 0 | -2.1944232 | 0.0000000 | 0.0000000 |
| chr9 | 712199 | 712498 | KANK1 | 0 | + | 712199 | 712498 | 0 | 1 | 299 | 0 | -1.9548430 | 0.0000000 | 0.0000005 |
| chr9 | 712549 | 712598 | KANK1 | 0 | + | 712549 | 712598 | 0 | 1 | 49 | 0 | -2.5788939 | 0.0091314 | 0.0790400 |
| chr9 | 712749 | 713397 | KANK1 | 0 | + | 712749 | 713397 | 0 | 1 | 648 | 0 | -1.3507619 | 0.0000000 | 0.0000000 |
| chr19 | 11192946 | 11193643 | KANK2 | 0 | - | 11192946 | 11193643 | 0 | 1 | 697 | 0 | -0.7231687 | 0.0014353 | 0.0207426 |
| chr19 | 11193843 | 11193992 | KANK2 | 0 | - | 11193843 | 11193992 | 0 | 1 | 149 | 0 | -1.7935127 | 0.0023800 | 0.0301167 |
| chr2 | 96593555 | 96593903 | KANSL3 | 0 | - | 96593555 | 96593903 | 0 | 1 | 348 | 0 | -1.3615154 | 0.0002830 | 0.0059251 |
| chr2 | 96594454 | 96595402 | KANSL3 | 0 | - | 96594454 | 96595402 | 0 | 1 | 948 | 0 | -0.7367668 | 0.0007371 | 0.0125445 |
| chr16 | 75631803 | 75635705 | KARS | 0 | - | 75631803 | 75635705 | 0 | 3 | 53,120,26 | 0,2370,3877 | -1.6812486 | 0.0000093 | 0.0003788 |
| chr8 | 41932125 | 41932574 | KAT6A | 0 | - | 41932125 | 41932574 | 0 | 1 | 449 | 0 | -1.9428195 | 0.0000001 | 0.0000081 |
| chr8 | 41933823 | 41933972 | KAT6A | 0 | - | 41933823 | 41933972 | 0 | 1 | 149 | 0 | -1.9780649 | 0.0019402 | 0.0258715 |
| chr8 | 42048530 | 42049228 | KAT6A | 0 | - | 42048530 | 42049228 | 0 | 1 | 698 | 0 | -1.2973421 | 0.0008601 | 0.0140814 |
| chr7 | 32869699 | 32870342 | KBTBD2 | 0 | - | 32869699 | 32870342 | 0 | 1 | 643 | 0 | -0.9669529 | 0.0000326 | 0.0010651 |
| chr11 | 47577921 | 47578868 | KBTBD4 | 0 | - | 47577921 | 47578868 | 0 | 2 | 108,41 | 0,907 | -1.4217878 | 0.0012527 | 0.0188252 |
| chr13 | 41131108 | 41131505 | KBTBD6 | 0 | - | 41131108 | 41131505 | 0 | 1 | 397 | 0 | -1.2601943 | 0.0012291 | 0.0185627 |
| chr1 | 233671334 | 233671432 | KCNK1 | 0 | + | 233671334 | 233671432 | 0 | 1 | 98 | 0 | -1.3441562 | 0.0002031 | 0.0044955 |
| chr6 | 36487613 | 36488209 | KCTD20 | 0 | + | 36487613 | 36488209 | 0 | 1 | 596 | 0 | -0.9406340 | 0.0017626 | 0.0240162 |
| chr11 | 78173492 | 78188579 | KCTD21 | 0 | - | 78173492 | 78188579 | 0 | 2 | 1092,7 | 0,15081 | -0.5992259 | 0.0052304 | 0.0537712 |
| chr22 | 38482700 | 38482948 | KDELR3 | 0 | + | 38482700 | 38482948 | 0 | 1 | 248 | 0 | -0.9378230 | 0.0003009 | 0.0062128 |
| chr1 | 202729782 | 202729981 | KDM5B | 0 | - | 202729782 | 202729981 | 0 | 1 | 199 | 0 | -1.2457854 | 0.0021402 | 0.0277011 |
| chr1 | 202808222 | 202808371 | KDM5B | 0 | - | 202808222 | 202808371 | 0 | 1 | 149 | 0 | 1.9386435 | 0.0001881 | 0.0042503 |
| chrX | 45063458 | 45063757 | KDM6A | 0 | + | 45063458 | 45063757 | 0 | 1 | 299 | 0 | -0.9722001 | 0.0052722 | 0.0539984 |
| chrX | 45069619 | 45070167 | KDM6A | 0 | + | 45069619 | 45070167 | 0 | 1 | 548 | 0 | -0.7219204 | 0.0003284 | 0.0066704 |
| chr14 | 24431585 | 24432081 | KHNYN | 0 | + | 24431585 | 24432081 | 0 | 1 | 496 | 0 | -0.6545302 | 0.0025990 | 0.0321548 |
| chr14 | 24437362 | 24437760 | KHNYN | 0 | + | 24437362 | 24437760 | 0 | 1 | 398 | 0 | -0.8169819 | 0.0027583 | 0.0333423 |
| chr19 | 6418019 | 6418068 | KHSRP | 0 | - | 6418019 | 6418068 | 0 | 1 | 49 | 0 | -2.5135474 | 0.0034485 | 0.0395854 |
| chr1 | 175160116 | 175160215 | KIAA0040 | 0 | - | 175160116 | 175160215 | 0 | 1 | 99 | 0 | -1.2555841 | 0.0081033 | 0.0726408 |
| chr1 | 175160364 | 175160761 | KIAA0040 | 0 | - | 175160364 | 175160761 | 0 | 1 | 397 | 0 | -0.7949357 | 0.0000105 | 0.0004157 |
| chr1 | 175177646 | 175192799 | KIAA0040 | 0 | - | 175177646 | 175192799 | 0 | 2 | 39,160 | 0,14994 | 0.8685851 | 0.0034669 | 0.0397385 |
| chr1 | 39412046 | 39412095 | KIAA0754 | 0 | + | 39412046 | 39412095 | 0 | 1 | 49 | 0 | -4.7273988 | 0.0105965 | 0.0876272 |
| chr5 | 176347501 | 176348009 | KIAA1191 | 0 | - | 176347501 | 176348009 | 0 | 2 | 308,89 | 0,420 | -0.6913761 | 0.0082291 | 0.0734056 |
| chr1 | 32773135 | 32773383 | KIAA1522 | 0 | + | 32773135 | 32773383 | 0 | 1 | 248 | 0 | -1.6375575 | 0.0000039 | 0.0001776 |
| chr7 | 138917229 | 138917328 | KIAA1549 | 0 | - | 138917229 | 138917328 | 0 | 1 | 99 | 0 | -1.9326690 | 0.0114528 | 0.0924595 |
| chr12 | 31981140 | 31981189 | KIAA1551 | 0 | + | 31981140 | 31981189 | 0 | 1 | 49 | 0 | 3.6355129 | 0.0067764 | 0.0644813 |
| chr22 | 44247072 | 44247221 | KIAA1644 | 0 | - | 44247072 | 44247221 | 0 | 1 | 149 | 0 | 1.1941337 | 0.0071160 | 0.0665740 |
| chr22 | 44249269 | 44296686 | KIAA1644 | 0 | - | 44249269 | 44296686 | 0 | 3 | 417,318,15 | 0,36159,47403 | -0.7323676 | 0.0077022 | 0.0701565 |
| chr22 | 25028724 | 25029022 | KIAA1671 | 0 | + | 25028724 | 25029022 | 0 | 1 | 298 | 0 | -1.5092397 | 0.0000714 | 0.0020035 |
| chr22 | 25038954 | 25039253 | KIAA1671 | 0 | + | 25038954 | 25039253 | 0 | 1 | 299 | 0 | -2.0958403 | 0.0000000 | 0.0000001 |
| chr22 | 25039304 | 25039653 | KIAA1671 | 0 | + | 25039304 | 25039653 | 0 | 1 | 349 | 0 | -1.2959774 | 0.0020496 | 0.0268527 |
| chr22 | 25041153 | 25041452 | KIAA1671 | 0 | + | 25041153 | 25041452 | 0 | 1 | 299 | 0 | -1.9400234 | 0.0000000 | 0.0000008 |
| chr6 | 17764189 | 17764935 | KIF13A | 0 | - | 17764189 | 17764935 | 0 | 1 | 746 | 0 | -0.7479353 | 0.0003920 | 0.0077082 |
| chr17 | 44926357 | 44926456 | KIF18B | 0 | - | 44926357 | 44926456 | 0 | 1 | 99 | 0 | 2.2922991 | 0.0052420 | 0.0538106 |
| chr5 | 138179780 | 138179829 | KIF20A | 0 | + | 138179780 | 138179829 | 0 | 1 | 49 | 0 | 2.5459729 | 0.0010972 | 0.0171087 |
| chr10 | 32055995 | 32056393 | KIF5B | 0 | - | 32055995 | 32056393 | 0 | 1 | 398 | 0 | 0.9513817 | 0.0007900 | 0.0132456 |
| chr8 | 102649975 | 102651280 | KLF10 | 0 | - | 102649975 | 102651280 | 0 | 2 | 417,132 | 0,1174 | -0.5447501 | 0.0088222 | 0.0771207 |
| chr8 | 102651581 | 102651879 | KLF10 | 0 | - | 102651581 | 102651879 | 0 | 1 | 298 | 0 | -1.2177619 | 0.0000564 | 0.0016421 |
| chr9 | 107487695 | 107487893 | KLF4 | 0 | - | 107487695 | 107487893 | 0 | 1 | 198 | 0 | -0.9263054 | 0.0002016 | 0.0044735 |
| chr13 | 73062431 | 73062480 | KLF5 | 0 | + | 73062431 | 73062480 | 0 | 1 | 49 | 0 | -2.4867579 | 0.0084077 | 0.0744484 |
| chr10 | 3781641 | 3784983 | KLF6 | 0 | - | 3781641 | 3784983 | 0 | 2 | 574,71 | 0,3272 | -0.6678967 | 0.0002568 | 0.0054961 |
| chr17 | 41854187 | 41854833 | KLHL11 | 0 | - | 41854187 | 41854833 | 0 | 1 | 646 | 0 | -0.8722990 | 0.0016460 | 0.0228754 |
| chrX | 118116532 | 118116780 | KLHL13 | 0 | - | 118116532 | 118116780 | 0 | 1 | 248 | 0 | 1.5923723 | 0.0000021 | 0.0001030 |
| chrX | 23987850 | 23988646 | KLHL15 | 0 | - | 23987850 | 23988646 | 0 | 1 | 796 | 0 | -0.6839815 | 0.0012993 | 0.0193173 |
| chrX | 23988697 | 23988796 | KLHL15 | 0 | - | 23988697 | 23988796 | 0 | 1 | 99 | 0 | -1.7777271 | 0.0039929 | 0.0440068 |
| chrX | 24006352 | 24006551 | KLHL15 | 0 | - | 24006352 | 24006551 | 0 | 1 | 199 | 0 | -1.6245067 | 0.0086366 | 0.0760810 |
| chr3 | 47344280 | 47344776 | KLHL18 | 0 | + | 47344280 | 47344776 | 0 | 1 | 496 | 0 | -1.1809818 | 0.0013311 | 0.0197089 |
| chr1 | 6593205 | 6601856 | KLHL21 | 0 | - | 6593205 | 6601856 | 0 | 4 | 454,73,406,60 | 0,2280,5842,8592 | -0.5979308 | 0.0004202 | 0.0081117 |
| chr22 | 20465041 | 20465238 | KLHL22 | 0 | - | 20465041 | 20465238 | 0 | 1 | 197 | 0 | -1.3405994 | 0.0026247 | 0.0323444 |
| chr2 | 169735864 | 169736112 | KLHL23 | 0 | + | 169735864 | 169736112 | 0 | 1 | 248 | 0 | -1.3649413 | 0.0066736 | 0.0640237 |
| chr15 | 85768073 | 85768271 | KLHL25 | 0 | - | 85768073 | 85768271 | 0 | 1 | 198 | 0 | -1.7707799 | 0.0004936 | 0.0091298 |
| chr16 | 84657051 | 84657299 | KLHL36 | 0 | + | 84657051 | 84657299 | 0 | 1 | 248 | 0 | -1.0897209 | 0.0004399 | 0.0084114 |
| chr7 | 152177690 | 152177889 | KMT2C | 0 | - | 152177690 | 152177889 | 0 | 1 | 199 | 0 | -1.8472934 | 0.0075874 | 0.0694504 |
| chr12 | 49026341 | 49026939 | KMT2D | 0 | - | 49026341 | 49026939 | 0 | 1 | 598 | 0 | -0.8688399 | 0.0006177 | 0.0109759 |
| chr12 | 49031276 | 49031825 | KMT2D | 0 | - | 49031276 | 49031825 | 0 | 1 | 549 | 0 | -0.9350311 | 0.0058568 | 0.0584000 |
| chr12 | 49037622 | 49038419 | KMT2D | 0 | - | 49037622 | 49038419 | 0 | 1 | 797 | 0 | -1.2429343 | 0.0000014 | 0.0000741 |
| chr12 | 49038870 | 49038919 | KMT2D | 0 | - | 49038870 | 49038919 | 0 | 1 | 49 | 0 | -2.1336721 | 0.0066767 | 0.0640237 |
| chr12 | 49050044 | 49050342 | KMT2D | 0 | - | 49050044 | 49050342 | 0 | 1 | 298 | 0 | -1.1029434 | 0.0119599 | 0.0949450 |
| chr7 | 105113191 | 105113389 | KMT2E | 0 | + | 105113191 | 105113389 | 0 | 1 | 198 | 0 | 1.1473795 | 0.0128834 | 0.0999540 |
| chr15 | 40393545 | 40394139 | KNSTRN | 0 | + | 40393545 | 40394139 | 0 | 1 | 594 | 0 | -0.5407696 | 0.0026226 | 0.0323444 |
| chr3 | 122426526 | 122426724 | KPNA1 | 0 | - | 122426526 | 122426724 | 0 | 1 | 198 | 0 | -0.8419440 | 0.0076914 | 0.0700935 |
| chr1 | 32172135 | 32172184 | KPNA6 | 0 | + | 32172135 | 32172184 | 0 | 1 | 49 | 0 | 2.4514529 | 0.0002256 | 0.0049032 |
| chr17 | 47680569 | 47682599 | KPNB1 | 0 | + | 47680569 | 47682599 | 0 | 2 | 101,196 | 0,1835 | -1.1179372 | 0.0000918 | 0.0024374 |
| chr19 | 10553184 | 10554021 | KRI1 | 0 | - | 10553184 | 10554021 | 0 | 1 | 837 | 0 | -0.6814799 | 0.0003235 | 0.0065918 |
| chr1 | 52032305 | 52032603 | KTI12 | 0 | - | 52032305 | 52032603 | 0 | 1 | 298 | 0 | -1.1683521 | 0.0002698 | 0.0057070 |
| chr19 | 18560528 | 18564921 | KXD1 | 0 | + | 18560528 | 18564921 | 0 | 3 | 23,122,53 | 0,1508,4341 | -0.9264528 | 0.0127062 | 0.0988761 |
| chr3 | 49122711 | 49122859 | LAMB2 | 0 | - | 49122711 | 49122859 | 0 | 1 | 148 | 0 | -1.7687812 | 0.0000000 | 0.0000003 |
| chr3 | 49131638 | 49131687 | LAMB2 | 0 | - | 49131638 | 49131687 | 0 | 1 | 49 | 0 | -1.5447891 | 0.0088279 | 0.0771207 |
| chr1 | 209652426 | 209652475 | LAMB3 | 0 | - | 209652426 | 209652475 | 0 | 1 | 49 | 0 | 1.8094688 | 0.0078529 | 0.0709938 |
| chr8 | 97775780 | 97776078 | LAPTM4B | 0 | + | 97775780 | 97776078 | 0 | 1 | 298 | 0 | 1.1318249 | 0.0002603 | 0.0055382 |
| chr3 | 45547452 | 45548193 | LARS2 | 0 | + | 45547452 | 45548193 | 0 | 1 | 741 | 0 | -0.5342521 | 0.0018026 | 0.0244761 |
| chr13 | 20988524 | 20988723 | LATS2 | 0 | - | 20988524 | 20988723 | 0 | 1 | 199 | 0 | -1.4435959 | 0.0000025 | 0.0001201 |
| chr16 | 67940174 | 67940319 | LCAT | 0 | - | 67940174 | 67940319 | 0 | 1 | 145 | 0 | -1.8305038 | 0.0000149 | 0.0005619 |
| chr11 | 18394536 | 18394634 | LDHA | 0 | + | 18394536 | 18394634 | 0 | 1 | 98 | 0 | 1.4227519 | 0.0001198 | 0.0030294 |
| chr19 | 11089561 | 11100267 | LDLR | 0 | + | 11089561 | 11100267 | 0 | 2 | 55,45 | 0,10662 | 1.2449307 | 0.0015387 | 0.0217505 |
| chr19 | 11132135 | 11132384 | LDLR | 0 | + | 11132135 | 11132384 | 0 | 1 | 249 | 0 | -1.5151246 | 0.0000000 | 0.0000002 |
| chr19 | 11133132 | 11133380 | LDLR | 0 | + | 11133132 | 11133380 | 0 | 1 | 248 | 0 | -0.7189451 | 0.0103330 | 0.0861465 |
| chr22 | 44495157 | 44496251 | LDOC1L | 0 | - | 44495157 | 44496251 | 0 | 1 | 1094 | 0 | -1.0074901 | 0.0000000 | 0.0000015 |
| chr12 | 65169921 | 65170869 | LEMD3 | 0 | + | 65169921 | 65170869 | 0 | 1 | 948 | 0 | -1.0066146 | 0.0000000 | 0.0000031 |
| chr12 | 65170920 | 65210926 | LEMD3 | 0 | + | 65170920 | 65210926 | 0 | 2 | 199,1 | 0,40006 | -1.1424085 | 0.0056382 | 0.0568151 |
| chr22 | 37679707 | 37679756 | LGALS1 | 0 | + | 37679707 | 37679756 | 0 | 1 | 49 | 0 | 1.3453074 | 0.0000859 | 0.0023205 |
| chr17 | 78971548 | 78971744 | LGALS3BP | 0 | - | 78971548 | 78971744 | 0 | 1 | 196 | 0 | -1.0772944 | 0.0000001 | 0.0000070 |
| chr17 | 78971842 | 78972579 | LGALS3BP | 0 | - | 78971842 | 78972579 | 0 | 1 | 737 | 0 | -0.8472566 | 0.0000001 | 0.0000044 |
| chr11 | 27472338 | 27472787 | LGR4 | 0 | - | 27472338 | 27472787 | 0 | 1 | 449 | 0 | 0.6943706 | 0.0046923 | 0.0495735 |
| chr12 | 50176821 | 50177613 | LIMA1 | 0 | - | 50176821 | 50177613 | 0 | 1 | 792 | 0 | -0.7666409 | 0.0053563 | 0.0547355 |
| chr12 | 50177911 | 50178010 | LIMA1 | 0 | - | 50177911 | 50178010 | 0 | 1 | 99 | 0 | -1.0621171 | 0.0112577 | 0.0912918 |
| chr4 | 41698552 | 41698601 | LIMCH1 | 0 | + | 41698552 | 41698601 | 0 | 1 | 49 | 0 | 1.5286910 | 0.0111205 | 0.0905845 |
| chr3 | 45594831 | 45595528 | LIMD1 | 0 | + | 45594831 | 45595528 | 0 | 1 | 697 | 0 | -0.8597481 | 0.0000137 | 0.0005239 |
| chr3 | 45595679 | 45596027 | LIMD1 | 0 | + | 45595679 | 45596027 | 0 | 1 | 348 | 0 | -0.9763977 | 0.0092763 | 0.0797982 |
| chr22 | 31276894 | 31277191 | LIMK2 | 0 | + | 31276894 | 31277191 | 0 | 1 | 297 | 0 | 1.1155233 | 0.0000243 | 0.0008295 |
| chr4 | 82984331 | 82984629 | LIN54 | 0 | - | 82984331 | 82984629 | 0 | 1 | 298 | 0 | -1.3323391 | 0.0010425 | 0.0164160 |
| chr6 | 165939788 | 165939837 | LINC00473 | 0 | - | 165939788 | 165939837 | 0 | 1 | 49 | 0 | -3.0240417 | 0.0063648 | 0.0618982 |
| chr20 | 36049862 | 36050960 | LINC00657 | 0 | - | 36049862 | 36050960 | 0 | 1 | 1098 | 0 | 0.8329737 | 0.0000000 | 0.0000001 |
| chr11 | 12979680 | 12979729 | LINC00958 | 0 | - | 12979680 | 12979729 | 0 | 1 | 49 | 0 | -1.3208675 | 0.0010939 | 0.0170767 |
| chr11 | 12979972 | 12980362 | LINC00958 | 0 | - | 12979972 | 12980362 | 0 | 1 | 390 | 0 | -0.8089181 | 0.0000053 | 0.0002309 |
| chr15 | 92882843 | 92883041 | LINC01578 | 0 | + | 92882843 | 92883041 | 0 | 1 | 198 | 0 | 1.2887547 | 0.0004527 | 0.0086019 |
| chr10 | 89245740 | 89251840 | LIPA | 0 | - | 89245740 | 89251840 | 0 | 3 | 54,112,134 | 0,1798,5967 | 1.0848558 | 0.0001750 | 0.0040306 |
| chr18 | 59355656 | 59359226 | LMAN1 | 0 | - | 59355656 | 59359226 | 0 | 2 | 3,196 | 0,3375 | 1.2398493 | 0.0000160 | 0.0005935 |
| chr22 | 50503147 | 50503546 | LMF2 | 0 | - | 50503147 | 50503546 | 0 | 1 | 399 | 0 | -1.0319777 | 0.0030923 | 0.0363370 |
| chr19 | 2430212 | 2430311 | LMNB2 | 0 | - | 2430212 | 2430311 | 0 | 1 | 99 | 0 | 1.3200227 | 0.0043976 | 0.0472911 |
| chr7 | 98192222 | 98192321 | LMTK2 | 0 | + | 98192222 | 98192321 | 0 | 1 | 99 | 0 | -2.1415047 | 0.0063981 | 0.0621214 |
| chr19 | 48498222 | 48498520 | LMTK3 | 0 | - | 48498222 | 48498520 | 0 | 1 | 298 | 0 | -1.1066259 | 0.0034841 | 0.0398419 |
| chr6 | 137824161 | 137824406 | LOC100130476 | 0 | - | 137824161 | 137824406 | 0 | 1 | 245 | 0 | 1.1763910 | 0.0012569 | 0.0188530 |
| chr2 | 87076071 | 87076120 | LOC285074 | 0 | - | 87076071 | 87076120 | 0 | 1 | 49 | 0 | 2.4041461 | 0.0057807 | 0.0578328 |
| chr2 | 87076217 | 87076266 | LOC285074 | 0 | - | 87076217 | 87076266 | 0 | 1 | 49 | 0 | 2.1860534 | 0.0116387 | 0.0934168 |
| chr20 | 41358742 | 41358990 | LPIN3 | 0 | + | 41358742 | 41358990 | 0 | 1 | 248 | 0 | 1.6902578 | 0.0053602 | 0.0547398 |
| chr3 | 188609529 | 188609678 | LPP | 0 | + | 188609529 | 188609678 | 0 | 1 | 149 | 0 | -1.9571615 | 0.0002269 | 0.0049262 |
| chr19 | 4537907 | 4538351 | LRG1 | 0 | - | 4537907 | 4538351 | 0 | 1 | 444 | 0 | -0.6404451 | 0.0013843 | 0.0202278 |
| chr14 | 22875546 | 22876345 | LRP10 | 0 | + | 22875546 | 22876345 | 0 | 1 | 799 | 0 | -1.0995067 | 0.0000002 | 0.0000138 |
| chr14 | 22877783 | 22877932 | LRP10 | 0 | + | 22877783 | 22877932 | 0 | 1 | 149 | 0 | -2.0574234 | 0.0000000 | 0.0000002 |
| chr1 | 46285402 | 46286001 | LRRC41 | 0 | - | 46285402 | 46286001 | 0 | 1 | 599 | 0 | -1.6221858 | 0.0000000 | 0.0000000 |
| chr17 | 50382715 | 50383110 | LRRC59 | 0 | - | 50382715 | 50383110 | 0 | 1 | 395 | 0 | -0.8092428 | 0.0028186 | 0.0338863 |
| chr9 | 128907743 | 128907842 | LRRC8A | 0 | + | 128907743 | 128907842 | 0 | 1 | 99 | 0 | -2.1331025 | 0.0000610 | 0.0017576 |
| chr19 | 7898678 | 7898877 | LRRC8E | 0 | + | 7898678 | 7898877 | 0 | 1 | 199 | 0 | -1.1946944 | 0.0076828 | 0.0700849 |
| chr7 | 102472916 | 102473115 | LRWD1 | 0 | + | 102472916 | 102473115 | 0 | 1 | 199 | 0 | -1.0245494 | 0.0017099 | 0.0234833 |
| chr3 | 194641433 | 194641780 | LSG1 | 0 | - | 194641433 | 194641780 | 0 | 1 | 347 | 0 | -0.8359679 | 0.0008070 | 0.0134411 |
| chr19 | 18306970 | 18307216 | LSM4 | 0 | - | 18306970 | 18307216 | 0 | 1 | 246 | 0 | 0.7551279 | 0.0072225 | 0.0670688 |
| chr17 | 50719664 | 50719713 | LUC7L3 | 0 | + | 50719664 | 50719713 | 0 | 1 | 49 | 0 | 1.1557360 | 0.0049269 | 0.0514218 |
| chr1 | 23088666 | 23091500 | LUZP1 | 0 | - | 23088666 | 23091500 | 0 | 2 | 388,311 | 0,2524 | -0.5302189 | 0.0071501 | 0.0666773 |
| chr22 | 20997401 | 20997996 | LZTR1 | 0 | + | 20997401 | 20997996 | 0 | 1 | 595 | 0 | -0.7329527 | 0.0076261 | 0.0697442 |
| chr1 | 39387422 | 39387521 | MACF1 | 0 | + | 39387422 | 39387521 | 0 | 1 | 99 | 0 | -3.1240557 | 0.0128220 | 0.0995204 |
| chr1 | 39388321 | 39422385 | MACF1 | 0 | + | 39388321 | 39422385 | 0 | 2 | 338,12 | 0,34053 | -1.4209584 | 0.0015927 | 0.0223227 |
| chr6 | 43640359 | 43640507 | MAD2L1BP | 0 | + | 43640359 | 43640507 | 0 | 1 | 148 | 0 | -0.6998124 | 0.0041388 | 0.0451750 |
| chr11 | 47285179 | 47285484 | MADD | 0 | + | 47285179 | 47285484 | 0 | 2 | 16,34 | 0,272 | -3.9741521 | 0.0000000 | 0.0000002 |
| chr17 | 81922531 | 81922729 | MAFG | 0 | - | 81922531 | 81922729 | 0 | 1 | 198 | 0 | 1.7330550 | 0.0000001 | 0.0000078 |
| chr17 | 81922779 | 81923405 | MAFG | 0 | - | 81922779 | 81923405 | 0 | 3 | 279,65,53 | 0,371,574 | 1.9841367 | 0.0000000 | 0.0000000 |
| chrX | 152701215 | 152701510 | MAGEA3 | 0 | + | 152701215 | 152701510 | 0 | 1 | 295 | 0 | -0.9751650 | 0.0001811 | 0.0041299 |
| chrX | 152766930 | 152769667 | MAGEA6 | 0 | - | 152766930 | 152769667 | 0 | 3 | 786,130,14 | 0,866,2724 | -0.7316473 | 0.0008195 | 0.0135657 |
| chr11 | 65500049 | 65501093 | MALAT1 | 0 | + | 65500049 | 65501093 | 0 | 1 | 1044 | 0 | 1.6354918 | 0.0000000 | 0.0000000 |
| chr11 | 65501292 | 65501391 | MALAT1 | 0 | + | 65501292 | 65501391 | 0 | 1 | 99 | 0 | 1.6289492 | 0.0000000 | 0.0000007 |
| chr11 | 65501491 | 65501888 | MALAT1 | 0 | + | 65501491 | 65501888 | 0 | 1 | 397 | 0 | 2.5894371 | 0.0000000 | 0.0000000 |
| chr11 | 65502585 | 65503081 | MALAT1 | 0 | + | 65502585 | 65503081 | 0 | 1 | 496 | 0 | 1.9427324 | 0.0000000 | 0.0000000 |
| chr11 | 65505767 | 65505915 | MALAT1 | 0 | + | 65505767 | 65505915 | 0 | 1 | 148 | 0 | 2.6857622 | 0.0000000 | 0.0000000 |
| chr5 | 179765395 | 179765991 | MAML1 | 0 | + | 179765395 | 179765991 | 0 | 1 | 596 | 0 | -0.7347292 | 0.0035589 | 0.0405316 |
| chr5 | 179766390 | 179766688 | MAML1 | 0 | + | 179766390 | 179766688 | 0 | 1 | 298 | 0 | -0.8669678 | 0.0086517 | 0.0761251 |
| chr1 | 117367862 | 117402260 | MAN1A2 | 0 | + | 117367862 | 117402260 | 0 | 2 | 624,75 | 0,34324 | -1.1633048 | 0.0000085 | 0.0003530 |
| chr15 | 90920198 | 90920247 | MAN2A2 | 0 | + | 90920198 | 90920247 | 0 | 1 | 49 | 0 | -1.7364123 | 0.0018978 | 0.0254560 |
| chr19 | 12666579 | 12666727 | MAN2B1 | 0 | - | 12666579 | 12666727 | 0 | 1 | 148 | 0 | 3.0562461 | 0.0000007 | 0.0000375 |
| chr20 | 37316579 | 37316628 | MANBAL | 0 | + | 37316579 | 37316628 | 0 | 1 | 49 | 0 | -1.9514417 | 0.0078172 | 0.0708333 |
| chr1 | 37799683 | 37800472 | MANEAL | 0 | + | 37799683 | 37800472 | 0 | 1 | 789 | 0 | -0.5750601 | 0.0013273 | 0.0196679 |
| chr19 | 17726506 | 17727104 | MAP1S | 0 | + | 17726506 | 17727104 | 0 | 1 | 598 | 0 | -1.4635574 | 0.0000000 | 0.0000000 |
| chr19 | 17727604 | 17727853 | MAP1S | 0 | + | 17727604 | 17727853 | 0 | 1 | 249 | 0 | -1.8041673 | 0.0000000 | 0.0000003 |
| chr15 | 66386923 | 66387370 | MAP2K1 | 0 | + | 66386923 | 66387370 | 0 | 1 | 447 | 0 | 1.0293147 | 0.0001089 | 0.0028041 |
| chr19 | 4090371 | 4090470 | MAP2K2 | 0 | - | 4090371 | 4090470 | 0 | 1 | 99 | 0 | 1.3117693 | 0.0026710 | 0.0327445 |
| chr19 | 40191993 | 40192689 | MAP3K10 | 0 | + | 40191993 | 40192689 | 0 | 1 | 696 | 0 | -1.0374527 | 0.0001626 | 0.0037953 |
| chr3 | 47916059 | 47918748 | MAP4 | 0 | - | 47916059 | 47918748 | 0 | 2 | 1116,30 | 0,2660 | -1.1827780 | 0.0000002 | 0.0000155 |
| chr2 | 101891542 | 101891691 | MAP4K4 | 0 | + | 101891542 | 101891691 | 0 | 1 | 149 | 0 | -1.7976560 | 0.0000003 | 0.0000172 |
| chr18 | 50663258 | 50664155 | MAPK4 | 0 | + | 50663258 | 50664155 | 0 | 1 | 897 | 0 | -1.2394580 | 0.0000000 | 0.0000000 |
| chr15 | 52046413 | 52046909 | MAPK6 | 0 | + | 52046413 | 52046909 | 0 | 1 | 496 | 0 | -0.7301884 | 0.0087867 | 0.0769387 |
| chr15 | 41823895 | 41824044 | MAPKBP1 | 0 | + | 41823895 | 41824044 | 0 | 1 | 149 | 0 | 1.7019437 | 0.0014981 | 0.0213084 |
| chr1 | 220748629 | 220748678 | MARC2 | 0 | + | 220748629 | 220748678 | 0 | 1 | 49 | 0 | -2.0463972 | 0.0021858 | 0.0281513 |
| chr19 | 8430864 | 8438675 | MARCH2 | 0 | + | 8430864 | 8438675 | 0 | 2 | 4,288 | 0,7524 | -1.0759717 | 0.0000720 | 0.0020172 |
| chr6 | 113860011 | 113860357 | MARCKS | 0 | + | 113860011 | 113860357 | 0 | 1 | 346 | 0 | -0.7905350 | 0.0079754 | 0.0718500 |
| chr1 | 32335048 | 32335995 | MARCKSL1 | 0 | - | 32335048 | 32335995 | 0 | 2 | 50,49 | 0,899 | 0.9647298 | 0.0067291 | 0.0643010 |
| chr5 | 67166797 | 67166896 | MAST4 | 0 | + | 67166797 | 67166896 | 0 | 1 | 99 | 0 | 1.8934290 | 0.0050710 | 0.0525615 |
| chr19 | 3785163 | 3785261 | MATK | 0 | - | 3785163 | 3785261 | 0 | 1 | 98 | 0 | 2.7839433 | 0.0000000 | 0.0000000 |
| chr20 | 3865849 | 3866148 | MAVS | 0 | + | 3865849 | 3866148 | 0 | 1 | 299 | 0 | 1.2665471 | 0.0000442 | 0.0013593 |
| chr16 | 29807572 | 29808594 | MAZ | 0 | + | 29807572 | 29808594 | 0 | 3 | 257,64,25 | 0,658,998 | 0.7805806 | 0.0002734 | 0.0057565 |
| chr3 | 129436949 | 129437756 | MBD4 | 0 | - | 129436949 | 129437756 | 0 | 2 | 360,37 | 0,771 | -0.7183887 | 0.0095988 | 0.0816802 |
| chr13 | 97275887 | 97276285 | MBNL2 | 0 | + | 97275887 | 97276285 | 0 | 1 | 398 | 0 | -0.7953304 | 0.0106333 | 0.0877607 |
| chr16 | 84101720 | 84116893 | MBTPS1 | 0 | - | 84101720 | 84116893 | 0 | 2 | 388,159 | 0,15015 | -0.6946470 | 0.0122239 | 0.0962392 |
| chr11 | 119308673 | 119310398 | MCAM | 0 | - | 119308673 | 119310398 | 0 | 2 | 1243,50 | 0,1676 | -0.5976988 | 0.0087117 | 0.0764296 |
| chr2 | 46903069 | 46903318 | MCFD2 | 0 | - | 46903069 | 46903318 | 0 | 1 | 249 | 0 | -1.1904570 | 0.0004883 | 0.0090647 |
| chr1 | 150575647 | 150575696 | MCL1 | 0 | - | 150575647 | 150575696 | 0 | 1 | 49 | 0 | -1.4104530 | 0.0004241 | 0.0081779 |
| chr3 | 127598406 | 127599399 | MCM2 | 0 | + | 127598406 | 127599399 | 0 | 2 | 67,82 | 0,912 | 1.6546071 | 0.0000000 | 0.0000004 |
| chr8 | 47960948 | 47960997 | MCM4 | 0 | + | 47960948 | 47960997 | 0 | 1 | 49 | 0 | 1.7961779 | 0.0114763 | 0.0924901 |
| chr12 | 68841613 | 68841662 | MDM2 | 0 | + | 68841613 | 68841662 | 0 | 1 | 49 | 0 | -1.3922754 | 0.0078055 | 0.0707776 |
| chr6 | 83407848 | 83431070 | ME1 | 0 | - | 83407848 | 83431070 | 0 | 2 | 54,194 | 0,23029 | 1.2573070 | 0.0012188 | 0.0184371 |
| chrX | 154030256 | 154030904 | MECP2 | 0 | - | 154030256 | 154030904 | 0 | 1 | 648 | 0 | -0.6513057 | 0.0000210 | 0.0007497 |
| chrX | 154031205 | 154031404 | MECP2 | 0 | - | 154031205 | 154031404 | 0 | 1 | 199 | 0 | -1.5328817 | 0.0000235 | 0.0008100 |
| chr17 | 39409084 | 39410482 | MED1 | 0 | - | 39409084 | 39410482 | 0 | 1 | 1398 | 0 | -0.7421616 | 0.0051703 | 0.0533612 |
| chr17 | 62010800 | 62011198 | MED13 | 0 | - | 62010800 | 62011198 | 0 | 1 | 398 | 0 | -1.2677523 | 0.0000029 | 0.0001369 |
| chr12 | 116008872 | 116009070 | MED13L | 0 | - | 116008872 | 116009070 | 0 | 1 | 198 | 0 | -1.5073360 | 0.0071032 | 0.0665413 |
| chr22 | 20586738 | 20586886 | MED15 | 0 | + | 20586738 | 20586886 | 0 | 1 | 148 | 0 | -1.3340493 | 0.0000163 | 0.0006025 |
| chr19 | 16576350 | 16576846 | MED26 | 0 | - | 16576350 | 16576846 | 0 | 1 | 496 | 0 | -0.7476902 | 0.0051372 | 0.0531262 |
| chr1 | 43384908 | 43385007 | MED8 | 0 | - | 43384908 | 43385007 | 0 | 1 | 99 | 0 | -1.2673190 | 0.0044863 | 0.0478436 |
| chr15 | 99645594 | 99645643 | MEF2A | 0 | + | 99645594 | 99645643 | 0 | 1 | 49 | 0 | 2.3972550 | 0.0120829 | 0.0955289 |
| chr7 | 100428840 | 100428938 | MEPCE | 0 | + | 100428840 | 100428938 | 0 | 1 | 98 | 0 | 2.5273086 | 0.0001657 | 0.0038461 |
| chr15 | 81001200 | 81001397 | MESDC1 | 0 | + | 81001200 | 81001397 | 0 | 1 | 197 | 0 | -1.1492110 | 0.0001412 | 0.0034096 |
| chr7 | 116699197 | 116699644 | MET | 0 | + | 116699197 | 116699644 | 0 | 1 | 447 | 0 | -0.7140057 | 0.0018359 | 0.0248279 |
| chr7 | 116699695 | 116700242 | MET | 0 | + | 116699695 | 116700242 | 0 | 1 | 547 | 0 | -1.1565204 | 0.0000426 | 0.0013171 |
| chr7 | 116796743 | 116796941 | MET | 0 | + | 116796743 | 116796941 | 0 | 1 | 198 | 0 | -1.3476938 | 0.0026592 | 0.0326395 |
| chr7 | 116797689 | 116797938 | MET | 0 | + | 116797689 | 116797938 | 0 | 1 | 249 | 0 | -0.9286009 | 0.0048645 | 0.0509763 |
| chr7 | 116797988 | 116798237 | MET | 0 | + | 116797988 | 116798237 | 0 | 1 | 249 | 0 | -1.1825452 | 0.0000391 | 0.0012306 |
| chr12 | 95513666 | 95514258 | METAP2 | 0 | + | 95513666 | 95514258 | 0 | 1 | 592 | 0 | -0.5861097 | 0.0110179 | 0.0899922 |
| chr17 | 62426326 | 62426474 | METTL2A | 0 | + | 62426326 | 62426474 | 0 | 1 | 148 | 0 | -1.7102562 | 0.0001833 | 0.0041750 |
| chr7 | 128479167 | 128479414 | METTL2B | 0 | + | 128479167 | 128479414 | 0 | 1 | 247 | 0 | -1.2098081 | 0.0005618 | 0.0101229 |
| chr15 | 43817417 | 43824606 | MFAP1 | 0 | - | 43817417 | 43824606 | 0 | 2 | 32,116 | 0,7074 | 2.4308063 | 0.0000008 | 0.0000463 |
| chr5 | 154053058 | 154053807 | MFAP3 | 0 | + | 154053058 | 154053807 | 0 | 1 | 749 | 0 | -0.6109253 | 0.0007684 | 0.0129194 |
| chr8 | 8891292 | 8891390 | MFHAS1 | 0 | - | 8891292 | 8891390 | 0 | 1 | 98 | 0 | -1.5531212 | 0.0004906 | 0.0090915 |
| chr3 | 197002683 | 197002881 | MFI2 | 0 | - | 197002683 | 197002881 | 0 | 1 | 198 | 0 | 0.9590075 | 0.0081515 | 0.0729649 |
| chr1 | 11980430 | 11982113 | MFN2 | 0 | + | 11980430 | 11982113 | 0 | 2 | 55,144 | 0,1540 | 1.1493311 | 0.0024457 | 0.0307546 |
| chr8 | 144509084 | 144509279 | MFSD3 | 0 | + | 144509084 | 144509279 | 0 | 1 | 195 | 0 | -1.3461763 | 0.0000488 | 0.0014665 |
| chr5 | 180791638 | 180792634 | MGAT1 | 0 | - | 180791638 | 180792634 | 0 | 1 | 996 | 0 | -0.9567687 | 0.0000000 | 0.0000003 |
| chr14 | 49621954 | 49622594 | MGAT2 | 0 | + | 49621954 | 49622594 | 0 | 1 | 640 | 0 | -0.8379176 | 0.0000022 | 0.0001092 |
| chr10 | 101798128 | 101800244 | MGEA5 | 0 | - | 101798128 | 101800244 | 0 | 3 | 27,614,3 | 0,714,2114 | -0.9072514 | 0.0000005 | 0.0000306 |
| chr22 | 17788848 | 17789047 | MICAL3 | 0 | - | 17788848 | 17789047 | 0 | 1 | 199 | 0 | -1.2791182 | 0.0096436 | 0.0819456 |
| chr22 | 17790046 | 17790294 | MICAL3 | 0 | - | 17790046 | 17790294 | 0 | 1 | 248 | 0 | -1.1794068 | 0.0069333 | 0.0655265 |
| chr22 | 17818312 | 17818411 | MICAL3 | 0 | - | 17818312 | 17818411 | 0 | 1 | 99 | 0 | -1.7728514 | 0.0018195 | 0.0246425 |
| chrX | 10449305 | 10454906 | MID1 | 0 | - | 10449305 | 10454906 | 0 | 2 | 412,37 | 0,5565 | -1.3255854 | 0.0000101 | 0.0004046 |
| chrX | 38804938 | 38805535 | MID1IP1 | 0 | + | 38804938 | 38805535 | 0 | 1 | 597 | 0 | -0.7860961 | 0.0000008 | 0.0000425 |
| chr19 | 1258146 | 1258245 | MIDN | 0 | + | 1258146 | 1258245 | 0 | 1 | 99 | 0 | -1.5826300 | 0.0002956 | 0.0061253 |
| chr22 | 39513567 | 39514360 | MIEF1 | 0 | + | 39513567 | 39514360 | 0 | 1 | 793 | 0 | -0.5922558 | 0.0000645 | 0.0018397 |
| chr17 | 39729344 | 39729393 | MIEN1 | 0 | - | 39729344 | 39729393 | 0 | 1 | 49 | 0 | -1.5425526 | 0.0001741 | 0.0040167 |
| chr17 | 5488914 | 5489512 | MIS12 | 0 | + | 5488914 | 5489512 | 0 | 1 | 598 | 0 | -0.7110946 | 0.0014992 | 0.0213084 |
| chr21 | 32278869 | 32279016 | MIS18A | 0 | - | 32278869 | 32279016 | 0 | 1 | 147 | 0 | 1.3060361 | 0.0092711 | 0.0797917 |
| chr19 | 751146 | 757699 | MISP | 0 | + | 751146 | 757699 | 0 | 2 | 26,810 | 0,5744 | -1.1459439 | 0.0000000 | 0.0000000 |
| chr19 | 757945 | 758191 | MISP | 0 | + | 757945 | 758191 | 0 | 1 | 246 | 0 | -1.9788429 | 0.0000000 | 0.0000000 |
| chr3 | 69959415 | 69965190 | MITF | 0 | + | 69959415 | 69965190 | 0 | 2 | 6,344 | 0,5432 | -1.1062677 | 0.0003383 | 0.0068370 |
| chr10 | 128098160 | 128098359 | MKI67 | 0 | - | 128098160 | 128098359 | 0 | 1 | 199 | 0 | -1.3290323 | 0.0078018 | 0.0707776 |
| chr10 | 128098910 | 128099059 | MKI67 | 0 | - | 128098910 | 128099059 | 0 | 1 | 149 | 0 | -1.3921221 | 0.0097805 | 0.0828368 |
| chr10 | 128104088 | 128104137 | MKI67 | 0 | - | 128104088 | 128104137 | 0 | 1 | 49 | 0 | -2.5683935 | 0.0055632 | 0.0561840 |
| chr10 | 128106337 | 128106386 | MKI67 | 0 | - | 128106337 | 128106386 | 0 | 1 | 49 | 0 | -2.6280656 | 0.0005283 | 0.0096425 |
| chr12 | 120696318 | 120696717 | MLEC | 0 | + | 120696318 | 120696717 | 0 | 1 | 399 | 0 | -0.8010931 | 0.0002494 | 0.0053643 |
| chr16 | 2208489 | 2209368 | MLST8 | 0 | + | 2208489 | 2209368 | 0 | 2 | 125,610 | 0,270 | -0.9524571 | 0.0000001 | 0.0000040 |
| chr12 | 122142216 | 122143012 | MLXIP | 0 | + | 122142216 | 122143012 | 0 | 1 | 796 | 0 | -1.0319038 | 0.0000152 | 0.0005697 |
| chr22 | 27796776 | 27798068 | MN1 | 0 | - | 27796776 | 27798068 | 0 | 1 | 1292 | 0 | -1.7006335 | 0.0000000 | 0.0000000 |
| chr22 | 27799709 | 27799907 | MN1 | 0 | - | 27799709 | 27799907 | 0 | 1 | 198 | 0 | -1.4675651 | 0.0043457 | 0.0470367 |
| chr22 | 27800306 | 27800653 | MN1 | 0 | - | 27800306 | 27800653 | 0 | 1 | 347 | 0 | -0.9475837 | 0.0000426 | 0.0013171 |
| chr17 | 2386606 | 2386854 | MNT | 0 | - | 2386606 | 2386854 | 0 | 1 | 248 | 0 | -1.4051494 | 0.0001904 | 0.0042840 |
| chr16 | 77198951 | 77199198 | MON1B | 0 | + | 77198951 | 77199198 | 0 | 1 | 247 | 0 | -1.1132240 | 0.0038245 | 0.0425919 |
| chr1 | 112700597 | 112700696 | MOV10 | 0 | + | 112700597 | 112700696 | 0 | 1 | 99 | 0 | -1.6605649 | 0.0011155 | 0.0172859 |
| chr14 | 67279192 | 67292512 | MPP5 | 0 | + | 67279192 | 67292512 | 0 | 2 | 346,2 | 0,13319 | -1.1185866 | 0.0000061 | 0.0002613 |
| chr4 | 6640785 | 6641181 | MRFAP1 | 0 | + | 6640785 | 6641181 | 0 | 1 | 396 | 0 | -0.8001239 | 0.0000391 | 0.0012306 |
| chr11 | 59806676 | 59806824 | MRPL16 | 0 | - | 59806676 | 59806824 | 0 | 1 | 148 | 0 | -1.1302294 | 0.0046102 | 0.0489056 |
| chr11 | 6681962 | 6682109 | MRPL17 | 0 | - | 6681962 | 6682109 | 0 | 1 | 147 | 0 | -0.8931815 | 0.0000001 | 0.0000087 |
| chr17 | 50367914 | 50368243 | MRPL27 | 0 | - | 50367914 | 50368243 | 0 | 1 | 329 | 0 | -0.5706364 | 0.0000074 | 0.0003111 |
| chr19 | 17306407 | 17306650 | MRPL34 | 0 | + | 17306407 | 17306650 | 0 | 1 | 243 | 0 | -0.7944800 | 0.0000004 | 0.0000260 |
| chr5 | 1798573 | 1798716 | MRPL36 | 0 | - | 1798573 | 1798716 | 0 | 1 | 143 | 0 | -0.5673321 | 0.0000287 | 0.0009540 |
| chr19 | 3767373 | 3767517 | MRPL54 | 0 | + | 3767373 | 3767517 | 0 | 1 | 144 | 0 | -1.2016211 | 0.0011003 | 0.0171174 |
| chr9 | 135503824 | 135504073 | MRPS2 | 0 | + | 135503824 | 135504073 | 0 | 1 | 249 | 0 | -1.3010203 | 0.0000030 | 0.0001417 |
| chr9 | 135504174 | 135504423 | MRPS2 | 0 | + | 135504174 | 135504423 | 0 | 1 | 249 | 0 | -0.7855564 | 0.0000754 | 0.0020970 |
| chr1 | 150293842 | 150294090 | MRPS21 | 0 | + | 150293842 | 150294090 | 0 | 1 | 248 | 0 | 1.1482062 | 0.0000000 | 0.0000000 |
| chr5 | 72219954 | 72220250 | MRPS27 | 0 | - | 72219954 | 72220250 | 0 | 1 | 296 | 0 | -1.0180147 | 0.0034674 | 0.0397385 |
| chr13 | 40766814 | 40767012 | MRPS31 | 0 | - | 40766814 | 40767012 | 0 | 1 | 198 | 0 | -1.3513060 | 0.0054617 | 0.0554060 |
| chr12 | 27710823 | 27714796 | MRPS35 | 0 | + | 27710823 | 27714796 | 0 | 2 | 133,17 | 0,3957 | 1.6187259 | 0.0002047 | 0.0045270 |
| chr2 | 95117928 | 95121814 | MRPS5 | 0 | - | 95117928 | 95121814 | 0 | 2 | 18,81 | 0,3806 | 2.0048910 | 0.0002782 | 0.0058416 |
| chr17 | 75266083 | 75266230 | MRPS7 | 0 | + | 75266083 | 75266230 | 0 | 1 | 147 | 0 | -1.3744829 | 0.0000784 | 0.0021614 |
| chr1 | 19251630 | 19251729 | MRTO4 | 0 | + | 19251630 | 19251729 | 0 | 1 | 99 | 0 | 1.8200091 | 0.0024208 | 0.0305050 |
| chr9 | 100442075 | 100442173 | MSANTD3 | 0 | + | 100442075 | 100442173 | 0 | 1 | 98 | 0 | -1.1042146 | 0.0069900 | 0.0658701 |
| chr2 | 47799124 | 47799671 | MSH6 | 0 | + | 47799124 | 47799671 | 0 | 1 | 547 | 0 | -0.7449824 | 0.0002248 | 0.0048932 |
| chr17 | 40122537 | 40131552 | MSL1 | 0 | + | 40122537 | 40131552 | 0 | 5 | 224,96,224,383,16 | 0,748,3646,6708,9000 | -0.5025226 | 0.0048168 | 0.0506520 |
| chr12 | 65464178 | 65464475 | MSRB3 | 0 | + | 65464178 | 65464475 | 0 | 1 | 297 | 0 | -1.9848247 | 0.0000000 | 0.0000039 |
| chr12 | 65464624 | 65464922 | MSRB3 | 0 | + | 65464624 | 65464922 | 0 | 1 | 298 | 0 | -1.2049681 | 0.0010323 | 0.0162967 |
| chr3 | 49902482 | 49902978 | MST1R | 0 | - | 49902482 | 49902978 | 0 | 1 | 496 | 0 | -1.0544572 | 0.0008257 | 0.0136545 |
| chr3 | 49903029 | 49903376 | MST1R | 0 | - | 49903029 | 49903376 | 0 | 1 | 347 | 0 | -1.2630081 | 0.0040627 | 0.0445594 |
| chr11 | 62601445 | 62601840 | MTA2 | 0 | - | 62601445 | 62601840 | 0 | 1 | 395 | 0 | 1.5061458 | 0.0000000 | 0.0000000 |
| chr9 | 21865424 | 21865721 | MTAP | 0 | + | 21865424 | 21865721 | 0 | 1 | 297 | 0 | 1.1043463 | 0.0039692 | 0.0437990 |
| chr18 | 8784269 | 8784668 | MTCL1 | 0 | + | 8784269 | 8784668 | 0 | 1 | 399 | 0 | -1.9377380 | 0.0000000 | 0.0000000 |
| chr8 | 97724635 | 97725033 | MTDH | 0 | + | 97724635 | 97725033 | 0 | 1 | 398 | 0 | -1.1840295 | 0.0000503 | 0.0015002 |
| chr20 | 62202108 | 62202505 | MTG2 | 0 | + | 62202108 | 62202505 | 0 | 1 | 397 | 0 | -1.3331678 | 0.0000357 | 0.0011503 |
| chr14 | 64388341 | 64400814 | MTHFD1 | 0 | + | 64388341 | 64400814 | 0 | 2 | 128,22 | 0,12452 | -1.0683045 | 0.0091170 | 0.0789911 |
| chr13 | 27440315 | 27440413 | MTIF3 | 0 | - | 27440315 | 27440413 | 0 | 1 | 98 | 0 | -1.4163271 | 0.0059901 | 0.0592723 |
| chr17 | 58495562 | 58496308 | MTMR4 | 0 | - | 58495562 | 58496308 | 0 | 1 | 746 | 0 | -0.6526923 | 0.0104378 | 0.0868454 |
| chr16 | 70663344 | 70664239 | MTSS1L | 0 | - | 70663344 | 70664239 | 0 | 1 | 895 | 0 | -0.6274066 | 0.0010585 | 0.0166389 |
| chr8 | 17753992 | 17754191 | MTUS1 | 0 | - | 17753992 | 17754191 | 0 | 1 | 199 | 0 | -1.4152526 | 0.0123505 | 0.0968208 |
| chr2 | 176269445 | 176269593 | MTX2 | 0 | + | 176269445 | 176269593 | 0 | 1 | 148 | 0 | 1.5594668 | 0.0002805 | 0.0058782 |
| chr19 | 8945598 | 8945747 | MUC16 | 0 | - | 8945598 | 8945747 | 0 | 1 | 149 | 0 | -2.3744170 | 0.0005233 | 0.0095723 |
| chr19 | 8953442 | 8953591 | MUC16 | 0 | - | 8953442 | 8953591 | 0 | 1 | 149 | 0 | -2.4688978 | 0.0000450 | 0.0013705 |
| chr19 | 8958638 | 8958837 | MUC16 | 0 | - | 8958638 | 8958837 | 0 | 1 | 199 | 0 | -1.1933858 | 0.0057099 | 0.0573934 |
| chr19 | 8960537 | 8960586 | MUC16 | 0 | - | 8960537 | 8960586 | 0 | 1 | 49 | 0 | -3.4085439 | 0.0043590 | 0.0471525 |
| chr19 | 8961286 | 8961485 | MUC16 | 0 | - | 8961286 | 8961485 | 0 | 1 | 199 | 0 | -2.0221178 | 0.0001801 | 0.0041135 |
| chr19 | 8966632 | 8966831 | MUC16 | 0 | - | 8966632 | 8966831 | 0 | 1 | 199 | 0 | -2.3999743 | 0.0000274 | 0.0009133 |
| chr1 | 20500185 | 20500627 | MUL1 | 0 | - | 20500185 | 20500627 | 0 | 1 | 442 | 0 | -0.8325049 | 0.0007963 | 0.0133234 |
| chr1 | 20501119 | 20501266 | MUL1 | 0 | - | 20501119 | 20501266 | 0 | 1 | 147 | 0 | -1.6450908 | 0.0000642 | 0.0018344 |
| chr19 | 1360340 | 1360389 | MUM1 | 0 | + | 1360340 | 1360389 | 0 | 1 | 49 | 0 | -1.9279448 | 0.0052319 | 0.0537712 |
| chr4 | 2250511 | 2251190 | MXD4 | 0 | - | 2250511 | 2251190 | 0 | 2 | 191,107 | 0,573 | 1.3637974 | 0.0000006 | 0.0000354 |
| chr19 | 53875638 | 53875986 | MYADM | 0 | + | 53875638 | 53875986 | 0 | 1 | 348 | 0 | -1.0829901 | 0.0000508 | 0.0015127 |
| chr17 | 4539541 | 4539888 | MYBBP1A | 0 | - | 4539541 | 4539888 | 0 | 1 | 347 | 0 | -0.9190199 | 0.0073542 | 0.0678746 |
| chr8 | 127736217 | 127736512 | MYC | 0 | + | 127736217 | 127736512 | 0 | 1 | 295 | 0 | 0.7435460 | 0.0044494 | 0.0476471 |
| chr19 | 4664896 | 4670355 | MYDGF | 0 | - | 4664896 | 4670355 | 0 | 3 | 42,51,195 | 0,3699,5265 | 0.5703659 | 0.0003074 | 0.0063173 |
| chr17 | 8504700 | 8504849 | MYH10 | 0 | - | 8504700 | 8504849 | 0 | 1 | 149 | 0 | -2.1873198 | 0.0000219 | 0.0007732 |
| chr17 | 8623087 | 8630711 | MYH10 | 0 | - | 8623087 | 8630711 | 0 | 2 | 191,58 | 0,7567 | -1.4991069 | 0.0002105 | 0.0046374 |
| chr22 | 36294183 | 36295014 | MYH9 | 0 | - | 36294183 | 36295014 | 0 | 2 | 116,83 | 0,749 | -2.9022667 | 0.0000000 | 0.0000000 |
| chr22 | 36296966 | 36298919 | MYH9 | 0 | - | 36296966 | 36298919 | 0 | 2 | 49,1 | 0,1953 | -1.9193586 | 0.0079269 | 0.0715552 |
| chr22 | 36306033 | 36306543 | MYH9 | 0 | - | 36306033 | 36306543 | 0 | 2 | 19,130 | 0,381 | -4.8884510 | 0.0000000 | 0.0000000 |
| chr22 | 36316586 | 36316635 | MYH9 | 0 | - | 36316586 | 36316635 | 0 | 1 | 49 | 0 | -6.8902281 | 0.0000000 | 0.0000000 |
| chr22 | 36318223 | 36318272 | MYH9 | 0 | - | 36318223 | 36318272 | 0 | 1 | 49 | 0 | -4.4979885 | 0.0000016 | 0.0000813 |
| chr22 | 36320220 | 36320318 | MYH9 | 0 | - | 36320220 | 36320318 | 0 | 1 | 98 | 0 | -2.7333725 | 0.0000000 | 0.0000020 |
| chr12 | 56161486 | 56161535 | MYL6 | 0 | + | 56161486 | 56161535 | 0 | 1 | 49 | 0 | 2.0072577 | 0.0005705 | 0.0102585 |
| chr5 | 16701070 | 16701468 | MYO10 | 0 | - | 16701070 | 16701468 | 0 | 1 | 398 | 0 | -1.0115210 | 0.0000009 | 0.0000515 |
| chr17 | 29073859 | 29073908 | MYO18A | 0 | - | 29073859 | 29073908 | 0 | 1 | 49 | 0 | -2.3161758 | 0.0000053 | 0.0002309 |
| chr17 | 29165951 | 29166050 | MYO18A | 0 | - | 29165951 | 29166050 | 0 | 1 | 99 | 0 | -1.8980470 | 0.0000046 | 0.0002051 |
| chr17 | 29166101 | 29166998 | MYO18A | 0 | - | 29166101 | 29166998 | 0 | 1 | 897 | 0 | -1.6336919 | 0.0000000 | 0.0000000 |
| chr17 | 1464386 | 1464934 | MYO1C | 0 | - | 1464386 | 1464934 | 0 | 1 | 548 | 0 | -1.5751197 | 0.0000069 | 0.0002929 |
| chr17 | 1465185 | 1467272 | MYO1C | 0 | - | 1465185 | 1467272 | 0 | 2 | 568,31 | 0,2057 | -1.3095528 | 0.0000382 | 0.0012134 |
| chr19 | 17075831 | 17102213 | MYO9B | 0 | + | 17075831 | 17102213 | 0 | 2 | 44,554 | 0,25829 | -2.2195588 | 0.0000000 | 0.0000000 |
| chr19 | 17195162 | 17195311 | MYO9B | 0 | + | 17195162 | 17195311 | 0 | 1 | 149 | 0 | -1.7476687 | 0.0026257 | 0.0323444 |
| chr19 | 17195361 | 17195460 | MYO9B | 0 | + | 17195361 | 17195460 | 0 | 1 | 99 | 0 | -2.2775624 | 0.0006067 | 0.0108242 |
| chr10 | 68121554 | 68122303 | MYPN | 0 | + | 68121554 | 68122303 | 0 | 1 | 749 | 0 | -1.6143926 | 0.0000000 | 0.0000000 |
| chr4 | 139388798 | 139388847 | NAA15 | 0 | + | 139388798 | 139388847 | 0 | 1 | 49 | 0 | -2.4129686 | 0.0043794 | 0.0472594 |
| chr4 | 139388897 | 139388996 | NAA15 | 0 | + | 139388897 | 139388996 | 0 | 1 | 99 | 0 | -2.5235041 | 0.0000387 | 0.0012236 |
| chr17 | 42543306 | 42543455 | NAGLU | 0 | + | 42543306 | 42543455 | 0 | 1 | 149 | 0 | -1.4935513 | 0.0106119 | 0.0876272 |
| chr17 | 42543704 | 42544051 | NAGLU | 0 | + | 42543704 | 42544051 | 0 | 1 | 347 | 0 | -0.8978100 | 0.0066962 | 0.0640540 |
| chr16 | 730002 | 730051 | NARFL | 0 | - | 730002 | 730051 | 0 | 1 | 49 | 0 | -2.3890881 | 0.0087943 | 0.0769683 |
| chr11 | 34143509 | 34146462 | NAT10 | 0 | + | 34143509 | 34146462 | 0 | 2 | 20,379 | 0,2575 | -0.7840542 | 0.0121015 | 0.0955669 |
| chr4 | 2061120 | 2064164 | NAT8L | 0 | + | 2061120 | 2064164 | 0 | 2 | 43,405 | 0,2640 | 0.7491932 | 0.0001739 | 0.0040159 |
| chr17 | 74771265 | 74771755 | NAT9 | 0 | - | 74771265 | 74771755 | 0 | 1 | 490 | 0 | -0.7703986 | 0.0001611 | 0.0037738 |
| chr1 | 35560444 | 35560642 | NCDN | 0 | + | 35560444 | 35560642 | 0 | 1 | 198 | 0 | -1.4188542 | 0.0012546 | 0.0188375 |
| chr12 | 49795512 | 49796011 | NCKAP5L | 0 | - | 49795512 | 49796011 | 0 | 1 | 499 | 0 | -0.7504568 | 0.0019872 | 0.0263237 |
| chr3 | 48674339 | 48674538 | NCKIPSD | 0 | - | 48674339 | 48674538 | 0 | 1 | 199 | 0 | -1.4162593 | 0.0023972 | 0.0303133 |
| chr20 | 47635940 | 47635989 | NCOA3 | 0 | + | 47635940 | 47635989 | 0 | 1 | 49 | 0 | -1.6417624 | 0.0085207 | 0.0753014 |
| chr10 | 46010275 | 46011120 | NCOA4 | 0 | - | 46010275 | 46011120 | 0 | 1 | 845 | 0 | -1.6966368 | 0.0000000 | 0.0000002 |
| chr10 | 46015255 | 46019404 | NCOA4 | 0 | - | 46015255 | 46019404 | 0 | 3 | 12,155,82 | 0,1285,4068 | 1.2832640 | 0.0000141 | 0.0005361 |
| chr6 | 125889322 | 125889520 | NCOA7 | 0 | + | 125889322 | 125889520 | 0 | 1 | 198 | 0 | -1.6089196 | 0.0053185 | 0.0544114 |
| chr17 | 16101311 | 16101610 | NCOR1 | 0 | - | 16101311 | 16101610 | 0 | 1 | 299 | 0 | -1.2573455 | 0.0000923 | 0.0024380 |
| chr12 | 124398121 | 124398170 | NCOR2 | 0 | - | 124398121 | 124398170 | 0 | 1 | 49 | 0 | 2.5066744 | 0.0061778 | 0.0607058 |
| chr5 | 150555418 | 150556116 | NDST1 | 0 | + | 150555418 | 150556116 | 0 | 1 | 698 | 0 | 0.6423038 | 0.0058260 | 0.0581568 |
| chr16 | 23587229 | 23596318 | NDUFAB1 | 0 | - | 23587229 | 23596318 | 0 | 2 | 91,196 | 0,8894 | 0.9621747 | 0.0066836 | 0.0640356 |
| chr3 | 179604787 | 179604932 | NDUFB5 | 0 | + | 179604787 | 179604932 | 0 | 1 | 145 | 0 | 1.0808927 | 0.0030362 | 0.0359573 |
| chr11 | 65423648 | 65423897 | NEAT1 | 0 | + | 65423648 | 65423897 | 0 | 1 | 249 | 0 | 1.2793973 | 0.0000003 | 0.0000190 |
| chr11 | 65423948 | 65424646 | NEAT1 | 0 | + | 65423948 | 65424646 | 0 | 1 | 698 | 0 | 1.6685809 | 0.0000000 | 0.0000000 |
| chr11 | 65424797 | 65424896 | NEAT1 | 0 | + | 65424797 | 65424896 | 0 | 1 | 99 | 0 | 2.6860274 | 0.0000010 | 0.0000541 |
| chr11 | 65425147 | 65425296 | NEAT1 | 0 | + | 65425147 | 65425296 | 0 | 1 | 149 | 0 | 2.9438816 | 0.0000000 | 0.0000000 |
| chr11 | 65425397 | 65425446 | NEAT1 | 0 | + | 65425397 | 65425446 | 0 | 1 | 49 | 0 | 5.3173031 | 0.0000001 | 0.0000044 |
| chr11 | 65425947 | 65425996 | NEAT1 | 0 | + | 65425947 | 65425996 | 0 | 1 | 49 | 0 | 3.4963211 | 0.0001545 | 0.0036662 |
| chr11 | 65426297 | 65426346 | NEAT1 | 0 | + | 65426297 | 65426346 | 0 | 1 | 49 | 0 | 2.1826586 | 0.0058044 | 0.0580157 |
| chr1 | 16459594 | 16459990 | NECAP2 | 0 | + | 16459594 | 16459990 | 0 | 1 | 396 | 0 | -1.0210932 | 0.0025962 | 0.0321548 |
| chr22 | 29490644 | 29491090 | NEFH | 0 | + | 29490644 | 29491090 | 0 | 1 | 446 | 0 | -0.5696436 | 0.0096311 | 0.0819160 |
| chr12 | 54292307 | 54292805 | NFE2 | 0 | - | 54292307 | 54292805 | 0 | 1 | 498 | 0 | -0.9567453 | 0.0024076 | 0.0303597 |
| chr17 | 48048423 | 48050965 | NFE2L1 | 0 | + | 48048423 | 48050965 | 0 | 2 | 40,359 | 0,2184 | -0.5000252 | 0.0048480 | 0.0508419 |
| chr17 | 48058562 | 48058859 | NFE2L1 | 0 | + | 48058562 | 48058859 | 0 | 1 | 297 | 0 | -0.7109606 | 0.0003026 | 0.0062410 |
| chr2 | 177231550 | 177231649 | NFE2L2 | 0 | - | 177231550 | 177231649 | 0 | 1 | 99 | 0 | -1.1290649 | 0.0080192 | 0.0721368 |
| chr19 | 3366593 | 3382136 | NFIC | 0 | + | 3366593 | 3382136 | 0 | 2 | 74,425 | 0,15119 | -0.8917124 | 0.0016044 | 0.0224688 |
| chr9 | 33294738 | 33301267 | NFX1 | 0 | + | 33294738 | 33301267 | 0 | 2 | 690,5 | 0,6525 | -0.8067495 | 0.0002238 | 0.0048895 |
| chr15 | 90271331 | 90272064 | NGRN | 0 | + | 90271331 | 90272064 | 0 | 1 | 733 | 0 | -1.3264696 | 0.0000000 | 0.0000000 |
| chr15 | 22838690 | 22838937 | NIPA2 | 0 | + | 22838690 | 22838937 | 0 | 1 | 247 | 0 | 0.9339021 | 0.0018625 | 0.0250746 |
| chr5 | 36975808 | 36976306 | NIPBL | 0 | + | 36975808 | 36976306 | 0 | 1 | 498 | 0 | -0.7940591 | 0.0003726 | 0.0074153 |
| chr5 | 36985030 | 36986077 | NIPBL | 0 | + | 36985030 | 36986077 | 0 | 1 | 1047 | 0 | -0.6336026 | 0.0020641 | 0.0269776 |
| chr22 | 29555356 | 29555554 | NIPSNAP1 | 0 | - | 29555356 | 29555554 | 0 | 1 | 198 | 0 | -0.8408497 | 0.0125117 | 0.0977405 |
| chr3 | 52487816 | 52487915 | NISCH | 0 | + | 52487816 | 52487915 | 0 | 1 | 99 | 0 | -2.4953423 | 0.0000601 | 0.0017362 |
| chr3 | 52487965 | 52488064 | NISCH | 0 | + | 52487965 | 52488064 | 0 | 1 | 99 | 0 | -2.6186663 | 0.0000249 | 0.0008460 |
| chr3 | 100334767 | 100339153 | NIT2 | 0 | + | 100334767 | 100339153 | 0 | 2 | 32,67 | 0,4320 | 1.4901363 | 0.0005300 | 0.0096559 |
| chrX | 119589485 | 119589833 | NKRF | 0 | - | 119589485 | 119589833 | 0 | 1 | 348 | 0 | -0.8944655 | 0.0059010 | 0.0587441 |
| chrX | 119590932 | 119591280 | NKRF | 0 | - | 119590932 | 119591280 | 0 | 1 | 348 | 0 | -1.0864994 | 0.0026807 | 0.0327540 |
| chr16 | 57025464 | 57025812 | NLRC5 | 0 | + | 57025464 | 57025812 | 0 | 1 | 348 | 0 | -1.3245739 | 0.0013986 | 0.0203793 |
| chr17 | 51153559 | 51154366 | NME1 | 0 | + | 51153559 | 51154366 | 0 | 2 | 104,142 | 0,666 | 2.4473608 | 0.0000000 | 0.0000000 |
| chr17 | 51161242 | 51161893 | NME1 | 0 | + | 51161242 | 51161893 | 0 | 2 | 31,166 | 0,486 | -1.0487486 | 0.0003119 | 0.0064036 |
| chr17 | 51153586 | 51154346 | NME1-NME2 | 0 | + | 51153586 | 51154346 | 0 | 2 | 77,122 | 0,639 | 2.5847087 | 0.0000000 | 0.0000000 |
| chr11 | 114295912 | 114296010 | NNMT | 0 | + | 114295912 | 114296010 | 0 | 1 | 98 | 0 | 2.2397620 | 0.0032873 | 0.0380972 |
| chr4 | 56976574 | 56977265 | NOA1 | 0 | - | 56976574 | 56977265 | 0 | 1 | 691 | 0 | -0.7403064 | 0.0002861 | 0.0059565 |
| chr1 | 944303 | 944452 | NOC2L | 0 | - | 944303 | 944452 | 0 | 1 | 149 | 0 | 1.1734799 | 0.0020071 | 0.0264241 |
| chr17 | 67717932 | 67719714 | NOL11 | 0 | + | 67717932 | 67719714 | 0 | 2 | 157,41 | 0,1742 | 0.9038403 | 0.0032705 | 0.0379392 |
| chr9 | 33461947 | 33463106 | NOL6 | 0 | - | 33461947 | 33463106 | 0 | 2 | 867,74 | 0,1086 | -0.6023282 | 0.0000347 | 0.0011240 |
| chr1 | 6524479 | 6524777 | NOL9 | 0 | - | 6524479 | 6524777 | 0 | 1 | 298 | 0 | -1.5938585 | 0.0001792 | 0.0040962 |
| chr7 | 156950371 | 156950569 | NOM1 | 0 | + | 156950371 | 156950569 | 0 | 1 | 198 | 0 | -1.9777643 | 0.0000745 | 0.0020777 |
| chrX | 71283638 | 71284991 | NONO | 0 | + | 71283638 | 71284991 | 0 | 3 | 84,57,8 | 0,759,1346 | 1.4452585 | 0.0000375 | 0.0011988 |
| chr12 | 6556920 | 6557565 | NOP2 | 0 | - | 6556920 | 6557565 | 0 | 1 | 645 | 0 | -0.9246068 | 0.0000001 | 0.0000040 |
| chr1 | 119915644 | 119915943 | NOTCH2 | 0 | - | 119915644 | 119915943 | 0 | 1 | 299 | 0 | -0.8552173 | 0.0126542 | 0.0985559 |
| chr1 | 119916443 | 119916641 | NOTCH2 | 0 | - | 119916443 | 119916641 | 0 | 1 | 198 | 0 | -1.3883741 | 0.0001513 | 0.0036107 |
| chr17 | 81952852 | 81953098 | NOTUM | 0 | - | 81952852 | 81953098 | 0 | 1 | 246 | 0 | 1.0135333 | 0.0113829 | 0.0921421 |
| chr3 | 132722110 | 132722359 | NPHP3-ACAD11 | 0 | - | 132722110 | 132722359 | 0 | 1 | 249 | 0 | 1.6283035 | 0.0068538 | 0.0650127 |
| chr17 | 81558324 | 81565553 | NPLOC4 | 0 | - | 81558324 | 81565553 | 0 | 2 | 1093,49 | 0,7181 | 0.5598041 | 0.0085283 | 0.0753316 |
| chr10 | 101782580 | 101783364 | NPM3 | 0 | - | 101782580 | 101783364 | 0 | 3 | 18,86,92 | 0,259,693 | 1.9453778 | 0.0000003 | 0.0000190 |
| chr16 | 69710591 | 69710640 | NQO1 | 0 | - | 69710591 | 69710640 | 0 | 1 | 49 | 0 | -2.5149989 | 0.0010819 | 0.0169173 |
| chr16 | 69711087 | 69711136 | NQO1 | 0 | - | 69711087 | 69711136 | 0 | 1 | 49 | 0 | -1.2782204 | 0.0091897 | 0.0793924 |
| chr19 | 19201459 | 19201653 | NR2C2AP | 0 | - | 19201459 | 19201653 | 0 | 1 | 194 | 0 | -1.5789023 | 0.0000154 | 0.0005761 |
| chr5 | 143314144 | 143403282 | NR3C1 | 0 | - | 143314144 | 143403282 | 0 | 3 | 25,1197,72 | 0,85512,89067 | -0.8803677 | 0.0000000 | 0.0000030 |
| chr12 | 52051436 | 52052512 | NR4A1 | 0 | + | 52051436 | 52052512 | 0 | 2 | 133,17 | 0,1060 | 1.3345569 | 0.0022930 | 0.0292196 |
| chr1 | 114708616 | 114709579 | NRAS | 0 | - | 114708616 | 114709579 | 0 | 2 | 39,11 | 0,953 | -2.3927567 | 0.0025031 | 0.0313242 |
| chr10 | 63153910 | 63154154 | NRBF2 | 0 | + | 63153910 | 63154154 | 0 | 1 | 244 | 0 | -0.7832876 | 0.0090113 | 0.0782994 |
| chr10 | 63154251 | 63154446 | NRBF2 | 0 | + | 63154251 | 63154446 | 0 | 1 | 195 | 0 | -0.7713667 | 0.0071214 | 0.0665740 |
| chr5 | 177294033 | 177294830 | NSD1 | 0 | + | 177294033 | 177294830 | 0 | 1 | 797 | 0 | -0.9174108 | 0.0006806 | 0.0117716 |
| chr5 | 177294881 | 177294930 | NSD1 | 0 | + | 177294881 | 177294930 | 0 | 1 | 49 | 0 | -2.1168935 | 0.0006657 | 0.0116135 |
| chr9 | 137448312 | 137448361 | NSMF | 0 | - | 137448312 | 137448361 | 0 | 1 | 49 | 0 | -1.6385565 | 0.0121545 | 0.0959013 |
| chr17 | 30185391 | 30185735 | NSRP1 | 0 | + | 30185391 | 30185735 | 0 | 1 | 344 | 0 | -0.9990436 | 0.0034385 | 0.0395080 |
| chr5 | 6632639 | 6633063 | NSUN2 | 0 | - | 6632639 | 6633063 | 0 | 2 | 118,180 | 0,245 | 1.8698271 | 0.0000000 | 0.0000008 |
| chr12 | 95786982 | 95790263 | NTN4 | 0 | - | 95786982 | 95790263 | 0 | 2 | 487,9 | 0,3273 | -1.0476449 | 0.0000325 | 0.0010650 |
| chr19 | 48913040 | 48918775 | NUCB1 | 0 | + | 48913040 | 48918775 | 0 | 3 | 157,91,50 | 0,434,5686 | -1.2489487 | 0.0000177 | 0.0006439 |
| chr4 | 87457874 | 87458071 | NUDT9 | 0 | + | 87457874 | 87458071 | 0 | 1 | 197 | 0 | -1.1984196 | 0.0008991 | 0.0146116 |
| chr17 | 29264021 | 29287683 | NUFIP2 | 0 | - | 29264021 | 29287683 | 0 | 3 | 571,33,1692 | 0,3477,21971 | -0.6160513 | 0.0000225 | 0.0007821 |
| chr11 | 72013037 | 72013485 | NUMA1 | 0 | - | 72013037 | 72013485 | 0 | 1 | 448 | 0 | -1.8954428 | 0.0000000 | 0.0000000 |
| chr11 | 72013885 | 72014333 | NUMA1 | 0 | - | 72013885 | 72014333 | 0 | 1 | 448 | 0 | -1.7002392 | 0.0000000 | 0.0000000 |
| chr11 | 72014983 | 72015032 | NUMA1 | 0 | - | 72014983 | 72015032 | 0 | 1 | 49 | 0 | -2.0563084 | 0.0077978 | 0.0707776 |
| chr11 | 72015482 | 72015681 | NUMA1 | 0 | - | 72015482 | 72015681 | 0 | 1 | 199 | 0 | -2.1261911 | 0.0000003 | 0.0000212 |
| chr14 | 73276458 | 73276707 | NUMB | 0 | - | 73276458 | 73276707 | 0 | 1 | 249 | 0 | -1.5130049 | 0.0000045 | 0.0001999 |
| chr9 | 131197401 | 131197950 | NUP214 | 0 | + | 131197401 | 131197950 | 0 | 1 | 549 | 0 | -1.4495702 | 0.0000000 | 0.0000020 |
| chr9 | 131198001 | 131198150 | NUP214 | 0 | + | 131198001 | 131198150 | 0 | 1 | 149 | 0 | -2.1330975 | 0.0007460 | 0.0126371 |
| chr22 | 45184937 | 45185630 | NUP50 | 0 | + | 45184937 | 45185630 | 0 | 1 | 693 | 0 | -0.6455650 | 0.0073357 | 0.0677732 |
| chr19 | 49908115 | 49908610 | NUP62 | 0 | - | 49908115 | 49908610 | 0 | 1 | 495 | 0 | -0.8273870 | 0.0000000 | 0.0000009 |
| chr19 | 49909008 | 49909205 | NUP62 | 0 | - | 49909008 | 49909205 | 0 | 1 | 197 | 0 | -1.6593998 | 0.0000000 | 0.0000000 |
| chr19 | 49909404 | 49927758 | NUP62 | 0 | - | 49909404 | 49927758 | 0 | 2 | 481,65 | 0,18290 | -1.2498268 | 0.0000000 | 0.0000000 |
| chr17 | 5419391 | 5419689 | NUP88 | 0 | - | 5419391 | 5419689 | 0 | 1 | 298 | 0 | 1.1339028 | 0.0000260 | 0.0008772 |
| chr11 | 3712595 | 3712644 | NUP98 | 0 | - | 3712595 | 3712644 | 0 | 1 | 49 | 0 | 2.5164029 | 0.0028402 | 0.0340600 |
| chr15 | 41332890 | 41342421 | NUSAP1 | 0 | + | 41332890 | 41342421 | 0 | 2 | 161,36 | 0,9496 | 1.0082827 | 0.0030434 | 0.0360197 |
| chr16 | 67846916 | 67865130 | NUTF2 | 0 | + | 67846916 | 67865130 | 0 | 2 | 70,29 | 0,18186 | -1.0086786 | 0.0027683 | 0.0334189 |
| chr20 | 23354140 | 23354577 | NXT1 | 0 | + | 23354140 | 23354577 | 0 | 1 | 437 | 0 | -0.5258512 | 0.0082971 | 0.0737933 |
| chr12 | 112941644 | 112941843 | OAS3 | 0 | + | 112941644 | 112941843 | 0 | 1 | 199 | 0 | -2.4806281 | 0.0000000 | 0.0000000 |
| chr12 | 112968017 | 112968066 | OAS3 | 0 | + | 112968017 | 112968066 | 0 | 1 | 49 | 0 | -1.1851219 | 0.0052288 | 0.0537712 |
| chr12 | 121020342 | 121020738 | OASL | 0 | - | 121020342 | 121020738 | 0 | 1 | 396 | 0 | 0.5612975 | 0.0044495 | 0.0476471 |
| chr19 | 2269535 | 2271421 | OAZ1 | 0 | + | 2269535 | 2271421 | 0 | 2 | 210,37 | 0,1850 | -0.6585205 | 0.0125717 | 0.0980822 |
| chr1 | 228360081 | 228360380 | OBSCN | 0 | + | 228360081 | 228360380 | 0 | 1 | 299 | 0 | -1.3564272 | 0.0074704 | 0.0687297 |
| chr19 | 17228825 | 17228874 | OCEL1 | 0 | + | 17228825 | 17228874 | 0 | 1 | 49 | 0 | -1.7699462 | 0.0001245 | 0.0031018 |
| chrX | 129540325 | 129540374 | OCRL | 0 | + | 129540325 | 129540374 | 0 | 1 | 49 | 0 | 2.2703851 | 0.0083322 | 0.0740329 |
| chr7 | 44708722 | 44709020 | OGDH | 0 | + | 44708722 | 44709020 | 0 | 1 | 298 | 0 | -1.1215148 | 0.0101272 | 0.0849391 |
| chr20 | 62804825 | 62804973 | OGFR | 0 | + | 62804825 | 62804973 | 0 | 1 | 148 | 0 | 0.9282945 | 0.0001599 | 0.0037661 |
| chr20 | 62812815 | 62813358 | OGFR | 0 | + | 62812815 | 62813358 | 0 | 1 | 543 | 0 | -0.6562724 | 0.0000128 | 0.0004970 |
| chr15 | 41298302 | 41298451 | OIP5-AS1 | 0 | + | 41298302 | 41298451 | 0 | 1 | 149 | 0 | 1.0142268 | 0.0114786 | 0.0924901 |
| chr15 | 41299299 | 41299497 | OIP5-AS1 | 0 | + | 41299299 | 41299497 | 0 | 1 | 198 | 0 | 1.5089086 | 0.0014271 | 0.0206574 |
| chr12 | 10159286 | 10159977 | OLR1 | 0 | - | 10159286 | 10159977 | 0 | 1 | 691 | 0 | -0.6241449 | 0.0021213 | 0.0275354 |
| chr1 | 241640201 | 241640349 | OPN3 | 0 | - | 241640201 | 241640349 | 0 | 1 | 148 | 0 | 1.1346032 | 0.0100172 | 0.0843375 |
| chr10 | 13108989 | 13109219 | OPTN | 0 | + | 13108989 | 13109219 | 0 | 2 | 27,123 | 0,108 | -0.5482795 | 0.0117086 | 0.0936894 |
| chr16 | 46697750 | 46697799 | ORC6 | 0 | + | 46697750 | 46697799 | 0 | 1 | 49 | 0 | 2.5026168 | 0.0120534 | 0.0953533 |
| chr11 | 59576292 | 59576924 | OSBP | 0 | - | 59576292 | 59576924 | 0 | 2 | 428,120 | 0,513 | -0.6876670 | 0.0001675 | 0.0038828 |
| chr8 | 89925493 | 89925591 | OSGIN2 | 0 | + | 89925493 | 89925591 | 0 | 1 | 98 | 0 | -2.2779919 | 0.0061913 | 0.0607058 |
| chr5 | 38846158 | 38846257 | OSMR | 0 | + | 38846158 | 38846257 | 0 | 1 | 99 | 0 | 1.1099051 | 0.0063584 | 0.0618692 |
| chr5 | 38885402 | 38886055 | OSMR | 0 | + | 38885402 | 38886055 | 0 | 2 | 83,17 | 0,637 | 1.1894951 | 0.0067955 | 0.0645605 |
| chr10 | 23440356 | 23440454 | OTUD1 | 0 | + | 23440356 | 23440454 | 0 | 1 | 98 | 0 | -1.0276048 | 0.0067077 | 0.0641306 |
| chr5 | 41870294 | 41870491 | OXCT1 | 0 | - | 41870294 | 41870491 | 0 | 1 | 197 | 0 | 0.9272029 | 0.0041093 | 0.0449341 |
| chr3 | 38253153 | 38253898 | OXSR1 | 0 | + | 38253153 | 38253898 | 0 | 1 | 745 | 0 | -0.5499758 | 0.0052944 | 0.0541950 |
| chr12 | 121228860 | 121229005 | P2RX4 | 0 | + | 121228860 | 121229005 | 0 | 2 | 7,43 | 0,103 | -2.1897587 | 0.0000904 | 0.0024098 |
| chr1 | 42766739 | 42766985 | P3H1 | 0 | - | 42766739 | 42766985 | 0 | 1 | 246 | 0 | 1.4909335 | 0.0014773 | 0.0211309 |
| chr17 | 41810902 | 41811147 | P3H4 | 0 | - | 41810902 | 41811147 | 0 | 2 | 133,16 | 0,230 | -0.7676477 | 0.0071079 | 0.0665512 |
| chr10 | 73074872 | 73077775 | P4HA1 | 0 | - | 73074872 | 73077775 | 0 | 2 | 44,55 | 0,2849 | 1.4278081 | 0.0063373 | 0.0617678 |
| chr11 | 47177621 | 47177919 | PACSIN3 | 0 | - | 47177621 | 47177919 | 0 | 1 | 298 | 0 | -0.7765001 | 0.0100091 | 0.0843375 |
| chr17 | 2593629 | 2593976 | PAFAH1B1 | 0 | + | 2593629 | 2593976 | 0 | 1 | 347 | 0 | 0.9446449 | 0.0000144 | 0.0005462 |
| chr19 | 746745 | 747141 | PALM | 0 | + | 746745 | 747141 | 0 | 1 | 396 | 0 | -0.8639813 | 0.0004034 | 0.0078545 |
| chr19 | 747241 | 747488 | PALM | 0 | + | 747241 | 747488 | 0 | 1 | 247 | 0 | -2.0196845 | 0.0000001 | 0.0000091 |
| chr9 | 110136448 | 110136597 | PALM2-AKAP2 | 0 | + | 110136448 | 110136597 | 0 | 1 | 149 | 0 | -0.9960953 | 0.0072564 | 0.0672460 |
| chr9 | 110137394 | 110137842 | PALM2-AKAP2 | 0 | + | 110137394 | 110137842 | 0 | 1 | 448 | 0 | -1.3465644 | 0.0000016 | 0.0000829 |
| chr9 | 110138041 | 110138090 | PALM2-AKAP2 | 0 | + | 110138041 | 110138090 | 0 | 1 | 49 | 0 | -2.9214513 | 0.0000019 | 0.0000952 |
| chr9 | 110138290 | 110138389 | PALM2-AKAP2 | 0 | + | 110138290 | 110138389 | 0 | 1 | 99 | 0 | -1.4518666 | 0.0004861 | 0.0090379 |
| chr11 | 94181378 | 94181823 | PANX1 | 0 | + | 94181378 | 94181823 | 0 | 1 | 445 | 0 | -0.6889748 | 0.0065764 | 0.0634107 |
| chr5 | 79612618 | 79612767 | PAPD4 | 0 | + | 79612618 | 79612767 | 0 | 1 | 149 | 0 | 1.1744963 | 0.0083535 | 0.0741491 |
| chr9 | 116187455 | 116187504 | PAPPA | 0 | + | 116187455 | 116187504 | 0 | 1 | 49 | 0 | -2.4793030 | 0.0047706 | 0.0502826 |
| chr9 | 116187555 | 116187903 | PAPPA | 0 | + | 116187555 | 116187903 | 0 | 1 | 348 | 0 | -1.2936151 | 0.0025449 | 0.0316725 |
| chr4 | 107701181 | 107720213 | PAPSS1 | 0 | - | 107701181 | 107720213 | 0 | 2 | 105,94 | 0,18939 | 1.2480651 | 0.0018484 | 0.0249035 |
| chr16 | 2971645 | 2971943 | PAQR4 | 0 | + | 2971645 | 2971943 | 0 | 1 | 298 | 0 | -0.6628198 | 0.0038033 | 0.0424835 |
| chr3 | 122555394 | 122555742 | PARP9 | 0 | - | 122555394 | 122555742 | 0 | 1 | 348 | 0 | -0.7454989 | 0.0014149 | 0.0205137 |
| chr1 | 54757693 | 54757991 | PARS2 | 0 | - | 54757693 | 54757991 | 0 | 1 | 298 | 0 | -1.1086555 | 0.0076598 | 0.0699348 |
| chr2 | 241126703 | 241126851 | PASK | 0 | - | 241126703 | 241126851 | 0 | 1 | 148 | 0 | -1.8671945 | 0.0063446 | 0.0617678 |
| chr2 | 70087453 | 70087902 | PCBP1 | 0 | + | 70087453 | 70087902 | 0 | 1 | 449 | 0 | -0.6560945 | 0.0000002 | 0.0000124 |
| chr2 | 70088303 | 70088702 | PCBP1 | 0 | + | 70088303 | 70088702 | 0 | 1 | 399 | 0 | -0.6656809 | 0.0000002 | 0.0000159 |
| chr3 | 51957503 | 51957748 | PCBP4 | 0 | - | 51957503 | 51957748 | 0 | 1 | 245 | 0 | -1.5236332 | 0.0004166 | 0.0080678 |
| chr5 | 141864534 | 141864583 | PCDH1 | 0 | - | 141864534 | 141864583 | 0 | 1 | 49 | 0 | -Inf | 0.0001898 | 0.0042759 |
| chr11 | 83157145 | 83157442 | PCF11 | 0 | + | 83157145 | 83157442 | 0 | 1 | 297 | 0 | 0.7946146 | 0.0004297 | 0.0082610 |
| chr14 | 24103588 | 24104033 | PCK2 | 0 | + | 24103588 | 24104033 | 0 | 1 | 445 | 0 | -0.7582827 | 0.0027543 | 0.0333423 |
| chr14 | 70977937 | 70978585 | PCNX | 0 | + | 70977937 | 70978585 | 0 | 1 | 648 | 0 | -0.7088099 | 0.0010660 | 0.0167073 |
| chr11 | 65618159 | 65619004 | PCNXL3 | 0 | + | 65618159 | 65619004 | 0 | 1 | 845 | 0 | -0.8644924 | 0.0000142 | 0.0005385 |
| chr11 | 65636667 | 65637290 | PCNXL3 | 0 | + | 65636667 | 65637290 | 0 | 2 | 23,525 | 0,99 | -0.6390402 | 0.0013863 | 0.0202278 |
| chr1 | 55039526 | 55040024 | PCSK9 | 0 | + | 55039526 | 55040024 | 0 | 1 | 498 | 0 | 1.2140412 | 0.0000000 | 0.0000000 |
| chr1 | 55063706 | 55064703 | PCSK9 | 0 | + | 55063706 | 55064703 | 0 | 1 | 997 | 0 | -0.5559715 | 0.0113594 | 0.0920341 |
| chr3 | 196270457 | 196283538 | PCYT1A | 0 | - | 196270457 | 196283538 | 0 | 2 | 85,64 | 0,13018 | 1.1561682 | 0.0095023 | 0.0810871 |
| chr10 | 103425019 | 103425515 | PDCD11 | 0 | + | 103425019 | 103425515 | 0 | 1 | 496 | 0 | -0.7962367 | 0.0001602 | 0.0037674 |
| chr2 | 100576397 | 100576495 | PDCL3 | 0 | + | 100576397 | 100576495 | 0 | 1 | 98 | 0 | -1.1914111 | 0.0015339 | 0.0216994 |
| chr3 | 57557053 | 57557401 | PDE12 | 0 | + | 57557053 | 57557401 | 0 | 1 | 348 | 0 | -1.5710073 | 0.0000027 | 0.0001287 |
| chr11 | 72577139 | 72577588 | PDE2A | 0 | - | 72577139 | 72577588 | 0 | 1 | 449 | 0 | 0.7764012 | 0.0070870 | 0.0664239 |
| chr5 | 58974037 | 58975761 | PDE4D | 0 | - | 58974037 | 58975761 | 0 | 2 | 1044,105 | 0,1620 | -0.8849141 | 0.0000000 | 0.0000000 |
| chr5 | 59768409 | 59893213 | PDE4D | 0 | - | 59768409 | 59893213 | 0 | 2 | 204,46 | 0,124759 | 1.3969875 | 0.0008066 | 0.0134411 |
| chr7 | 149003652 | 149005173 | PDIA4 | 0 | - | 149003652 | 149005173 | 0 | 2 | 558,33 | 0,1489 | -0.7415506 | 0.0001109 | 0.0028352 |
| chr8 | 93922359 | 93922804 | PDP1 | 0 | + | 93922359 | 93922804 | 0 | 1 | 445 | 0 | -0.6261179 | 0.0054477 | 0.0553214 |
| chr4 | 39825211 | 39825460 | PDS5A | 0 | - | 39825211 | 39825460 | 0 | 1 | 249 | 0 | -1.0050885 | 0.0096435 | 0.0819456 |
| chr21 | 43761310 | 43762157 | PDXK | 0 | + | 43761310 | 43762157 | 0 | 1 | 847 | 0 | 0.9725135 | 0.0000811 | 0.0022122 |
| chr12 | 118136130 | 118138070 | PEBP1 | 0 | + | 118136130 | 118138070 | 0 | 2 | 215,32 | 0,1909 | 1.1759023 | 0.0065435 | 0.0631949 |
| chr1 | 31635271 | 31635517 | PEF1 | 0 | - | 31635271 | 31635517 | 0 | 1 | 246 | 0 | 1.1918799 | 0.0019266 | 0.0257082 |
| chr7 | 94666356 | 94666405 | PEG10 | 0 | + | 94666356 | 94666405 | 0 | 1 | 49 | 0 | -2.1215792 | 0.0009954 | 0.0158531 |
| chr7 | 94666555 | 94666604 | PEG10 | 0 | + | 94666555 | 94666604 | 0 | 1 | 49 | 0 | -2.4075243 | 0.0043940 | 0.0472911 |
| chr11 | 66475906 | 66476103 | PELI3 | 0 | + | 66475906 | 66476103 | 0 | 1 | 197 | 0 | -1.3587158 | 0.0081852 | 0.0730859 |
| chr5 | 52801479 | 52801924 | PELO | 0 | + | 52801479 | 52801924 | 0 | 1 | 445 | 0 | -0.7459815 | 0.0001454 | 0.0034960 |
| chr6 | 42974965 | 42978843 | PEX6 | 0 | - | 42974965 | 42978843 | 0 | 2 | 74,575 | 0,3304 | -0.6932335 | 0.0044362 | 0.0475891 |
| chr10 | 3067569 | 3068636 | PFKP | 0 | + | 3067569 | 3068636 | 0 | 2 | 139,10 | 0,1058 | 1.1117086 | 0.0037356 | 0.0419365 |
| chr10 | 97426270 | 97430497 | PGAM1 | 0 | + | 97426270 | 97430497 | 0 | 2 | 177,119 | 0,4109 | 0.8785166 | 0.0032884 | 0.0380972 |
| chrX | 77969540 | 77969589 | PGAM4 | 0 | - | 77969540 | 77969589 | 0 | 1 | 49 | 0 | 1.2579134 | 0.0027994 | 0.0336819 |
| chr1 | 63593276 | 63593720 | PGM1 | 0 | + | 63593276 | 63593720 | 0 | 1 | 444 | 0 | 0.8825511 | 0.0037398 | 0.0419584 |
| chr4 | 37826710 | 37830008 | PGM2 | 0 | + | 37826710 | 37830008 | 0 | 2 | 104,45 | 0,3254 | 0.9780726 | 0.0030176 | 0.0358073 |
| chr16 | 2213245 | 2213493 | PGP | 0 | - | 2213245 | 2213493 | 0 | 1 | 248 | 0 | -0.8956413 | 0.0113422 | 0.0919357 |
| chr5 | 126603725 | 126608403 | PHAX | 0 | + | 126603725 | 126608403 | 0 | 2 | 459,40 | 0,4639 | -0.6184341 | 0.0008749 | 0.0142596 |
| chr6 | 169710296 | 169712388 | PHF10 | 0 | - | 169710296 | 169712388 | 0 | 2 | 96,3 | 0,2090 | 1.9939334 | 0.0000417 | 0.0012923 |
| chr17 | 28924109 | 28950096 | PHF12 | 0 | - | 28924109 | 28950096 | 0 | 3 | 194,73,32 | 0,2882,25956 | 0.8502854 | 0.0051717 | 0.0533612 |
| chr17 | 7236109 | 7236505 | PHF23 | 0 | - | 7236109 | 7236505 | 0 | 1 | 396 | 0 | -0.8537006 | 0.0000000 | 0.0000005 |
| chr6 | 63685450 | 63685698 | PHF3 | 0 | + | 63685450 | 63685698 | 0 | 1 | 248 | 0 | -1.3457354 | 0.0005132 | 0.0094255 |
| chr6 | 63713184 | 63713731 | PHF3 | 0 | + | 63713184 | 63713731 | 0 | 1 | 547 | 0 | -1.8312850 | 0.0000000 | 0.0000000 |
| chr22 | 41459910 | 41460346 | PHF5A | 0 | - | 41459910 | 41460346 | 0 | 1 | 436 | 0 | -0.7700539 | 0.0000397 | 0.0012434 |
| chr11 | 118628051 | 118628547 | PHLDB1 | 0 | + | 118628051 | 118628547 | 0 | 1 | 496 | 0 | -0.7646322 | 0.0118459 | 0.0942885 |
| chr3 | 111859832 | 111884613 | PHLDB2 | 0 | + | 111859832 | 111884613 | 0 | 2 | 47,550 | 0,24232 | -0.7520814 | 0.0005892 | 0.0105423 |
| chr3 | 111884764 | 111913665 | PHLDB2 | 0 | + | 111884764 | 111913665 | 0 | 2 | 649,347 | 0,28555 | -0.7376666 | 0.0000015 | 0.0000785 |
| chr16 | 71648869 | 71649318 | PHLPP2 | 0 | - | 71648869 | 71649318 | 0 | 1 | 449 | 0 | -1.1079048 | 0.0019233 | 0.0257023 |
| chr1 | 151315647 | 151324772 | PI4KB | 0 | - | 151315647 | 151324772 | 0 | 2 | 863,28 | 0,9098 | -0.7437945 | 0.0000886 | 0.0023718 |
| chr11 | 85958666 | 85959015 | PICALM | 0 | - | 85958666 | 85959015 | 0 | 1 | 349 | 0 | -1.0926727 | 0.0056494 | 0.0568957 |
| chr11 | 86068662 | 86069731 | PICALM | 0 | - | 86068662 | 86069731 | 0 | 2 | 436,12 | 0,1058 | 0.7108963 | 0.0009093 | 0.0147536 |
| chr18 | 10670645 | 10671193 | PIEZO2 | 0 | - | 10670645 | 10671193 | 0 | 1 | 548 | 0 | -0.9466706 | 0.0027569 | 0.0333423 |
| chr17 | 28561629 | 28563479 | PIGS | 0 | - | 28561629 | 28563479 | 0 | 2 | 1,49 | 0,1802 | -1.8452212 | 0.0001863 | 0.0042284 |
| chr22 | 31283175 | 31283224 | PIK3IP1 | 0 | - | 31283175 | 31283224 | 0 | 1 | 49 | 0 | -3.2360861 | 0.0072919 | 0.0675045 |
| chr19 | 18169641 | 18169889 | PIK3R2 | 0 | + | 18169641 | 18169889 | 0 | 1 | 248 | 0 | -1.7912582 | 0.0000001 | 0.0000055 |
| chr3 | 130735970 | 130745001 | PIK3R4 | 0 | - | 130735970 | 130745001 | 0 | 2 | 33,516 | 0,8516 | -1.2292600 | 0.0000320 | 0.0010572 |
| chr22 | 31618888 | 31619234 | PISD | 0 | - | 31618888 | 31619234 | 0 | 1 | 346 | 0 | -1.0530375 | 0.0011416 | 0.0175189 |
| chr1 | 23778435 | 23778681 | PITHD1 | 0 | + | 23778435 | 23778681 | 0 | 1 | 246 | 0 | 1.4643274 | 0.0008515 | 0.0140040 |
| chr16 | 2109881 | 2110329 | PKD1 | 0 | - | 2109881 | 2110329 | 0 | 1 | 448 | 0 | -0.7220843 | 0.0091996 | 0.0794401 |
| chr16 | 2110480 | 2111678 | PKD1 | 0 | - | 2110480 | 2111678 | 0 | 1 | 1198 | 0 | -1.3411260 | 0.0000000 | 0.0000000 |
| chr15 | 72231039 | 72231088 | PKM | 0 | - | 72231039 | 72231088 | 0 | 1 | 49 | 0 | 2.4665487 | 0.0034051 | 0.0391485 |
| chr1 | 88833142 | 88833575 | PKN2 | 0 | + | 88833142 | 88833575 | 0 | 2 | 16,331 | 0,103 | -0.8855587 | 0.0001882 | 0.0042503 |
| chr9 | 26905847 | 26905945 | PLAA | 0 | - | 26905847 | 26905945 | 0 | 1 | 98 | 0 | -1.2039159 | 0.0113880 | 0.0921421 |
| chr6 | 143948160 | 143948458 | PLAGL1 | 0 | - | 143948160 | 143948458 | 0 | 1 | 298 | 0 | -0.9395064 | 0.0048110 | 0.0506200 |
| chr17 | 45118860 | 45118909 | PLCD3 | 0 | - | 45118860 | 45118909 | 0 | 1 | 49 | 0 | -1.4037945 | 0.0081126 | 0.0726886 |
| chr3 | 17011393 | 17011789 | PLCL2 | 0 | + | 17011393 | 17011789 | 0 | 1 | 396 | 0 | -0.7530206 | 0.0074389 | 0.0684970 |
| chr19 | 40370019 | 40371784 | PLD3 | 0 | + | 40370019 | 40371784 | 0 | 3 | 10,128,112 | 0,91,1654 | -0.8828163 | 0.0013684 | 0.0200959 |
| chr8 | 143915446 | 143915545 | PLEC | 0 | - | 143915446 | 143915545 | 0 | 1 | 99 | 0 | -1.9270300 | 0.0000125 | 0.0004881 |
| chr8 | 143916294 | 143916892 | PLEC | 0 | - | 143916294 | 143916892 | 0 | 1 | 598 | 0 | -1.6026781 | 0.0000000 | 0.0000000 |
| chr8 | 143916943 | 143917042 | PLEC | 0 | - | 143916943 | 143917042 | 0 | 1 | 99 | 0 | -3.2534139 | 0.0000000 | 0.0000001 |
| chr8 | 143917591 | 143917640 | PLEC | 0 | - | 143917591 | 143917640 | 0 | 1 | 49 | 0 | -2.1027615 | 0.0000728 | 0.0020382 |
| chr8 | 143917691 | 143918788 | PLEC | 0 | - | 143917691 | 143918788 | 0 | 1 | 1097 | 0 | -1.8307407 | 0.0000000 | 0.0000000 |
| chr8 | 143918839 | 143918987 | PLEC | 0 | - | 143918839 | 143918987 | 0 | 1 | 148 | 0 | -2.0486358 | 0.0000031 | 0.0001450 |
| chr8 | 143919138 | 143919486 | PLEC | 0 | - | 143919138 | 143919486 | 0 | 1 | 348 | 0 | -2.7877609 | 0.0000000 | 0.0000000 |
| chr8 | 143919537 | 143920883 | PLEC | 0 | - | 143919537 | 143920883 | 0 | 1 | 1346 | 0 | -2.1681282 | 0.0000000 | 0.0000000 |
| chr8 | 143921134 | 143922130 | PLEC | 0 | - | 143921134 | 143922130 | 0 | 1 | 996 | 0 | -2.3278722 | 0.0000000 | 0.0000000 |
| chr8 | 143922888 | 143923236 | PLEC | 0 | - | 143922888 | 143923236 | 0 | 1 | 348 | 0 | -1.7401425 | 0.0000000 | 0.0000006 |
| chr8 | 143923985 | 143924284 | PLEC | 0 | - | 143923985 | 143924284 | 0 | 1 | 299 | 0 | -2.6647453 | 0.0000000 | 0.0000000 |
| chr19 | 29674980 | 29675029 | PLEKHF1 | 0 | + | 29674980 | 29675029 | 0 | 1 | 49 | 0 | -1.5112431 | 0.0081799 | 0.0730739 |
| chr14 | 64741127 | 64742024 | PLEKHG3 | 0 | + | 64741127 | 64742024 | 0 | 1 | 897 | 0 | -1.5553920 | 0.0000000 | 0.0000001 |
| chr17 | 42676686 | 42677030 | PLEKHH3 | 0 | - | 42676686 | 42677030 | 0 | 1 | 344 | 0 | 0.6177792 | 0.0039623 | 0.0437703 |
| chr19 | 4838981 | 4844709 | PLIN3 | 0 | - | 4838981 | 4844709 | 0 | 2 | 556,42 | 0,5687 | -0.5486856 | 0.0102260 | 0.0855693 |
| chr1 | 11934766 | 11949839 | PLOD1 | 0 | + | 11934766 | 11949839 | 0 | 3 | 90,92,67 | 0,13210,15007 | 1.2281257 | 0.0000512 | 0.0015168 |
| chr3 | 146070579 | 146071094 | PLOD2 | 0 | - | 146070579 | 146071094 | 0 | 2 | 294,53 | 0,463 | -1.2559645 | 0.0000003 | 0.0000196 |
| chr3 | 126988694 | 126989141 | PLXNA1 | 0 | + | 126988694 | 126989141 | 0 | 1 | 447 | 0 | -1.0056386 | 0.0003927 | 0.0077111 |
| chr3 | 126989341 | 126989390 | PLXNA1 | 0 | + | 126989341 | 126989390 | 0 | 1 | 49 | 0 | -2.9565445 | 0.0001126 | 0.0028687 |
| chr1 | 208210460 | 208217081 | PLXNA2 | 0 | - | 208210460 | 208217081 | 0 | 2 | 3,347 | 0,6275 | -0.9980917 | 0.0050651 | 0.0525607 |
| chr1 | 208217682 | 208243770 | PLXNA2 | 0 | - | 208217682 | 208243770 | 0 | 2 | 321,128 | 0,25961 | -0.8455597 | 0.0108948 | 0.0893893 |
| chr22 | 50289580 | 50307572 | PLXNB2 | 0 | - | 50289580 | 50307572 | 0 | 3 | 1018,60,20 | 0,5139,17973 | -0.7379145 | 0.0000010 | 0.0000557 |
| chr11 | 818951 | 819862 | PNPLA2 | 0 | + | 818951 | 819862 | 0 | 2 | 8,289 | 0,623 | 0.8391035 | 0.0035281 | 0.0402567 |
| chr11 | 824829 | 825125 | PNPLA2 | 0 | + | 824829 | 825125 | 0 | 1 | 296 | 0 | -0.7903334 | 0.0109018 | 0.0894066 |
| chr7 | 131510944 | 131511392 | PODXL | 0 | - | 131510944 | 131511392 | 0 | 1 | 448 | 0 | -0.9355181 | 0.0000357 | 0.0011503 |
| chr3 | 127660606 | 127660802 | PODXL2 | 0 | + | 127660606 | 127660802 | 0 | 1 | 196 | 0 | -1.4407721 | 0.0000000 | 0.0000033 |
| chr1 | 166849201 | 166850237 | POGK | 0 | + | 166849201 | 166850237 | 0 | 1 | 1036 | 0 | -1.1725927 | 0.0000000 | 0.0000000 |
| chr1 | 151404866 | 151405662 | POGZ | 0 | - | 151404866 | 151405662 | 0 | 1 | 796 | 0 | -0.7961791 | 0.0014824 | 0.0211869 |
| chr7 | 44114729 | 44114926 | POLD2 | 0 | - | 44114729 | 44114926 | 0 | 1 | 197 | 0 | -0.8622489 | 0.0116761 | 0.0935950 |
| chr22 | 42603026 | 42614912 | POLDIP3 | 0 | - | 42603026 | 42614912 | 0 | 2 | 135,114 | 0,11773 | 1.5491798 | 0.0008103 | 0.0134606 |
| chr15 | 89328688 | 89328787 | POLG | 0 | - | 89328688 | 89328787 | 0 | 1 | 99 | 0 | -1.2816611 | 0.0026779 | 0.0327445 |
| chr15 | 89333787 | 89334795 | POLG | 0 | - | 89333787 | 89334795 | 0 | 2 | 127,123 | 0,886 | 1.0630725 | 0.0006770 | 0.0117491 |
| chr13 | 27622889 | 27622938 | POLR1D | 0 | + | 27622889 | 27622938 | 0 | 1 | 49 | 0 | -1.3879561 | 0.0005748 | 0.0103153 |
| chr13 | 27622989 | 27623236 | POLR1D | 0 | + | 27622989 | 27623236 | 0 | 1 | 247 | 0 | -0.8163368 | 0.0000188 | 0.0006805 |
| chr2 | 127846662 | 127846958 | POLR2D | 0 | - | 127846662 | 127846958 | 0 | 1 | 296 | 0 | -0.6895775 | 0.0007107 | 0.0121636 |
| chr3 | 184368349 | 184368497 | POLR2H | 0 | + | 184368349 | 184368497 | 0 | 1 | 148 | 0 | -0.9295958 | 0.0123512 | 0.0968208 |
| chr7 | 72946241 | 72946539 | POM121 | 0 | + | 72946241 | 72946539 | 0 | 1 | 298 | 0 | -0.8307839 | 0.0057529 | 0.0577026 |
| chr7 | 75418519 | 75418817 | POM121C | 0 | - | 75418519 | 75418817 | 0 | 1 | 298 | 0 | -1.1830027 | 0.0083059 | 0.0738360 |
| chr3 | 43080261 | 43080359 | POMGNT2 | 0 | - | 43080261 | 43080359 | 0 | 1 | 98 | 0 | -1.9129101 | 0.0000261 | 0.0008772 |
| chr7 | 95405008 | 95405446 | PON2 | 0 | - | 95405008 | 95405446 | 0 | 1 | 438 | 0 | -1.1732991 | 0.0068782 | 0.0651751 |
| chr7 | 100706912 | 100707059 | POP7 | 0 | + | 100706912 | 100707059 | 0 | 1 | 147 | 0 | -0.8912108 | 0.0005371 | 0.0097363 |
| chr7 | 100707157 | 100707304 | POP7 | 0 | + | 100707157 | 100707304 | 0 | 1 | 147 | 0 | -0.6108932 | 0.0017039 | 0.0234426 |
| chr19 | 10106371 | 10106557 | PPAN | 0 | + | 10106371 | 10106557 | 0 | 2 | 42,57 | 0,130 | 1.1686420 | 0.0089042 | 0.0774063 |
| chr1 | 56496475 | 56496524 | PPAP2B | 0 | - | 56496475 | 56496524 | 0 | 1 | 49 | 0 | -1.8890525 | 0.0050429 | 0.0524205 |
| chr19 | 49149638 | 49150531 | PPFIA3 | 0 | + | 49149638 | 49150531 | 0 | 3 | 81,72,296 | 0,442,598 | 0.5615899 | 0.0117887 | 0.0939985 |
| chr22 | 21920466 | 21920615 | PPM1F | 0 | - | 21920466 | 21920615 | 0 | 1 | 149 | 0 | 1.5420625 | 0.0018034 | 0.0244761 |
| chr2 | 28751948 | 28752097 | PPP1CB | 0 | + | 28751948 | 28752097 | 0 | 1 | 149 | 0 | 1.1798471 | 0.0024563 | 0.0308450 |
| chr12 | 79934753 | 79935001 | PPP1R12A | 0 | - | 79934753 | 79935001 | 0 | 1 | 248 | 0 | 1.7808670 | 0.0000058 | 0.0002478 |
| chr19 | 48874553 | 48874751 | PPP1R15A | 0 | + | 48874553 | 48874751 | 0 | 1 | 198 | 0 | -0.9722460 | 0.0100342 | 0.0844325 |
| chr1 | 204405454 | 204405503 | PPP1R15B | 0 | - | 204405454 | 204405503 | 0 | 1 | 49 | 0 | -1.5843690 | 0.0091662 | 0.0792276 |
| chr1 | 204406052 | 204410274 | PPP1R15B | 0 | - | 204406052 | 204410274 | 0 | 2 | 262,783 | 0,3440 | -1.1268272 | 0.0000000 | 0.0000000 |
| chr1 | 204410524 | 204411021 | PPP1R15B | 0 | - | 204410524 | 204411021 | 0 | 1 | 497 | 0 | -0.9741200 | 0.0000000 | 0.0000001 |
| chr1 | 204411171 | 204411568 | PPP1R15B | 0 | - | 204411171 | 204411568 | 0 | 1 | 397 | 0 | -0.9833558 | 0.0002003 | 0.0044509 |
| chr1 | 204411718 | 204411817 | PPP1R15B | 0 | - | 204411718 | 204411817 | 0 | 1 | 99 | 0 | -1.1866631 | 0.0030488 | 0.0360602 |
| chr17 | 50134872 | 50135267 | PPP1R9B | 0 | - | 50134872 | 50135267 | 0 | 1 | 395 | 0 | -0.9126970 | 0.0027233 | 0.0331627 |
| chr17 | 50149776 | 50150319 | PPP1R9B | 0 | - | 50149776 | 50150319 | 0 | 1 | 543 | 0 | -1.8306008 | 0.0000000 | 0.0000001 |
| chr5 | 134197355 | 134199093 | PPP2CA | 0 | - | 134197355 | 134199093 | 0 | 2 | 490,8 | 0,1731 | -0.6022094 | 0.0027630 | 0.0333768 |
| chr5 | 134225811 | 134226109 | PPP2CA | 0 | - | 134225811 | 134226109 | 0 | 1 | 298 | 0 | 0.6783514 | 0.0059883 | 0.0592723 |
| chr19 | 52189999 | 52206040 | PPP2R1A | 0 | + | 52189999 | 52206040 | 0 | 3 | 176,91,78 | 0,11945,15964 | 1.4367804 | 0.0000002 | 0.0000152 |
| chr6 | 43011802 | 43011999 | PPP2R5D | 0 | + | 43011802 | 43011999 | 0 | 1 | 197 | 0 | -1.0877407 | 0.0114477 | 0.0924595 |
| chr2 | 55617428 | 55617725 | PPP4R3B | 0 | - | 55617428 | 55617725 | 0 | 1 | 297 | 0 | 0.9823569 | 0.0024589 | 0.0308567 |
| chr10 | 102133044 | 102137870 | PPRC1 | 0 | + | 102133044 | 102137870 | 0 | 2 | 178,21 | 0,4806 | 1.2756076 | 0.0038407 | 0.0427192 |
| chr10 | 102139429 | 102139876 | PPRC1 | 0 | + | 102139429 | 102139876 | 0 | 1 | 447 | 0 | -1.1880610 | 0.0000003 | 0.0000215 |
| chr10 | 102140026 | 102140324 | PPRC1 | 0 | + | 102140026 | 102140324 | 0 | 1 | 298 | 0 | -1.3104761 | 0.0000135 | 0.0005191 |
| chr10 | 102140474 | 102140523 | PPRC1 | 0 | + | 102140474 | 102140523 | 0 | 1 | 49 | 0 | -2.4112965 | 0.0016748 | 0.0231856 |
| chr6 | 105402875 | 105403024 | PREP | 0 | - | 105402875 | 105403024 | 0 | 1 | 149 | 0 | 1.0913972 | 0.0106446 | 0.0877944 |
| chr1 | 147167926 | 147172490 | PRKAB2 | 0 | - | 147167926 | 147172490 | 0 | 3 | 8,179,62 | 0,4063,4503 | 1.4757458 | 0.0000136 | 0.0005194 |
| chr12 | 49013090 | 49018552 | PRKAG1 | 0 | - | 49013090 | 49018552 | 0 | 2 | 21,29 | 0,5434 | 1.4158730 | 0.0005455 | 0.0098679 |
| chr8 | 47774155 | 47776869 | PRKDC | 0 | - | 47774155 | 47776869 | 0 | 2 | 223,26 | 0,2689 | -1.0553474 | 0.0017214 | 0.0236085 |
| chr1 | 107056989 | 107057136 | PRMT6 | 0 | + | 107056989 | 107057136 | 0 | 1 | 147 | 0 | -1.2703606 | 0.0032607 | 0.0379200 |
| chr20 | 4700057 | 4700304 | PRNP | 0 | + | 4700057 | 4700304 | 0 | 1 | 247 | 0 | -1.2367440 | 0.0000203 | 0.0007292 |
| chr20 | 4700601 | 4700748 | PRNP | 0 | + | 4700601 | 4700748 | 0 | 1 | 147 | 0 | -1.3850057 | 0.0001531 | 0.0036482 |
| chr10 | 11869691 | 11869889 | PROSER2 | 0 | + | 11869691 | 11869889 | 0 | 1 | 198 | 0 | -0.9397227 | 0.0084051 | 0.0744484 |
| chr9 | 113291469 | 113292013 | PRPF4 | 0 | + | 113291469 | 113292013 | 0 | 1 | 544 | 0 | -0.6772259 | 0.0016937 | 0.0233831 |
| chr19 | 49597033 | 49597432 | PRR12 | 0 | + | 49597033 | 49597432 | 0 | 1 | 399 | 0 | -1.1864368 | 0.0004044 | 0.0078660 |
| chr11 | 36451274 | 36462874 | PRR5L | 0 | + | 36451274 | 36462874 | 0 | 2 | 62,533 | 0,11068 | 0.6587939 | 0.0105303 | 0.0873488 |
| chr9 | 131474466 | 131476508 | PRRC2B | 0 | + | 131474466 | 131476508 | 0 | 1 | 2042 | 0 | -1.5713903 | 0.0000000 | 0.0000001 |
| chr1 | 171540702 | 171541549 | PRRC2C | 0 | + | 171540702 | 171541549 | 0 | 1 | 847 | 0 | -0.6397800 | 0.0033376 | 0.0385095 |
| chr1 | 171541800 | 171542049 | PRRC2C | 0 | + | 171541800 | 171542049 | 0 | 1 | 249 | 0 | -1.5225609 | 0.0002712 | 0.0057162 |
| chr11 | 86808280 | 86808479 | PRSS23 | 0 | + | 86808280 | 86808479 | 0 | 1 | 199 | 0 | -0.7929473 | 0.0000000 | 0.0000002 |
| chr2 | 232520463 | 232522020 | PRSS56 | 0 | + | 232520463 | 232522020 | 0 | 4 | 233,108,51,50 | 0,858,1353,1508 | 1.0016718 | 0.0007233 | 0.0123211 |
| chr1 | 151033924 | 151034619 | PRUNE | 0 | + | 151033924 | 151034619 | 0 | 1 | 695 | 0 | -0.6986298 | 0.0031795 | 0.0372405 |
| chr17 | 32480542 | 32480641 | PSMD11 | 0 | + | 32480542 | 32480641 | 0 | 1 | 99 | 0 | -2.2099647 | 0.0009278 | 0.0149732 |
| chr17 | 39980818 | 39984417 | PSMD3 | 0 | + | 39980818 | 39984417 | 0 | 2 | 373,124 | 0,3476 | 1.2085019 | 0.0000043 | 0.0001935 |
| chr16 | 74296824 | 74296873 | PSMD7 | 0 | + | 74296824 | 74296873 | 0 | 1 | 49 | 0 | 1.3691759 | 0.0122147 | 0.0962085 |
| chr16 | 74305601 | 74305699 | PSMD7 | 0 | + | 74305601 | 74305699 | 0 | 1 | 98 | 0 | -1.0671074 | 0.0103291 | 0.0861465 |
| chr17 | 42839322 | 42841637 | PSME3 | 0 | + | 42839322 | 42841637 | 0 | 2 | 59,140 | 0,2176 | 0.9289266 | 0.0067854 | 0.0645326 |
| chr19 | 797491 | 804063 | PTBP1 | 0 | + | 797491 | 804063 | 0 | 4 | 15,31,76,28 | 0,1922,6070,6545 | -1.1191241 | 0.0030759 | 0.0362145 |
| chr7 | 99419169 | 99420111 | PTCD1 | 0 | - | 99419169 | 99420111 | 0 | 1 | 942 | 0 | -0.6008608 | 0.0001957 | 0.0043804 |
| chr8 | 96334009 | 96334058 | PTDSS1 | 0 | + | 96334009 | 96334058 | 0 | 1 | 49 | 0 | 2.3712826 | 0.0016175 | 0.0225656 |
| chr1 | 116949272 | 116966929 | PTGFRN | 0 | + | 116949272 | 116966929 | 0 | 3 | 301,426,19 | 0,11971,17639 | -0.5392140 | 0.0006625 | 0.0115911 |
| chr1 | 116967029 | 116967276 | PTGFRN | 0 | + | 116967029 | 116967276 | 0 | 1 | 247 | 0 | -0.9112878 | 0.0072297 | 0.0671019 |
| chr11 | 47571519 | 47571767 | PTPMT1 | 0 | + | 47571519 | 47571767 | 0 | 1 | 248 | 0 | -0.8146735 | 0.0000001 | 0.0000077 |
| chr12 | 112504750 | 112505927 | PTPN11 | 0 | + | 112504750 | 112505927 | 0 | 2 | 47,103 | 0,1075 | -1.2375899 | 0.0127357 | 0.0990201 |
| chr1 | 43622076 | 43622325 | PTPRF | 0 | + | 43622076 | 43622325 | 0 | 1 | 249 | 0 | -0.9703359 | 0.0066789 | 0.0640237 |
| chr19 | 5244356 | 5256109 | PTPRS | 0 | - | 5244356 | 5256109 | 0 | 3 | 127,270,2 | 0,1420,11752 | -0.8560818 | 0.0096857 | 0.0821494 |
| chr17 | 42404092 | 42404390 | PTRF | 0 | - | 42404092 | 42404390 | 0 | 1 | 298 | 0 | -1.3554438 | 0.0000000 | 0.0000015 |
| chr17 | 42404440 | 42405136 | PTRF | 0 | - | 42404440 | 42405136 | 0 | 1 | 696 | 0 | -0.9794690 | 0.0000010 | 0.0000535 |
| chr1 | 31059281 | 31065717 | PUM1 | 0 | - | 31059281 | 31065717 | 0 | 2 | 297,102 | 0,6335 | -0.8558691 | 0.0045038 | 0.0480012 |
| chr7 | 44884736 | 44885135 | PURB | 0 | - | 44884736 | 44885135 | 0 | 1 | 399 | 0 | 0.6752882 | 0.0116257 | 0.0933974 |
| chr12 | 131932291 | 131943564 | PUS1 | 0 | + | 131932291 | 131943564 | 0 | 4 | 22,103,692,26 | 0,6882,9001,11248 | -0.5641164 | 0.0000322 | 0.0010594 |
| chr19 | 44662563 | 44662612 | PVR | 0 | + | 44662563 | 44662612 | 0 | 1 | 49 | 0 | -1.4599151 | 0.0071749 | 0.0668240 |
| chr19 | 44662963 | 44663462 | PVR | 0 | + | 44662963 | 44663462 | 0 | 1 | 499 | 0 | -0.7768255 | 0.0000132 | 0.0005089 |
| chr10 | 132405408 | 132405654 | PWWP2B | 0 | + | 132405408 | 132405654 | 0 | 1 | 246 | 0 | -0.6793383 | 0.0123116 | 0.0967191 |
| chr6 | 3751564 | 3751713 | PXDC1 | 0 | - | 3751564 | 3751713 | 0 | 1 | 149 | 0 | 0.8151094 | 0.0004750 | 0.0088765 |
| chr2 | 1633387 | 1633786 | PXDN | 0 | - | 1633387 | 1633786 | 0 | 1 | 399 | 0 | -1.8689238 | 0.0000004 | 0.0000253 |
| chr2 | 1644704 | 1649471 | PXDN | 0 | - | 1644704 | 1649471 | 0 | 2 | 49,1300 | 0,3468 | -0.6356213 | 0.0000991 | 0.0025942 |
| chr3 | 49104391 | 49104733 | QARS | 0 | - | 49104391 | 49104733 | 0 | 2 | 81,117 | 0,226 | 2.0496911 | 0.0000000 | 0.0000031 |
| chr16 | 29679120 | 29694913 | QPRT | 0 | + | 29679120 | 29694913 | 0 | 2 | 91,250 | 0,15544 | 0.6602140 | 0.0001918 | 0.0043096 |
| chr16 | 29697653 | 29697750 | QPRT | 0 | + | 29697653 | 29697750 | 0 | 1 | 97 | 0 | -1.5940161 | 0.0000165 | 0.0006075 |
| chr3 | 49029907 | 49030203 | QRICH1 | 0 | - | 49029907 | 49030203 | 0 | 1 | 296 | 0 | -0.6143459 | 0.0108518 | 0.0892796 |
| chr3 | 49056907 | 49057550 | QRICH1 | 0 | - | 49056907 | 49057550 | 0 | 1 | 643 | 0 | -0.7165242 | 0.0000046 | 0.0002050 |
| chr1 | 180194384 | 180196995 | QSOX1 | 0 | + | 180194384 | 180196995 | 0 | 2 | 9,734 | 0,1878 | -0.5991343 | 0.0001038 | 0.0026916 |
| chr1 | 180197391 | 180197688 | QSOX1 | 0 | + | 180197391 | 180197688 | 0 | 1 | 297 | 0 | -1.1603228 | 0.0000000 | 0.0000014 |
| chr2 | 26034059 | 26034556 | RAB10 | 0 | + | 26034059 | 26034556 | 0 | 1 | 497 | 0 | 0.9776673 | 0.0008579 | 0.0140584 |
| chr8 | 37877382 | 37877481 | RAB11FIP1 | 0 | - | 37877382 | 37877481 | 0 | 1 | 99 | 0 | -3.2930376 | 0.0000098 | 0.0003962 |
| chr2 | 73075079 | 73075627 | RAB11FIP5 | 0 | - | 73075079 | 73075627 | 0 | 1 | 548 | 0 | -0.9825088 | 0.0001171 | 0.0029686 |
| chr16 | 2153028 | 2153372 | RAB26 | 0 | + | 2153028 | 2153372 | 0 | 3 | 18,77,54 | 0,118,291 | 2.2288906 | 0.0000443 | 0.0013593 |
| chr16 | 2153421 | 2154062 | RAB26 | 0 | + | 2153421 | 2154062 | 0 | 1 | 641 | 0 | 0.6342401 | 0.0000517 | 0.0015308 |
| chr6 | 146544028 | 146544077 | RAB32 | 0 | + | 146544028 | 146544077 | 0 | 1 | 49 | 0 | 2.1755646 | 0.0007399 | 0.0125795 |
| chr15 | 63189628 | 63189677 | RAB8B | 0 | + | 63189628 | 63189677 | 0 | 1 | 49 | 0 | 1.3024663 | 0.0111973 | 0.0909652 |
| chr9 | 136833734 | 136833983 | RABL6 | 0 | + | 136833734 | 136833983 | 0 | 1 | 249 | 0 | -1.0862390 | 0.0109618 | 0.0897767 |
| chr7 | 6402619 | 6403218 | RAC1 | 0 | + | 6402619 | 6403218 | 0 | 1 | 599 | 0 | -1.1382302 | 0.0007024 | 0.0120634 |
| chr17 | 17794768 | 17795663 | RAI1 | 0 | + | 17794768 | 17795663 | 0 | 1 | 895 | 0 | -1.4275231 | 0.0000000 | 0.0000000 |
| chr17 | 17795912 | 17796110 | RAI1 | 0 | + | 17795912 | 17796110 | 0 | 1 | 198 | 0 | -2.3814988 | 0.0000002 | 0.0000154 |
| chr17 | 17797056 | 17797155 | RAI1 | 0 | + | 17797056 | 17797155 | 0 | 1 | 99 | 0 | -1.5538090 | 0.0038101 | 0.0424835 |
| chr17 | 17797205 | 17797354 | RAI1 | 0 | + | 17797205 | 17797354 | 0 | 1 | 149 | 0 | -1.7540568 | 0.0088230 | 0.0771207 |
| chr17 | 17798001 | 17798199 | RAI1 | 0 | + | 17798001 | 17798199 | 0 | 1 | 198 | 0 | -2.2823116 | 0.0000167 | 0.0006132 |
| chr18 | 9536031 | 9536129 | RALBP1 | 0 | + | 9536031 | 9536129 | 0 | 1 | 98 | 0 | -2.0193834 | 0.0009200 | 0.0148743 |
| chr12 | 53211312 | 53211411 | RARG | 0 | - | 53211312 | 53211411 | 0 | 1 | 99 | 0 | -1.1021113 | 0.0091282 | 0.0790400 |
| chr19 | 48720634 | 48720831 | RASIP1 | 0 | - | 48720634 | 48720831 | 0 | 1 | 197 | 0 | -0.9684764 | 0.0059099 | 0.0587805 |
| chr12 | 64610513 | 64684805 | RASSF3 | 0 | + | 64610513 | 64684805 | 0 | 2 | 231,19 | 0,74274 | 1.2827430 | 0.0001250 | 0.0031042 |
| chr19 | 10316313 | 10316412 | RAVER1 | 0 | - | 10316313 | 10316412 | 0 | 1 | 99 | 0 | -1.4506312 | 0.0060106 | 0.0594432 |
| chrX | 47145268 | 47145366 | RBM10 | 0 | + | 47145268 | 47145366 | 0 | 1 | 98 | 0 | 2.4629193 | 0.0016384 | 0.0227862 |
| chr20 | 35652946 | 35653094 | RBM12 | 0 | - | 35652946 | 35653094 | 0 | 1 | 148 | 0 | -1.3769681 | 0.0022047 | 0.0283345 |
| chr11 | 66617022 | 66643805 | RBM14-RBM4 | 0 | + | 66617022 | 66643805 | 0 | 2 | 36,356 | 0,26428 | -1.1862468 | 0.0000000 | 0.0000005 |
| chr1 | 110340864 | 110341212 | RBM15 | 0 | + | 110340864 | 110341212 | 0 | 1 | 348 | 0 | -1.3538349 | 0.0000001 | 0.0000070 |
| chr1 | 110341560 | 110341609 | RBM15 | 0 | + | 110341560 | 110341609 | 0 | 1 | 49 | 0 | -1.7358694 | 0.0021138 | 0.0274586 |
| chr3 | 51392216 | 51393513 | RBM15B | 0 | + | 51392216 | 51393513 | 0 | 1 | 1297 | 0 | -1.0306087 | 0.0000000 | 0.0000000 |
| chr7 | 155745249 | 155763856 | RBM33 | 0 | + | 155745249 | 155763856 | 0 | 2 | 354,45 | 0,18563 | -1.0085608 | 0.0039030 | 0.0432532 |
| chr3 | 49962643 | 49968673 | RBM6 | 0 | + | 49962643 | 49968673 | 0 | 2 | 43,1204 | 0,4827 | -0.5275648 | 0.0077658 | 0.0705936 |
| chr1 | 145926876 | 145927486 | RBM8A | 0 | - | 145926876 | 145927486 | 0 | 3 | 11,60,127 | 0,142,484 | 0.6822863 | 0.0060791 | 0.0599892 |
| chrX | 136875155 | 136875293 | RBMX | 0 | - | 136875155 | 136875293 | 0 | 2 | 14,36 | 0,103 | 1.9279477 | 0.0000805 | 0.0022012 |
| chrX | 136879032 | 136879340 | RBMX | 0 | - | 136879032 | 136879340 | 0 | 2 | 127,22 | 0,287 | 1.7263551 | 0.0002197 | 0.0048103 |
| chrX | 136879439 | 136880631 | RBMX | 0 | - | 136879439 | 136880631 | 0 | 2 | 15,35 | 0,1158 | 1.9601071 | 0.0008744 | 0.0142596 |
| chr1 | 88982548 | 88983046 | RBMXL1 | 0 | - | 88982548 | 88983046 | 0 | 1 | 498 | 0 | -0.5811954 | 0.0067584 | 0.0644116 |
| chr1 | 28538403 | 28538749 | RCC1 | 0 | + | 28538403 | 28538749 | 0 | 1 | 346 | 0 | -0.8616452 | 0.0002002 | 0.0044509 |
| chr1 | 17407052 | 17407250 | RCC2 | 0 | - | 17407052 | 17407250 | 0 | 1 | 198 | 0 | 1.2148046 | 0.0040498 | 0.0444987 |
| chr15 | 90960456 | 90961742 | RCCD1 | 0 | + | 90960456 | 90961742 | 0 | 3 | 43,30,125 | 0,569,1162 | 2.4633264 | 0.0000000 | 0.0000014 |
| chr11 | 32091030 | 32097166 | RCN1 | 0 | + | 32091030 | 32097166 | 0 | 2 | 421,23 | 0,6114 | 0.8798903 | 0.0001414 | 0.0034097 |
| chr14 | 67677344 | 67677393 | RDH11 | 0 | - | 67677344 | 67677393 | 0 | 1 | 49 | 0 | -1.4465784 | 0.0102612 | 0.0857846 |
| chr14 | 67677443 | 67677640 | RDH11 | 0 | - | 67677443 | 67677640 | 0 | 1 | 197 | 0 | -1.3298622 | 0.0003167 | 0.0064790 |
| chr14 | 67678183 | 67685070 | RDH11 | 0 | - | 67678183 | 67685070 | 0 | 2 | 241,56 | 0,6832 | -1.2132178 | 0.0000793 | 0.0021811 |
| chr11 | 110233335 | 110236103 | RDX | 0 | - | 110233335 | 110236103 | 0 | 2 | 145,5 | 0,2764 | -1.1212461 | 0.0062944 | 0.0615108 |
| chr17 | 75661054 | 75662910 | RECQL5 | 0 | - | 75661054 | 75662910 | 0 | 3 | 13,103,432 | 0,552,1425 | -0.8726968 | 0.0007976 | 0.0133234 |
| chr11 | 65654141 | 65654190 | RELA | 0 | - | 65654141 | 65654190 | 0 | 1 | 49 | 0 | 3.0008532 | 0.0004448 | 0.0084787 |
| chr11 | 65654290 | 65654785 | RELA | 0 | - | 65654290 | 65654785 | 0 | 1 | 495 | 0 | 0.7380287 | 0.0000001 | 0.0000040 |
| chr7 | 150371630 | 150372024 | REPIN1 | 0 | + | 150371630 | 150372024 | 0 | 1 | 394 | 0 | -0.6026343 | 0.0071776 | 0.0668240 |
| chr7 | 150372369 | 150372467 | REPIN1 | 0 | + | 150372369 | 150372467 | 0 | 1 | 98 | 0 | -1.7217763 | 0.0003003 | 0.0062075 |
| chr7 | 150372763 | 150373108 | REPIN1 | 0 | + | 150372763 | 150373108 | 0 | 1 | 345 | 0 | -0.5385177 | 0.0056946 | 0.0572880 |
| chr4 | 56930623 | 56930722 | REST | 0 | + | 56930623 | 56930722 | 0 | 1 | 99 | 0 | -2.6207645 | 0.0025862 | 0.0320754 |
| chr2 | 85342993 | 85343091 | RETSAT | 0 | - | 85342993 | 85343091 | 0 | 1 | 98 | 0 | -0.9932438 | 0.0080950 | 0.0726385 |
| chr19 | 1827888 | 1827987 | REXO1 | 0 | - | 1827888 | 1827987 | 0 | 1 | 99 | 0 | -2.0080753 | 0.0030894 | 0.0363338 |
| chr17 | 82048050 | 82048099 | RFNG | 0 | - | 82048050 | 82048099 | 0 | 1 | 49 | 0 | -1.3541918 | 0.0046883 | 0.0495599 |
| chr16 | 74652095 | 74661250 | RFWD3 | 0 | - | 74652095 | 74661250 | 0 | 2 | 28,319 | 0,8837 | -0.7543474 | 0.0010606 | 0.0166421 |
| chr5 | 98779616 | 98780013 | RGMB | 0 | + | 98779616 | 98780013 | 0 | 1 | 397 | 0 | -0.8897153 | 0.0105677 | 0.0875388 |
| chr12 | 2888195 | 2888441 | RHNO1 | 0 | + | 2888195 | 2888441 | 0 | 1 | 246 | 0 | -0.7336647 | 0.0013152 | 0.0195044 |
| chr2 | 20447516 | 20447861 | RHOB | 0 | + | 20447516 | 20447861 | 0 | 1 | 345 | 0 | -1.4387666 | 0.0000000 | 0.0000000 |
| chr2 | 20448504 | 20448850 | RHOB | 0 | + | 20448504 | 20448850 | 0 | 1 | 346 | 0 | -0.5826062 | 0.0000233 | 0.0008043 |
| chr11 | 3827320 | 3827369 | RHOG | 0 | - | 3827320 | 3827369 | 0 | 1 | 49 | 0 | -1.4125410 | 0.0004394 | 0.0084104 |
| chr11 | 3827954 | 3828003 | RHOG | 0 | - | 3827954 | 3828003 | 0 | 1 | 49 | 0 | -1.7118351 | 0.0000681 | 0.0019228 |
| chr1 | 228743498 | 228743696 | RHOU | 0 | + | 228743498 | 228743696 | 0 | 1 | 198 | 0 | -0.8663553 | 0.0069565 | 0.0656874 |
| chr19 | 32979984 | 32980033 | RHPN2 | 0 | - | 32979984 | 32980033 | 0 | 1 | 49 | 0 | -2.2093296 | 0.0038938 | 0.0431774 |
| chr6 | 3113999 | 3114246 | RIPK1 | 0 | + | 3113999 | 3114246 | 0 | 1 | 247 | 0 | -1.1928672 | 0.0101253 | 0.0849391 |
| chr8 | 89757896 | 89758094 | RIPK2 | 0 | + | 89757896 | 89758094 | 0 | 1 | 198 | 0 | 1.1247506 | 0.0095156 | 0.0811242 |
| chr8 | 89789425 | 89790368 | RIPK2 | 0 | + | 89789425 | 89790368 | 0 | 2 | 58,290 | 0,654 | -0.8666090 | 0.0005523 | 0.0099706 |
| chrX | 74591559 | 74591758 | RLIM | 0 | - | 74591559 | 74591758 | 0 | 1 | 199 | 0 | -1.1860697 | 0.0044761 | 0.0478055 |
| chr6 | 151445356 | 151445698 | RMND1 | 0 | - | 151445356 | 151445698 | 0 | 1 | 342 | 0 | -1.3625012 | 0.0021021 | 0.0273456 |
| chr14 | 20699605 | 20699704 | RNASE4 | 0 | + | 20699605 | 20699704 | 0 | 1 | 99 | 0 | -2.9097972 | 0.0009705 | 0.0155665 |
| chr1 | 51270043 | 51271414 | RNF11 | 0 | + | 51270043 | 51271414 | 0 | 2 | 83,264 | 0,1108 | -0.6616581 | 0.0019745 | 0.0262514 |
| chr20 | 49953293 | 49953392 | RNF114 | 0 | + | 49953293 | 49953392 | 0 | 1 | 99 | 0 | 4.3548863 | 0.0000000 | 0.0000000 |
| chr20 | 49953639 | 49953688 | RNF114 | 0 | + | 49953639 | 49953688 | 0 | 1 | 49 | 0 | 4.4133564 | 0.0000000 | 0.0000004 |
| chr3 | 49720876 | 49721100 | RNF123 | 0 | + | 49720876 | 49721100 | 0 | 2 | 19,81 | 0,144 | -2.4129476 | 0.0015562 | 0.0219464 |
| chr3 | 196488681 | 196503069 | RNF168 | 0 | - | 196488681 | 196503069 | 0 | 2 | 3,197 | 0,14192 | -1.4475129 | 0.0017013 | 0.0234426 |
| chr11 | 74835687 | 74836334 | RNF169 | 0 | + | 74835687 | 74836334 | 0 | 1 | 647 | 0 | -1.4007462 | 0.0000004 | 0.0000234 |
| chr11 | 74836933 | 74836982 | RNF169 | 0 | + | 74836933 | 74836982 | 0 | 1 | 49 | 0 | -2.9586770 | 0.0000002 | 0.0000105 |
| chr1 | 32936794 | 32937291 | RNF19B | 0 | - | 32936794 | 32937291 | 0 | 1 | 497 | 0 | -0.6007442 | 0.0043155 | 0.0467654 |
| chr1 | 32964615 | 32964763 | RNF19B | 0 | - | 32964615 | 32964763 | 0 | 1 | 148 | 0 | 2.1505859 | 0.0000001 | 0.0000104 |
| chr17 | 80332028 | 80332227 | RNF213 | 0 | + | 80332028 | 80332227 | 0 | 1 | 199 | 0 | -1.3933523 | 0.0024248 | 0.0305136 |
| chr17 | 80339305 | 80339803 | RNF213 | 0 | + | 80339305 | 80339803 | 0 | 1 | 498 | 0 | -1.2188376 | 0.0012899 | 0.0192564 |
| chr17 | 80345269 | 80345468 | RNF213 | 0 | + | 80345269 | 80345468 | 0 | 1 | 199 | 0 | -1.4712402 | 0.0064109 | 0.0622130 |
| chr17 | 80348114 | 80348262 | RNF213 | 0 | + | 80348114 | 80348262 | 0 | 1 | 148 | 0 | -1.6253248 | 0.0011398 | 0.0175189 |
| chr7 | 5620991 | 5621090 | RNF216 | 0 | - | 5620991 | 5621090 | 0 | 1 | 99 | 0 | -1.8944653 | 0.0043980 | 0.0472911 |
| chr7 | 5741226 | 5741525 | RNF216 | 0 | - | 5741226 | 5741525 | 0 | 1 | 299 | 0 | -1.4236626 | 0.0012745 | 0.0190582 |
| chr13 | 26214313 | 26214412 | RNF6 | 0 | - | 26214313 | 26214412 | 0 | 1 | 99 | 0 | -1.3588681 | 0.0065435 | 0.0631949 |
| chr17 | 791820 | 792258 | RNMTL1 | 0 | + | 791820 | 792258 | 0 | 1 | 438 | 0 | -1.1204411 | 0.0000000 | 0.0000007 |
| chr9 | 91723246 | 91723894 | ROR2 | 0 | - | 91723246 | 91723894 | 0 | 1 | 648 | 0 | -1.0134966 | 0.0000100 | 0.0004016 |
| chr9 | 91724195 | 91724444 | ROR2 | 0 | - | 91724195 | 91724444 | 0 | 1 | 249 | 0 | -1.2375361 | 0.0008458 | 0.0139230 |
| chr9 | 91724545 | 91724743 | ROR2 | 0 | - | 91724545 | 91724743 | 0 | 1 | 198 | 0 | -1.4650894 | 0.0004185 | 0.0080966 |
| chr1 | 151807208 | 151807554 | RORC | 0 | - | 151807208 | 151807554 | 0 | 1 | 346 | 0 | 0.7229159 | 0.0076866 | 0.0700849 |
| chrX | 46837061 | 46853483 | RP2 | 0 | + | 46837061 | 46853483 | 0 | 2 | 142,8 | 0,16415 | 1.2059279 | 0.0028921 | 0.0345220 |
| chr10 | 102455946 | 102456194 | RPARP-AS1 | 0 | + | 102455946 | 102456194 | 0 | 1 | 248 | 0 | 1.3049299 | 0.0033031 | 0.0382189 |
| chr9 | 127447721 | 127447920 | RPL12 | 0 | - | 127447721 | 127447920 | 0 | 2 | 6,44 | 0,156 | 2.1843528 | 0.0004980 | 0.0091835 |
| chr17 | 28723597 | 28723872 | RPL23A | 0 | + | 28723597 | 28723872 | 0 | 2 | 44,6 | 0,270 | 2.9240737 | 0.0011368 | 0.0175189 |
| chr3 | 101681137 | 101681186 | RPL24 | 0 | - | 101681137 | 101681186 | 0 | 1 | 49 | 0 | 2.2573415 | 0.0073399 | 0.0677767 |
| chr17 | 42998429 | 43000003 | RPL27 | 0 | + | 42998429 | 43000003 | 0 | 3 | 43,83,71 | 0,320,1504 | -1.4923387 | 0.0029907 | 0.0355116 |
| chr17 | 43002768 | 43002905 | RPL27 | 0 | + | 43002768 | 43002905 | 0 | 2 | 16,34 | 0,104 | 1.4427148 | 0.0088529 | 0.0771784 |
| chr3 | 51995893 | 51995942 | RPL29 | 0 | - | 51995893 | 51995942 | 0 | 1 | 49 | 0 | -1.1969447 | 0.0005482 | 0.0099068 |
| chr22 | 39312932 | 39312981 | RPL3 | 0 | - | 39312932 | 39312981 | 0 | 1 | 49 | 0 | 1.3926586 | 0.0101165 | 0.0849276 |
| chr22 | 39316829 | 39317596 | RPL3 | 0 | - | 39316829 | 39317596 | 0 | 2 | 13,136 | 0,632 | -2.4868139 | 0.0001612 | 0.0037738 |
| chr3 | 197950181 | 197954007 | RPL35A | 0 | + | 197950181 | 197954007 | 0 | 4 | 41,43,153,5 | 0,755,978,3822 | 0.8581411 | 0.0031305 | 0.0367379 |
| chr14 | 49618832 | 49618881 | RPL36AL | 0 | - | 49618832 | 49618881 | 0 | 1 | 49 | 0 | -0.7266589 | 0.0013004 | 0.0193173 |
| chr2 | 216498797 | 216499284 | RPL37A | 0 | + | 216498797 | 216499284 | 0 | 2 | 81,15 | 0,473 | -1.7557821 | 0.0021688 | 0.0280313 |
| chrX | 119786550 | 119786645 | RPL39 | 0 | - | 119786550 | 119786645 | 0 | 1 | 95 | 0 | 1.2347041 | 0.0063386 | 0.0617678 |
| chr15 | 66499365 | 66500115 | RPL4 | 0 | - | 66499365 | 66500115 | 0 | 2 | 288,60 | 0,691 | -0.7859442 | 0.0025529 | 0.0317494 |
| chr9 | 133351327 | 133351376 | RPL7A | 0 | + | 133351327 | 133351376 | 0 | 1 | 49 | 0 | 2.0560411 | 0.0014419 | 0.0208043 |
| chr15 | 69455450 | 69455499 | RPLP1 | 0 | + | 69455450 | 69455499 | 0 | 1 | 49 | 0 | 1.5103929 | 0.0033380 | 0.0385095 |
| chr11 | 812778 | 812827 | RPLP2 | 0 | + | 812778 | 812827 | 0 | 1 | 49 | 0 | 1.2975751 | 0.0040683 | 0.0445939 |
| chr15 | 74955449 | 74956044 | RPP25 | 0 | - | 74955449 | 74956044 | 0 | 1 | 595 | 0 | -0.7829679 | 0.0000000 | 0.0000006 |
| chr9 | 34610915 | 34611108 | RPP25L | 0 | - | 34610915 | 34611108 | 0 | 1 | 193 | 0 | -0.7264908 | 0.0120520 | 0.0953533 |
| chr1 | 150472829 | 150473326 | RPRD2 | 0 | + | 150472829 | 150473326 | 0 | 1 | 497 | 0 | -0.8214409 | 0.0003661 | 0.0073001 |
| chr19 | 49499564 | 49499662 | RPS11 | 0 | + | 49499564 | 49499662 | 0 | 1 | 98 | 0 | 1.6615419 | 0.0001219 | 0.0030654 |
| chr5 | 150444278 | 150444327 | RPS14 | 0 | - | 150444278 | 150444327 | 0 | 1 | 49 | 0 | 1.5603808 | 0.0014496 | 0.0208499 |
| chr19 | 1438462 | 1440108 | RPS15 | 0 | + | 1438462 | 1440108 | 0 | 3 | 119,86,90 | 0,345,1557 | -1.2680067 | 0.0000040 | 0.0001837 |
| chr16 | 18789061 | 18790334 | RPS15A | 0 | - | 18789061 | 18790334 | 0 | 2 | 58,91 | 0,1183 | -1.2042761 | 0.0053627 | 0.0547398 |
| chr20 | 62388330 | 62388471 | RPS21 | 0 | + | 62388330 | 62388471 | 0 | 2 | 35,15 | 0,127 | 1.7406447 | 0.0000142 | 0.0005396 |
| chr1 | 153991632 | 153992112 | RPS27 | 0 | + | 153991632 | 153992112 | 0 | 2 | 45,48 | 0,433 | 1.6091436 | 0.0007441 | 0.0126288 |
| chrX | 72275661 | 72277291 | RPS4X | 0 | - | 72275661 | 72277291 | 0 | 3 | 64,78,99 | 0,496,1532 | -2.2962702 | 0.0000000 | 0.0000000 |
| chr19 | 58394706 | 58394755 | RPS5 | 0 | + | 58394706 | 58394755 | 0 | 1 | 49 | 0 | 2.0139024 | 0.0010403 | 0.0163953 |
| chr17 | 59946638 | 59947235 | RPS6KB1 | 0 | + | 59946638 | 59947235 | 0 | 1 | 597 | 0 | -0.5593309 | 0.0101003 | 0.0848707 |
| chr17 | 80965281 | 80965976 | RPTOR | 0 | + | 80965281 | 80965976 | 0 | 1 | 695 | 0 | -0.6728993 | 0.0019453 | 0.0259208 |
| chr9 | 19049724 | 19050123 | RRAGA | 0 | + | 19049724 | 19050123 | 0 | 1 | 399 | 0 | -1.2236235 | 0.0000000 | 0.0000000 |
| chr20 | 17613728 | 17614126 | RRBP1 | 0 | - | 17613728 | 17614126 | 0 | 1 | 398 | 0 | -0.9339370 | 0.0001360 | 0.0033234 |
| chr20 | 17660064 | 17660362 | RRBP1 | 0 | - | 17660064 | 17660362 | 0 | 1 | 298 | 0 | -0.8791267 | 0.0017767 | 0.0241716 |
| chr6 | 7230471 | 7231017 | RREB1 | 0 | + | 7230471 | 7231017 | 0 | 1 | 546 | 0 | -1.9602488 | 0.0000000 | 0.0000010 |
| chr6 | 7231565 | 7231763 | RREB1 | 0 | + | 7231565 | 7231763 | 0 | 1 | 198 | 0 | -1.8894724 | 0.0001502 | 0.0035940 |
| chr6 | 7240543 | 7246860 | RREB1 | 0 | + | 7240543 | 7246860 | 0 | 2 | 60,437 | 0,5881 | -0.8171319 | 0.0051332 | 0.0531156 |
| chr11 | 4094844 | 4094992 | RRM1 | 0 | + | 4094844 | 4094992 | 0 | 1 | 148 | 0 | 0.9913331 | 0.0105236 | 0.0873333 |
| chr11 | 4138279 | 4138477 | RRM1 | 0 | + | 4138279 | 4138477 | 0 | 1 | 198 | 0 | -0.9566340 | 0.0031048 | 0.0364598 |
| chr1 | 156728520 | 156729161 | RRNAD1 | 0 | + | 156728520 | 156729161 | 0 | 1 | 641 | 0 | -0.7592667 | 0.0017029 | 0.0234426 |
| chr22 | 42511112 | 42511161 | RRP7A | 0 | - | 42511112 | 42511161 | 0 | 1 | 49 | 0 | -3.2655999 | 0.0001249 | 0.0031042 |
| chr22 | 42511955 | 42512401 | RRP7A | 0 | - | 42511955 | 42512401 | 0 | 1 | 446 | 0 | -1.1157332 | 0.0000478 | 0.0014423 |
| chr8 | 66429905 | 66430002 | RRS1 | 0 | + | 66429905 | 66430002 | 0 | 1 | 97 | 0 | -0.9511969 | 0.0049104 | 0.0513383 |
| chr12 | 122522269 | 122526964 | RSRC2 | 0 | - | 122522269 | 122526964 | 0 | 2 | 31,117 | 0,4579 | 1.3179003 | 0.0005230 | 0.0095723 |
| chr20 | 56518488 | 56518786 | RTFDC1 | 0 | + | 56518488 | 56518786 | 0 | 1 | 298 | 0 | -1.5704807 | 0.0001119 | 0.0028550 |
| chr9 | 35539056 | 35547171 | RUSC2 | 0 | + | 35539056 | 35547171 | 0 | 2 | 53,742 | 0,7374 | -0.9014803 | 0.0002076 | 0.0045782 |
| chr9 | 35548263 | 35548362 | RUSC2 | 0 | + | 35548263 | 35548362 | 0 | 1 | 99 | 0 | -2.1994137 | 0.0008148 | 0.0135237 |
| chr1 | 152033676 | 152037035 | S100A11 | 0 | - | 152033676 | 152037035 | 0 | 2 | 125,123 | 0,3237 | 0.6037366 | 0.0039467 | 0.0436309 |
| chr1 | 153607570 | 153607619 | S100A16 | 0 | - | 153607570 | 153607619 | 0 | 1 | 49 | 0 | -1.0133516 | 0.0022448 | 0.0287679 |
| chr1 | 153535312 | 153535998 | S100A6 | 0 | - | 153535312 | 153535998 | 0 | 2 | 49,50 | 0,637 | 1.0784905 | 0.0000000 | 0.0000000 |
| chr1 | 32826381 | 32826779 | S100PBP | 0 | + | 32826381 | 32826779 | 0 | 1 | 398 | 0 | -1.1869166 | 0.0005753 | 0.0103153 |
| chr19 | 47203737 | 47209451 | SAE1 | 0 | + | 47203737 | 47209451 | 0 | 2 | 4,293 | 0,5422 | -1.3550194 | 0.0001223 | 0.0030716 |
| chr20 | 51790941 | 51791090 | SALL4 | 0 | - | 51790941 | 51791090 | 0 | 1 | 149 | 0 | -2.0008871 | 0.0103985 | 0.0865726 |
| chr20 | 51791887 | 51792135 | SALL4 | 0 | - | 51791887 | 51792135 | 0 | 1 | 248 | 0 | -0.8695945 | 0.0117069 | 0.0936894 |
| chr14 | 54702206 | 54737046 | SAMD4A | 0 | + | 54702206 | 54737046 | 0 | 2 | 375,23 | 0,34818 | -0.8734414 | 0.0023014 | 0.0292861 |
| chr14 | 54789741 | 54789839 | SAMD4A | 0 | + | 54789741 | 54789839 | 0 | 1 | 98 | 0 | 1.9039486 | 0.0007575 | 0.0127837 |
| chr7 | 93100109 | 93100158 | SAMD9 | 0 | - | 93100109 | 93100158 | 0 | 1 | 49 | 0 | -Inf | 0.0117788 | 0.0939985 |
| chr17 | 75707567 | 75707665 | SAP30BP | 0 | + | 75707567 | 75707665 | 0 | 1 | 98 | 0 | -2.0322551 | 0.0011793 | 0.0179904 |
| chr9 | 137062227 | 137062375 | SAPCD2 | 0 | - | 137062227 | 137062375 | 0 | 1 | 148 | 0 | -1.6727701 | 0.0000339 | 0.0011033 |
| chr9 | 137062525 | 137062574 | SAPCD2 | 0 | - | 137062525 | 137062574 | 0 | 1 | 49 | 0 | -1.6644225 | 0.0048867 | 0.0511497 |
| chr9 | 137062625 | 137063172 | SAPCD2 | 0 | - | 137062625 | 137063172 | 0 | 1 | 547 | 0 | -1.3099055 | 0.0000001 | 0.0000045 |
| chr8 | 30069719 | 30073976 | SARAF | 0 | - | 30069719 | 30073976 | 0 | 2 | 341,100 | 0,4158 | -1.1863776 | 0.0000099 | 0.0003980 |
| chr1 | 109213893 | 109215058 | SARS | 0 | + | 109213893 | 109215058 | 0 | 2 | 236,512 | 0,654 | 1.3158786 | 0.0000000 | 0.0000000 |
| chr14 | 50665202 | 50665451 | SAV1 | 0 | - | 50665202 | 50665451 | 0 | 1 | 249 | 0 | -1.0613966 | 0.0099358 | 0.0838384 |
| chr7 | 66988238 | 66988335 | SBDS | 0 | - | 66988238 | 66988335 | 0 | 1 | 97 | 0 | -2.4320693 | 0.0004005 | 0.0078158 |
| chr19 | 49651307 | 49651505 | SCAF1 | 0 | + | 49651307 | 49651505 | 0 | 1 | 198 | 0 | -1.2490870 | 0.0000549 | 0.0016100 |
| chr19 | 49651704 | 49652000 | SCAF1 | 0 | + | 49651704 | 49652000 | 0 | 1 | 296 | 0 | -0.9726463 | 0.0001091 | 0.0028065 |
| chr19 | 49652447 | 49652793 | SCAF1 | 0 | + | 49652447 | 49652793 | 0 | 1 | 346 | 0 | -1.5405972 | 0.0000000 | 0.0000000 |
| chr19 | 49653042 | 49653587 | SCAF1 | 0 | + | 49653042 | 49653587 | 0 | 1 | 545 | 0 | -1.5917931 | 0.0000000 | 0.0000000 |
| chr21 | 31731970 | 31732068 | SCAF4 | 0 | - | 31731970 | 31732068 | 0 | 1 | 98 | 0 | 1.9920957 | 0.0067365 | 0.0643012 |
| chr8 | 27658727 | 27659174 | SCARA3 | 0 | + | 27658727 | 27659174 | 0 | 1 | 447 | 0 | -1.4612059 | 0.0000000 | 0.0000000 |
| chr10 | 100363042 | 100363191 | SCD | 0 | + | 100363042 | 100363191 | 0 | 1 | 149 | 0 | -1.3975660 | 0.0025648 | 0.0318758 |
| chr2 | 238098522 | 238098620 | SCLY | 0 | + | 238098522 | 238098620 | 0 | 1 | 98 | 0 | 4.4321429 | 0.0000511 | 0.0015158 |
| chr17 | 10697470 | 10697519 | SCO1 | 0 | - | 10697470 | 10697519 | 0 | 1 | 49 | 0 | 1.7204424 | 0.0039451 | 0.0436309 |
| chr17 | 56978157 | 56981102 | SCPEP1 | 0 | + | 56978157 | 56981102 | 0 | 2 | 79,21 | 0,2925 | 1.7926353 | 0.0013601 | 0.0199912 |
| chr12 | 100338605 | 100339049 | SCYL2 | 0 | + | 100338605 | 100339049 | 0 | 1 | 444 | 0 | -1.0475768 | 0.0004321 | 0.0082882 |
| chr2 | 20201640 | 20201937 | SDC1 | 0 | - | 20201640 | 20201937 | 0 | 1 | 297 | 0 | -0.6969794 | 0.0075345 | 0.0690905 |
| chr1 | 30869913 | 30870112 | SDC3 | 0 | - | 30869913 | 30870112 | 0 | 1 | 199 | 0 | -1.3208399 | 0.0011079 | 0.0172053 |
| chr20 | 45326915 | 45326964 | SDC4 | 0 | - | 45326915 | 45326964 | 0 | 1 | 49 | 0 | -1.5880252 | 0.0000010 | 0.0000524 |
| chr20 | 45327013 | 45327112 | SDC4 | 0 | - | 45327013 | 45327112 | 0 | 1 | 99 | 0 | -1.5180974 | 0.0000000 | 0.0000000 |
| chr20 | 45327161 | 45327358 | SDC4 | 0 | - | 45327161 | 45327358 | 0 | 1 | 197 | 0 | -1.0196306 | 0.0000069 | 0.0002930 |
| chr8 | 58581974 | 58582023 | SDCBP | 0 | + | 58581974 | 58582023 | 0 | 1 | 49 | 0 | -2.9814285 | 0.0021079 | 0.0274014 |
| chr1 | 225985275 | 225988340 | SDE2 | 0 | - | 225985275 | 225988340 | 0 | 2 | 249,445 | 0,2621 | -0.8115165 | 0.0000043 | 0.0001962 |
| chr1 | 17044762 | 17054070 | SDHB | 0 | - | 17044762 | 17054070 | 0 | 2 | 127,123 | 0,9186 | 1.2494481 | 0.0000016 | 0.0000821 |
| chr11 | 112086873 | 112087918 | SDHD | 0 | + | 112086873 | 112087918 | 0 | 2 | 87,62 | 0,984 | 1.4448204 | 0.0008946 | 0.0145681 |
| chr8 | 56320085 | 56320134 | SDR16C5 | 0 | - | 56320085 | 56320134 | 0 | 1 | 49 | 0 | 1.4659907 | 0.0044726 | 0.0478055 |
| chr9 | 136440644 | 136440792 | SEC16A | 0 | - | 136440644 | 136440792 | 0 | 1 | 148 | 0 | -1.4922888 | 0.0051563 | 0.0532629 |
| chr9 | 136474541 | 136474889 | SEC16A | 0 | - | 136474541 | 136474889 | 0 | 1 | 348 | 0 | -1.6141524 | 0.0000638 | 0.0018255 |
| chr9 | 136474939 | 136475187 | SEC16A | 0 | - | 136474939 | 136475187 | 0 | 1 | 248 | 0 | -1.1569132 | 0.0086937 | 0.0763464 |
| chr9 | 136475487 | 136475586 | SEC16A | 0 | - | 136475487 | 136475586 | 0 | 1 | 99 | 0 | -2.5448155 | 0.0006218 | 0.0110390 |
| chr9 | 136476234 | 136477378 | SEC16A | 0 | - | 136476234 | 136477378 | 0 | 1 | 1144 | 0 | -0.6395339 | 0.0073018 | 0.0675284 |
| chr4 | 109449485 | 109463473 | SEC24B | 0 | + | 109449485 | 109463473 | 0 | 2 | 25,573 | 0,13416 | -1.1174301 | 0.0000669 | 0.0018956 |
| chr3 | 128070887 | 128071334 | SEC61A1 | 0 | + | 128070887 | 128071334 | 0 | 1 | 447 | 0 | -1.0711930 | 0.0000004 | 0.0000227 |
| chr22 | 31104974 | 31105023 | SELM | 0 | - | 31104974 | 31105023 | 0 | 1 | 49 | 0 | -0.8051636 | 0.0094861 | 0.0809876 |
| chr17 | 7563393 | 7563791 | SENP3 | 0 | + | 7563393 | 7563791 | 0 | 1 | 398 | 0 | -1.1028387 | 0.0010171 | 0.0160999 |
| chr17 | 7563387 | 7563783 | SENP3-EIF4A1 | 0 | + | 7563387 | 7563783 | 0 | 1 | 396 | 0 | -1.0903075 | 0.0011414 | 0.0175189 |
| chr3 | 196885976 | 196886621 | SENP5 | 0 | + | 196885976 | 196886621 | 0 | 1 | 645 | 0 | -0.9417376 | 0.0003585 | 0.0071883 |
| chr17 | 77376351 | 77402358 | SEPT9 | 0 | + | 77376351 | 77402358 | 0 | 2 | 48,300 | 0,25708 | -0.7467523 | 0.0008450 | 0.0139230 |
| chr6 | 2839382 | 2840433 | SERPINB1 | 0 | - | 2839382 | 2840433 | 0 | 2 | 85,15 | 0,1037 | -2.7917641 | 0.0000000 | 0.0000000 |
| chr18 | 63503595 | 63503644 | SERPINB5 | 0 | + | 63503595 | 63503644 | 0 | 1 | 49 | 0 | -2.5005254 | 0.0000375 | 0.0011982 |
| chr6 | 2948507 | 2948969 | SERPINB6 | 0 | - | 2948507 | 2948969 | 0 | 2 | 193,56 | 0,407 | -1.1598190 | 0.0000063 | 0.0002677 |
| chr11 | 75572064 | 75572261 | SERPINH1 | 0 | + | 75572064 | 75572261 | 0 | 1 | 197 | 0 | -0.9464033 | 0.0000336 | 0.0010965 |
| chr11 | 75572459 | 75572656 | SERPINH1 | 0 | + | 75572459 | 75572656 | 0 | 1 | 197 | 0 | -1.2054145 | 0.0000015 | 0.0000777 |
| chr2 | 64635620 | 64636468 | SERTAD2 | 0 | - | 64635620 | 64636468 | 0 | 1 | 848 | 0 | -0.8217882 | 0.0000755 | 0.0020970 |
| chr2 | 64636569 | 64636868 | SERTAD2 | 0 | - | 64636569 | 64636868 | 0 | 1 | 299 | 0 | -1.3028229 | 0.0000872 | 0.0023402 |
| chr16 | 30979400 | 30979699 | SETD1A | 0 | + | 30979400 | 30979699 | 0 | 1 | 299 | 0 | -1.6265168 | 0.0000176 | 0.0006430 |
| chr12 | 121809789 | 121809938 | SETD1B | 0 | + | 121809789 | 121809938 | 0 | 1 | 149 | 0 | -2.7559106 | 0.0000010 | 0.0000540 |
| chr12 | 121809989 | 121810038 | SETD1B | 0 | + | 121809989 | 121810038 | 0 | 1 | 49 | 0 | -4.1316991 | 0.0000000 | 0.0000004 |
| chr12 | 121810339 | 121810788 | SETD1B | 0 | + | 121810339 | 121810788 | 0 | 1 | 449 | 0 | -1.1775407 | 0.0002060 | 0.0045501 |
| chr14 | 99398196 | 99398345 | SETD3 | 0 | - | 99398196 | 99398345 | 0 | 1 | 149 | 0 | -1.8631584 | 0.0005288 | 0.0096425 |
| chr1 | 149926529 | 149927173 | SF3B4 | 0 | - | 149926529 | 149927173 | 0 | 2 | 390,8 | 0,637 | -0.7230341 | 0.0120011 | 0.0951889 |
| chr6 | 144095028 | 144095077 | SF3B5 | 0 | - | 144095028 | 144095077 | 0 | 1 | 49 | 0 | -0.8016796 | 0.0000077 | 0.0003214 |
| chr10 | 7163452 | 7163501 | SFMBT2 | 0 | - | 7163452 | 7163501 | 0 | 1 | 49 | 0 | -Inf | 0.0106699 | 0.0879290 |
| chr1 | 26863629 | 26863775 | SFN | 0 | + | 26863629 | 26863775 | 0 | 1 | 146 | 0 | -1.1692752 | 0.0000000 | 0.0000000 |
| chr1 | 26863823 | 26863872 | SFN | 0 | + | 26863823 | 26863872 | 0 | 1 | 49 | 0 | -1.2219282 | 0.0000001 | 0.0000054 |
| chr6 | 134169692 | 134170088 | SGK1 | 0 | - | 134169692 | 134170088 | 0 | 1 | 396 | 0 | -0.7684909 | 0.0026217 | 0.0323444 |
| chr6 | 134174046 | 134174868 | SGK1 | 0 | - | 134174046 | 134174868 | 0 | 3 | 35,76,137 | 0,465,686 | 0.7126474 | 0.0120399 | 0.0953533 |
| chr14 | 63686245 | 63686492 | SGPP1 | 0 | - | 63686245 | 63686492 | 0 | 1 | 247 | 0 | -1.2146802 | 0.0085713 | 0.0756382 |
| chr14 | 63727700 | 63728095 | SGPP1 | 0 | - | 63727700 | 63728095 | 0 | 1 | 395 | 0 | 0.7391381 | 0.0031815 | 0.0372405 |
| chr19 | 2755256 | 2755551 | SGTA | 0 | - | 2755256 | 2755551 | 0 | 1 | 295 | 0 | -0.7704841 | 0.0035887 | 0.0406917 |
| chr8 | 19314018 | 19319627 | SH2D4A | 0 | + | 19314018 | 19319627 | 0 | 2 | 62,284 | 0,5326 | -0.7113508 | 0.0042850 | 0.0464907 |
| chr1 | 26280061 | 26281232 | SH3BGRL3 | 0 | + | 26280061 | 26281232 | 0 | 3 | 143,168,175 | 0,704,997 | -0.5816722 | 0.0088601 | 0.0771784 |
| chr4 | 151175048 | 151175444 | SH3D19 | 0 | - | 151175048 | 151175444 | 0 | 1 | 396 | 0 | -1.4589807 | 0.0000098 | 0.0003961 |
| chr10 | 103600975 | 103602824 | SH3PXD2A | 0 | - | 103600975 | 103602824 | 0 | 1 | 1849 | 0 | -0.5066788 | 0.0000196 | 0.0007064 |
| chr10 | 103602875 | 103603024 | SH3PXD2A | 0 | - | 103602875 | 103603024 | 0 | 1 | 149 | 0 | -1.9009602 | 0.0000019 | 0.0000953 |
| chr10 | 103603175 | 103603773 | SH3PXD2A | 0 | - | 103603175 | 103603773 | 0 | 1 | 598 | 0 | -0.8469070 | 0.0000224 | 0.0007821 |
| chr5 | 172337936 | 172338383 | SH3PXD2B | 0 | - | 172337936 | 172338383 | 0 | 1 | 447 | 0 | -1.0404308 | 0.0000362 | 0.0011643 |
| chr5 | 172339232 | 172339779 | SH3PXD2B | 0 | - | 172339232 | 172339779 | 0 | 1 | 547 | 0 | -1.0245040 | 0.0026781 | 0.0327445 |
| chr3 | 72750276 | 72750825 | SHQ1 | 0 | - | 72750276 | 72750825 | 0 | 1 | 549 | 0 | -0.9375352 | 0.0032671 | 0.0379236 |
| chr4 | 76739016 | 76739364 | SHROOM3 | 0 | + | 76739016 | 76739364 | 0 | 1 | 348 | 0 | -1.5655790 | 0.0000259 | 0.0008766 |
| chr9 | 34635366 | 34635811 | SIGMAR1 | 0 | - | 34635366 | 34635811 | 0 | 1 | 445 | 0 | -0.8057999 | 0.0000447 | 0.0013643 |
| chr15 | 75371426 | 75375679 | SIN3A | 0 | - | 75371426 | 75375679 | 0 | 2 | 784,15 | 0,4239 | -0.6309642 | 0.0012637 | 0.0189271 |
| chr1 | 232514101 | 232514299 | SIPA1L2 | 0 | - | 232514101 | 232514299 | 0 | 1 | 198 | 0 | -2.0790471 | 0.0003144 | 0.0064472 |
| chr11 | 216308 | 216653 | SIRT3 | 0 | - | 216308 | 216653 | 0 | 1 | 345 | 0 | -0.8650659 | 0.0026981 | 0.0329444 |
| chr16 | 67945828 | 67945956 | SLC12A4 | 0 | - | 67945828 | 67945956 | 0 | 2 | 44,6 | 0,123 | -1.9121474 | 0.0030604 | 0.0361503 |
| chr16 | 67968496 | 67968644 | SLC12A4 | 0 | - | 67968496 | 67968644 | 0 | 1 | 148 | 0 | 1.4594568 | 0.0024803 | 0.0310598 |
| chr1 | 112913735 | 112917988 | SLC16A1 | 0 | - | 112913735 | 112917988 | 0 | 2 | 431,811 | 0,3443 | -0.6359953 | 0.0001293 | 0.0031780 |
| chr1 | 112929338 | 112955658 | SLC16A1 | 0 | - | 112929338 | 112955658 | 0 | 2 | 15,383 | 0,25938 | 1.1027665 | 0.0000784 | 0.0021614 |
| chr17 | 68269053 | 68271137 | SLC16A6 | 0 | - | 68269053 | 68271137 | 0 | 2 | 294,299 | 0,1786 | -0.6612119 | 0.0090413 | 0.0785231 |
| chr5 | 36687045 | 36687094 | SLC1A3 | 0 | + | 36687045 | 36687094 | 0 | 1 | 49 | 0 | -1.5250354 | 0.0001855 | 0.0042143 |
| chr19 | 46787550 | 46788585 | SLC1A5 | 0 | - | 46787550 | 46788585 | 0 | 1 | 1035 | 0 | -0.9188378 | 0.0000005 | 0.0000301 |
| chr2 | 112659047 | 112659652 | SLC20A1 | 0 | + | 112659047 | 112659652 | 0 | 2 | 48,449 | 0,157 | -0.9639712 | 0.0012999 | 0.0193173 |
| chr5 | 132370300 | 132370349 | SLC22A5 | 0 | + | 132370300 | 132370349 | 0 | 1 | 49 | 0 | 2.2774054 | 0.0014872 | 0.0211891 |
| chr1 | 108200009 | 108200308 | SLC25A24 | 0 | - | 108200009 | 108200308 | 0 | 1 | 299 | 0 | 1.6184113 | 0.0000000 | 0.0000002 |
| chrX | 119468400 | 119469965 | SLC25A5 | 0 | + | 119468400 | 119469965 | 0 | 2 | 227,305 | 0,1261 | -0.9616259 | 0.0049238 | 0.0514189 |
| chr5 | 149960936 | 149977783 | SLC26A2 | 0 | + | 149960936 | 149977783 | 0 | 2 | 44,156 | 0,16692 | 1.1441142 | 0.0117541 | 0.0939293 |
| chr9 | 133471294 | 133472042 | SLC2A6 | 0 | - | 133471294 | 133472042 | 0 | 1 | 748 | 0 | -0.5451991 | 0.0074963 | 0.0688907 |
| chr1 | 211575383 | 211575826 | SLC30A1 | 0 | - | 211575383 | 211575826 | 0 | 1 | 443 | 0 | -0.6789992 | 0.0000393 | 0.0012361 |
| chr1 | 211576171 | 211578019 | SLC30A1 | 0 | - | 211576171 | 211578019 | 0 | 2 | 119,29 | 0,1820 | -1.3584643 | 0.0000054 | 0.0002349 |
| chr9 | 113260909 | 113261058 | SLC31A1 | 0 | + | 113260909 | 113261058 | 0 | 1 | 149 | 0 | -1.4281827 | 0.0030750 | 0.0362145 |
| chr6 | 87511460 | 87511704 | SLC35A1 | 0 | + | 87511460 | 87511704 | 0 | 1 | 244 | 0 | 1.3667468 | 0.0030199 | 0.0358114 |
| chr6 | 44255036 | 44256368 | SLC35B2 | 0 | - | 44255036 | 44256368 | 0 | 2 | 609,31 | 0,1302 | -0.9115478 | 0.0009114 | 0.0147756 |
| chr11 | 45811623 | 45811871 | SLC35C1 | 0 | + | 45811623 | 45811871 | 0 | 1 | 248 | 0 | -1.0084055 | 0.0076624 | 0.0699348 |
| chr19 | 16552433 | 16552482 | SLC35E1 | 0 | - | 16552433 | 16552482 | 0 | 1 | 49 | 0 | -2.4978742 | 0.0080922 | 0.0726385 |
| chr17 | 81245061 | 81245210 | SLC38A10 | 0 | - | 81245061 | 81245210 | 0 | 1 | 149 | 0 | -1.2394473 | 0.0062638 | 0.0612967 |
| chr12 | 46360779 | 46360978 | SLC38A2 | 0 | - | 46360779 | 46360978 | 0 | 1 | 199 | 0 | -0.9065912 | 0.0082682 | 0.0736091 |
| chr12 | 46361029 | 46361177 | SLC38A2 | 0 | - | 46361029 | 46361177 | 0 | 1 | 148 | 0 | -1.1693648 | 0.0008551 | 0.0140386 |
| chr12 | 46371186 | 46372812 | SLC38A2 | 0 | - | 46371186 | 46372812 | 0 | 2 | 194,304 | 0,1323 | 0.6844404 | 0.0024695 | 0.0309462 |
| chr18 | 36126303 | 36126651 | SLC39A6 | 0 | - | 36126303 | 36126651 | 0 | 1 | 348 | 0 | -0.8075084 | 0.0004849 | 0.0090251 |
| chr1 | 205800978 | 205810383 | SLC41A1 | 0 | - | 205800978 | 205810383 | 0 | 2 | 83,314 | 0,9092 | -0.8936063 | 0.0002988 | 0.0061843 |
| chr1 | 205810434 | 205810532 | SLC41A1 | 0 | - | 205810434 | 205810532 | 0 | 1 | 98 | 0 | -2.0956347 | 0.0000000 | 0.0000002 |
| chr1 | 205810682 | 205812898 | SLC41A1 | 0 | - | 205810682 | 205812898 | 0 | 2 | 406,91 | 0,2126 | -0.7243687 | 0.0109574 | 0.0897767 |
| chr11 | 57407352 | 57407799 | SLC43A3 | 0 | - | 57407352 | 57407799 | 0 | 1 | 447 | 0 | -0.7260452 | 0.0000169 | 0.0006170 |
| chr19 | 10643376 | 10643425 | SLC44A2 | 0 | + | 10643376 | 10643425 | 0 | 1 | 49 | 0 | -2.0327185 | 0.0110059 | 0.0899598 |
| chr19 | 10643474 | 10644017 | SLC44A2 | 0 | + | 10643474 | 10644017 | 0 | 1 | 543 | 0 | -0.5787150 | 0.0072362 | 0.0671272 |
| chr17 | 28404696 | 28404995 | SLC46A1 | 0 | - | 28404696 | 28404995 | 0 | 1 | 299 | 0 | -0.7190710 | 0.0020706 | 0.0270138 |
| chr2 | 27663916 | 27664411 | SLC4A1AP | 0 | + | 27663916 | 27664411 | 0 | 1 | 495 | 0 | -1.3022706 | 0.0001648 | 0.0038322 |
| chr1 | 155138269 | 155138466 | SLC50A1 | 0 | + | 155138269 | 155138466 | 0 | 1 | 197 | 0 | -1.0801874 | 0.0002578 | 0.0055110 |
| chr8 | 144360841 | 144361187 | SLC52A2 | 0 | + | 144360841 | 144361187 | 0 | 1 | 346 | 0 | -0.8541045 | 0.0041567 | 0.0453132 |
| chr13 | 29510813 | 29510962 | SLC7A1 | 0 | - | 29510813 | 29510962 | 0 | 1 | 149 | 0 | -1.7479437 | 0.0009364 | 0.0150852 |
| chr16 | 68298497 | 68298893 | SLC7A6 | 0 | + | 68298497 | 68298893 | 0 | 1 | 396 | 0 | 0.8221439 | 0.0111868 | 0.0909610 |
| chr3 | 57757256 | 57757704 | SLMAP | 0 | + | 57757256 | 57757704 | 0 | 1 | 448 | 0 | -1.4709481 | 0.0000000 | 0.0000000 |
| chr15 | 67065857 | 67065906 | SMAD3 | 0 | + | 67065857 | 67065906 | 0 | 1 | 49 | 0 | 3.0575260 | 0.0038467 | 0.0427595 |
| chr15 | 66702780 | 66702878 | SMAD6 | 0 | + | 66702780 | 66702878 | 0 | 1 | 98 | 0 | -1.8545862 | 0.0001274 | 0.0031396 |
| chr12 | 51245646 | 51246042 | SMAGP | 0 | - | 51245646 | 51246042 | 0 | 1 | 396 | 0 | -0.6067113 | 0.0038905 | 0.0431676 |
| chr11 | 93479252 | 93499307 | SMCO4 | 0 | - | 93479252 | 93499307 | 0 | 2 | 18,32 | 0,20024 | 1.7295300 | 0.0066273 | 0.0637654 |
| chr17 | 18316325 | 18316473 | SMCR8 | 0 | + | 18316325 | 18316473 | 0 | 1 | 148 | 0 | -1.2173147 | 0.0075901 | 0.0694504 |
| chr17 | 18317320 | 18318116 | SMCR8 | 0 | + | 18317320 | 18318116 | 0 | 1 | 796 | 0 | -1.3132693 | 0.0000000 | 0.0000000 |
| chr17 | 18322882 | 18323627 | SMCR8 | 0 | + | 18322882 | 18323627 | 0 | 1 | 745 | 0 | -0.7140935 | 0.0003318 | 0.0067288 |
| chr17 | 18323678 | 18323876 | SMCR8 | 0 | + | 18323678 | 18323876 | 0 | 1 | 198 | 0 | -1.5933863 | 0.0002590 | 0.0055203 |
| chr1 | 156265950 | 156266346 | SMG5 | 0 | - | 156265950 | 156266346 | 0 | 1 | 396 | 0 | -1.2976645 | 0.0000157 | 0.0005827 |
| chr17 | 59211000 | 59211246 | SMG8 | 0 | + | 59211000 | 59211246 | 0 | 1 | 246 | 0 | -1.0850931 | 0.0001067 | 0.0027600 |
| chr17 | 59213157 | 59213453 | SMG8 | 0 | + | 59213157 | 59213453 | 0 | 1 | 296 | 0 | -0.9005455 | 0.0002632 | 0.0055801 |
| chrX | 134991879 | 134992126 | SMIM10 | 0 | + | 134991879 | 134992126 | 0 | 1 | 247 | 0 | -0.7987130 | 0.0019147 | 0.0256104 |
| chr6 | 11135294 | 11135742 | SMIM13 | 0 | + | 11135294 | 11135742 | 0 | 1 | 448 | 0 | -0.7738019 | 0.0047161 | 0.0497713 |
| chr14 | 69879397 | 69879746 | SMOC1 | 0 | + | 69879397 | 69879746 | 0 | 1 | 349 | 0 | 0.9210635 | 0.0003608 | 0.0072278 |
| chr2 | 130151640 | 130152682 | SMPD4 | 0 | - | 130151640 | 130152682 | 0 | 1 | 1042 | 0 | -0.6056504 | 0.0008054 | 0.0134411 |
| chr12 | 54181495 | 54181742 | SMUG1 | 0 | - | 54181495 | 54181742 | 0 | 1 | 247 | 0 | 0.6730979 | 0.0051851 | 0.0534688 |
| chr17 | 1800167 | 1800465 | SMYD4 | 0 | - | 1800167 | 1800465 | 0 | 1 | 298 | 0 | -1.4040989 | 0.0026195 | 0.0323444 |
| chr2 | 73225845 | 73226240 | SMYD5 | 0 | + | 73225845 | 73226240 | 0 | 1 | 395 | 0 | -1.2354776 | 0.0009421 | 0.0151368 |
| chr1 | 227747977 | 227759033 | SNAP47 | 0 | + | 227747977 | 227759033 | 0 | 2 | 257,39 | 0,11018 | -0.6272388 | 0.0087630 | 0.0767781 |
| chr19 | 7922904 | 7922953 | SNAPC2 | 0 | + | 7922904 | 7922953 | 0 | 1 | 49 | 0 | -1.8694660 | 0.0036126 | 0.0408853 |
| chr1 | 37537073 | 37537122 | SNIP1 | 0 | - | 37537073 | 37537122 | 0 | 1 | 49 | 0 | -3.9838934 | 0.0044149 | 0.0473893 |
| chr20 | 1305233 | 1305831 | SNPH | 0 | + | 1305233 | 1305831 | 0 | 1 | 598 | 0 | -0.8717163 | 0.0043889 | 0.0472911 |
| chr19 | 49085439 | 49086504 | SNRNP70 | 0 | + | 49085439 | 49086504 | 0 | 2 | 198,100 | 0,966 | 0.6963075 | 0.0094081 | 0.0804349 |
| chr11 | 130914595 | 130914743 | SNX19 | 0 | - | 130914595 | 130914743 | 0 | 1 | 148 | 0 | -1.7808742 | 0.0008531 | 0.0140174 |
| chr11 | 130915143 | 130915341 | SNX19 | 0 | - | 130915143 | 130915341 | 0 | 1 | 198 | 0 | -1.7335504 | 0.0034780 | 0.0398356 |
| chr6 | 108212096 | 108212145 | SNX3 | 0 | - | 108212096 | 108212145 | 0 | 1 | 49 | 0 | -1.9111997 | 0.0037191 | 0.0417780 |
| chr15 | 75650201 | 75650449 | SNX33 | 0 | + | 75650201 | 75650449 | 0 | 1 | 248 | 0 | -1.7037995 | 0.0000001 | 0.0000070 |
| chr15 | 75658130 | 75658229 | SNX33 | 0 | + | 75658130 | 75658229 | 0 | 1 | 99 | 0 | -1.2778998 | 0.0065177 | 0.0630459 |
| chr17 | 78357772 | 78358518 | SOCS3 | 0 | - | 78357772 | 78358518 | 0 | 1 | 746 | 0 | -0.6324534 | 0.0002702 | 0.0057091 |
| chr18 | 70325502 | 70326301 | SOCS6 | 0 | + | 70325502 | 70326301 | 0 | 1 | 799 | 0 | -0.8376312 | 0.0001609 | 0.0037738 |
| chr6 | 159684939 | 159693272 | SOD2 | 0 | - | 159684939 | 159693272 | 0 | 4 | 95,117,203,128 | 0,3187,7722,8206 | 0.7021884 | 0.0001540 | 0.0036637 |
| chr13 | 36168808 | 36168907 | SOHLH2 | 0 | - | 36168808 | 36168907 | 0 | 1 | 99 | 0 | -1.4301710 | 0.0017407 | 0.0238071 |
| chr21 | 33550671 | 33551019 | SON | 0 | + | 33550671 | 33551019 | 0 | 1 | 348 | 0 | -0.9240022 | 0.0012942 | 0.0193042 |
| chr21 | 33552962 | 33553559 | SON | 0 | + | 33552962 | 33553559 | 0 | 1 | 597 | 0 | -0.9006332 | 0.0004202 | 0.0081117 |
| chr21 | 33553709 | 33554007 | SON | 0 | + | 33553709 | 33554007 | 0 | 1 | 298 | 0 | -1.0225850 | 0.0049890 | 0.0519501 |
| chr21 | 33554257 | 33554655 | SON | 0 | + | 33554257 | 33554655 | 0 | 1 | 398 | 0 | -1.1437839 | 0.0000173 | 0.0006320 |
| chr2 | 109614433 | 109616363 | SOWAHC | 0 | + | 109614433 | 109616363 | 0 | 1 | 1930 | 0 | -0.5848377 | 0.0000097 | 0.0003939 |
| chr1 | 204073118 | 204073366 | SOX13 | 0 | + | 204073118 | 204073366 | 0 | 1 | 248 | 0 | 1.5782473 | 0.0000154 | 0.0005761 |
| chr6 | 21596130 | 21596179 | SOX4 | 0 | + | 21596130 | 21596179 | 0 | 1 | 49 | 0 | -2.8729464 | 0.0033612 | 0.0387429 |
| chr12 | 53382360 | 53382409 | SP1 | 0 | + | 53382360 | 53382409 | 0 | 1 | 49 | 0 | -2.3518585 | 0.0017495 | 0.0238709 |
| chr12 | 53382460 | 53383509 | SP1 | 0 | + | 53382460 | 53383509 | 0 | 1 | 1049 | 0 | -1.7339641 | 0.0000000 | 0.0000000 |
| chr2 | 173955530 | 173955779 | SP3 | 0 | - | 173955530 | 173955779 | 0 | 1 | 249 | 0 | -0.8595358 | 0.0022719 | 0.0290330 |
| chr17 | 28592445 | 28592544 | SPAG5 | 0 | - | 28592445 | 28592544 | 0 | 1 | 99 | 0 | -1.4490847 | 0.0014667 | 0.0210623 |
| chr17 | 28592594 | 28592890 | SPAG5 | 0 | - | 28592594 | 28592890 | 0 | 1 | 296 | 0 | -0.8074307 | 0.0069195 | 0.0654296 |
| chr17 | 4959616 | 4959830 | SPAG7 | 0 | - | 4959616 | 4959830 | 0 | 2 | 28,71 | 0,144 | -1.3985634 | 0.0040554 | 0.0445065 |
| chr17 | 51120566 | 51120865 | SPAG9 | 0 | - | 51120566 | 51120865 | 0 | 1 | 299 | 0 | 1.3450545 | 0.0006607 | 0.0115707 |
| chr15 | 45403158 | 45405352 | SPATA5L1 | 0 | + | 45403158 | 45405352 | 0 | 2 | 361,33 | 0,2162 | -0.8920940 | 0.0040295 | 0.0443566 |
| chr15 | 45421074 | 45421123 | SPATA5L1 | 0 | + | 45421074 | 45421123 | 0 | 1 | 49 | 0 | -2.1701828 | 0.0092492 | 0.0796622 |
| chr19 | 11155666 | 11155762 | SPC24 | 0 | - | 11155666 | 11155762 | 0 | 1 | 96 | 0 | 1.7941102 | 0.0001983 | 0.0044275 |
| chr17 | 20204463 | 20205059 | SPECC1 | 0 | + | 20204463 | 20205059 | 0 | 1 | 596 | 0 | -0.9458575 | 0.0000000 | 0.0000008 |
| chr17 | 20205209 | 20205755 | SPECC1 | 0 | + | 20205209 | 20205755 | 0 | 1 | 546 | 0 | -0.9814317 | 0.0000152 | 0.0005697 |
| chr22 | 24321520 | 24322067 | SPECC1L | 0 | + | 24321520 | 24322067 | 0 | 1 | 547 | 0 | -0.8314906 | 0.0109335 | 0.0896259 |
| chr22 | 24321671 | 24322070 | SPECC1L-ADORA2A | 0 | + | 24321671 | 24322070 | 0 | 1 | 399 | 0 | -1.0014896 | 0.0046159 | 0.0489374 |
| chr1 | 15873028 | 15876392 | SPEN | 0 | + | 15873028 | 15876392 | 0 | 2 | 109,191 | 0,3174 | -1.4945115 | 0.0010494 | 0.0165100 |
| chr1 | 15928532 | 15928831 | SPEN | 0 | + | 15928532 | 15928831 | 0 | 1 | 299 | 0 | -1.5519142 | 0.0037712 | 0.0422315 |
| chr2 | 72888592 | 72891626 | SPR | 0 | + | 72888592 | 72891626 | 0 | 2 | 13,280 | 0,2755 | -0.5095982 | 0.0086777 | 0.0762565 |
| chr9 | 128577374 | 128577473 | SPTAN1 | 0 | + | 128577374 | 128577473 | 0 | 1 | 99 | 0 | -5.1968795 | 0.0000000 | 0.0000000 |
| chr9 | 128585831 | 128585880 | SPTAN1 | 0 | + | 128585831 | 128585880 | 0 | 1 | 49 | 0 | -4.5312493 | 0.0000000 | 0.0000000 |
| chr9 | 128591611 | 128593017 | SPTAN1 | 0 | + | 128591611 | 128593017 | 0 | 2 | 15,35 | 0,1372 | -3.1847915 | 0.0000818 | 0.0022204 |
| chr9 | 128594299 | 128594348 | SPTAN1 | 0 | + | 128594299 | 128594348 | 0 | 1 | 49 | 0 | -3.2545826 | 0.0005154 | 0.0094557 |
| chr9 | 128607693 | 128607939 | SPTAN1 | 0 | + | 128607693 | 128607939 | 0 | 2 | 11,88 | 0,159 | -4.6325471 | 0.0000000 | 0.0000000 |
| chr9 | 128607990 | 128608218 | SPTAN1 | 0 | + | 128607990 | 128608218 | 0 | 2 | 60,89 | 0,140 | -5.8530185 | 0.0000000 | 0.0000000 |
| chr2 | 54558496 | 54558795 | SPTBN1 | 0 | + | 54558496 | 54558795 | 0 | 1 | 299 | 0 | -0.8593035 | 0.0088609 | 0.0771784 |
| chr2 | 54624825 | 54624874 | SPTBN1 | 0 | + | 54624825 | 54624874 | 0 | 1 | 49 | 0 | -4.7468728 | 0.0000000 | 0.0000000 |
| chr2 | 54624925 | 54625943 | SPTBN1 | 0 | + | 54624925 | 54625943 | 0 | 2 | 38,12 | 0,1007 | -3.4223018 | 0.0000002 | 0.0000158 |
| chr2 | 54629386 | 54629635 | SPTBN1 | 0 | + | 54629386 | 54629635 | 0 | 1 | 249 | 0 | -1.8149890 | 0.0000050 | 0.0002174 |
| chr2 | 54631197 | 54631595 | SPTBN1 | 0 | + | 54631197 | 54631595 | 0 | 1 | 398 | 0 | -1.3389098 | 0.0000008 | 0.0000441 |
| chr2 | 54644465 | 54644514 | SPTBN1 | 0 | + | 54644465 | 54644514 | 0 | 1 | 49 | 0 | -3.2539378 | 0.0001332 | 0.0032587 |
| chr11 | 66700636 | 66701183 | SPTBN2 | 0 | - | 66700636 | 66701183 | 0 | 1 | 547 | 0 | -1.1609547 | 0.0007007 | 0.0120501 |
| chr8 | 124999026 | 124999573 | SQLE | 0 | + | 124999026 | 124999573 | 0 | 1 | 547 | 0 | -0.5332405 | 0.0014727 | 0.0211135 |
| chr15 | 45690997 | 45691145 | SQRDL | 0 | + | 45690997 | 45691145 | 0 | 1 | 148 | 0 | -0.8522397 | 0.0023470 | 0.0297615 |
| chr16 | 30737985 | 30739580 | SRCAP | 0 | + | 30737985 | 30739580 | 0 | 1 | 1595 | 0 | -0.7496103 | 0.0000955 | 0.0025095 |
| chr6 | 43171295 | 43171840 | SRF | 0 | + | 43171295 | 43171840 | 0 | 1 | 545 | 0 | -0.9534350 | 0.0057550 | 0.0577026 |
| chr4 | 56467646 | 56469702 | SRP72 | 0 | + | 56467646 | 56469702 | 0 | 2 | 99,50 | 0,2007 | 1.1605993 | 0.0009682 | 0.0155423 |
| chr22 | 26490124 | 26491888 | SRRD | 0 | + | 26490124 | 26491888 | 0 | 3 | 75,46,426 | 0,901,1339 | -0.6819798 | 0.0017050 | 0.0234426 |
| chr16 | 2763860 | 2764009 | SRRM2 | 0 | + | 2763860 | 2764009 | 0 | 1 | 149 | 0 | -1.7546803 | 0.0000023 | 0.0001116 |
| chr16 | 2764209 | 2764258 | SRRM2 | 0 | + | 2764209 | 2764258 | 0 | 1 | 49 | 0 | -2.8680884 | 0.0064330 | 0.0623935 |
| chr16 | 2765154 | 2765203 | SRRM2 | 0 | + | 2765154 | 2765203 | 0 | 1 | 49 | 0 | -2.1536866 | 0.0032652 | 0.0379236 |
| chr16 | 2766945 | 2766994 | SRRM2 | 0 | + | 2766945 | 2766994 | 0 | 1 | 49 | 0 | -2.8252269 | 0.0058086 | 0.0580157 |
| chr16 | 2767094 | 2767541 | SRRM2 | 0 | + | 2767094 | 2767541 | 0 | 1 | 447 | 0 | -1.7283396 | 0.0000000 | 0.0000000 |
| chr7 | 100875227 | 100875276 | SRRT | 0 | + | 100875227 | 100875276 | 0 | 1 | 49 | 0 | 2.1905688 | 0.0049743 | 0.0518269 |
| chr2 | 38751296 | 38751345 | SRSF7 | 0 | - | 38751296 | 38751345 | 0 | 1 | 49 | 0 | 0.9843394 | 0.0011554 | 0.0177010 |
| chr20 | 647819 | 648814 | SRXN1 | 0 | - | 647819 | 648814 | 0 | 1 | 995 | 0 | -0.5467039 | 0.0000950 | 0.0024993 |
| chr2 | 181916094 | 181916393 | SSFA2 | 0 | + | 181916094 | 181916393 | 0 | 1 | 299 | 0 | -0.6789088 | 0.0095260 | 0.0811747 |
| chr17 | 29631868 | 29632315 | SSH2 | 0 | - | 29631868 | 29632315 | 0 | 1 | 447 | 0 | -1.4059551 | 0.0006315 | 0.0111634 |
| chr7 | 100359472 | 100367631 | STAG3L5P-PVRIG2P-PILRB | 0 | + | 100359472 | 100367631 | 0 | 2 | 66,283 | 0,7877 | -0.7121972 | 0.0110659 | 0.0902620 |
| chr2 | 96208289 | 96208785 | STARD7 | 0 | - | 96208289 | 96208785 | 0 | 1 | 496 | 0 | 0.8826127 | 0.0014379 | 0.0207636 |
| chr12 | 57096199 | 57096881 | STAT6 | 0 | - | 57096199 | 57096881 | 0 | 2 | 563,32 | 0,651 | -0.6404104 | 0.0001553 | 0.0036767 |
| chr20 | 49179149 | 49179198 | STAU1 | 0 | - | 49179149 | 49179198 | 0 | 1 | 49 | 0 | 2.7086988 | 0.0006038 | 0.0107826 |
| chr20 | 2116818 | 2143831 | STK35 | 0 | + | 2116818 | 2143831 | 0 | 2 | 598,48 | 0,26966 | -0.7991317 | 0.0013018 | 0.0193220 |
| chr14 | 81270847 | 81277074 | STON2 | 0 | - | 81270847 | 81277074 | 0 | 2 | 26,174 | 0,6054 | -1.3068292 | 0.0044767 | 0.0478055 |
| chr16 | 682470 | 682618 | STUB1 | 0 | + | 682470 | 682618 | 0 | 1 | 148 | 0 | -1.3037603 | 0.0001226 | 0.0030717 |
| chr1 | 180976194 | 180976591 | STX6 | 0 | - | 180976194 | 180976591 | 0 | 1 | 397 | 0 | -0.6348379 | 0.0043775 | 0.0472594 |
| chr5 | 32585548 | 32591610 | SUB1 | 0 | + | 32585548 | 32591610 | 0 | 3 | 78,73,48 | 0,2964,6015 | 0.7874095 | 0.0002854 | 0.0059549 |
| chr19 | 19008423 | 19010147 | SUGP2 | 0 | - | 19008423 | 19010147 | 0 | 2 | 6,293 | 0,1432 | -1.2674476 | 0.0001100 | 0.0028165 |
| chr19 | 19024751 | 19025049 | SUGP2 | 0 | - | 19024751 | 19025049 | 0 | 1 | 298 | 0 | -1.9623677 | 0.0000000 | 0.0000019 |
| chr19 | 19025597 | 19026095 | SUGP2 | 0 | - | 19025597 | 19026095 | 0 | 1 | 498 | 0 | -1.6800450 | 0.0000000 | 0.0000031 |
| chr9 | 133331351 | 133331995 | SURF6 | 0 | - | 133331351 | 133331995 | 0 | 1 | 644 | 0 | -0.7999390 | 0.0000000 | 0.0000022 |
| chr14 | 69708802 | 69709001 | SUSD6 | 0 | + | 69708802 | 69709001 | 0 | 1 | 199 | 0 | -1.6334553 | 0.0079542 | 0.0717281 |
| chrX | 48700147 | 48700590 | SUV39H1 | 0 | + | 48700147 | 48700590 | 0 | 1 | 443 | 0 | -1.2076248 | 0.0001954 | 0.0043788 |
| chr19 | 55347188 | 55347286 | SUV420H2 | 0 | + | 55347188 | 55347286 | 0 | 1 | 98 | 0 | 2.1001430 | 0.0049939 | 0.0519710 |
| chr19 | 15113805 | 15114594 | SYDE1 | 0 | + | 15113805 | 15114594 | 0 | 1 | 789 | 0 | -0.8651807 | 0.0000000 | 0.0000005 |
| chr15 | 99130875 | 99131073 | SYNM | 0 | + | 99130875 | 99131073 | 0 | 1 | 198 | 0 | -2.0792436 | 0.0000000 | 0.0000039 |
| chr5 | 150648956 | 150649204 | SYNPO | 0 | + | 150648956 | 150649204 | 0 | 1 | 248 | 0 | -1.7215479 | 0.0000000 | 0.0000000 |
| chr5 | 150649353 | 150649651 | SYNPO | 0 | + | 150649353 | 150649651 | 0 | 1 | 298 | 0 | -2.5580107 | 0.0000000 | 0.0000000 |
| chr5 | 150650099 | 150650148 | SYNPO | 0 | + | 150650099 | 150650148 | 0 | 1 | 49 | 0 | -1.4168295 | 0.0045813 | 0.0486561 |
| chr11 | 65127820 | 65128115 | SYVN1 | 0 | - | 65127820 | 65128115 | 0 | 1 | 295 | 0 | -0.7682023 | 0.0081628 | 0.0730295 |
| chr6 | 149379153 | 149379451 | TAB2 | 0 | + | 149379153 | 149379451 | 0 | 1 | 298 | 0 | -0.9661182 | 0.0027116 | 0.0330647 |
| chr10 | 122086436 | 122086784 | TACC2 | 0 | + | 122086436 | 122086784 | 0 | 1 | 348 | 0 | -2.8089821 | 0.0000000 | 0.0000000 |
| chr10 | 122087235 | 122087533 | TACC2 | 0 | + | 122087235 | 122087533 | 0 | 1 | 298 | 0 | -2.1796205 | 0.0000000 | 0.0000025 |
| chr10 | 122087584 | 122087783 | TACC2 | 0 | + | 122087584 | 122087783 | 0 | 1 | 199 | 0 | -1.8867666 | 0.0015174 | 0.0215336 |
| chr10 | 122087834 | 122087883 | TACC2 | 0 | + | 122087834 | 122087883 | 0 | 1 | 49 | 0 | -2.5196386 | 0.0000240 | 0.0008202 |
| chr10 | 122210702 | 122211600 | TACC2 | 0 | + | 122210702 | 122211600 | 0 | 1 | 898 | 0 | -0.5095942 | 0.0028715 | 0.0343445 |
| chr5 | 141319837 | 141319936 | TAF7 | 0 | - | 141319837 | 141319936 | 0 | 1 | 99 | 0 | -0.8966812 | 0.0014866 | 0.0211891 |
| chr2 | 161137054 | 161179644 | TANK | 0 | + | 161137054 | 161179644 | 0 | 3 | 10,59,31 | 0,23374,42560 | 1.2462253 | 0.0041590 | 0.0453132 |
| chr3 | 10260381 | 10270138 | TATDN2 | 0 | + | 10260381 | 10270138 | 0 | 2 | 290,8 | 0,9750 | -0.7919018 | 0.0066015 | 0.0635853 |
| chr3 | 10270485 | 10270832 | TATDN2 | 0 | + | 10270485 | 10270832 | 0 | 1 | 347 | 0 | -1.0381985 | 0.0005705 | 0.0102585 |
| chr17 | 80010169 | 80010367 | TBC1D16 | 0 | - | 80010169 | 80010367 | 0 | 1 | 198 | 0 | -1.7049747 | 0.0001226 | 0.0030717 |
| chrX | 48560431 | 48560530 | TBC1D25 | 0 | + | 48560431 | 48560530 | 0 | 1 | 99 | 0 | -2.2563961 | 0.0021321 | 0.0276386 |
| chr5 | 179862816 | 179863064 | TBC1D9B | 0 | - | 179862816 | 179863064 | 0 | 1 | 248 | 0 | -1.8583097 | 0.0000000 | 0.0000038 |
| chr7 | 73569942 | 73570141 | TBL2 | 0 | - | 73569942 | 73570141 | 0 | 1 | 199 | 0 | -1.1714358 | 0.0099483 | 0.0839045 |
| chr7 | 143863379 | 143863577 | TCAF1 | 0 | - | 143863379 | 143863577 | 0 | 1 | 198 | 0 | -1.6771844 | 0.0012677 | 0.0189715 |
| chrX | 103586998 | 103587244 | TCEAL4 | 0 | + | 103586998 | 103587244 | 0 | 1 | 246 | 0 | -1.0320002 | 0.0002865 | 0.0059565 |
| chrX | 103253576 | 103253964 | TCEAL8 | 0 | - | 103253576 | 103253964 | 0 | 1 | 388 | 0 | -0.6007622 | 0.0001770 | 0.0040616 |
| chr1 | 23751117 | 23752491 | TCEB3 | 0 | + | 23751117 | 23752491 | 0 | 2 | 914,85 | 0,1290 | -0.9264601 | 0.0000000 | 0.0000021 |
| chr19 | 1610435 | 1610533 | TCF3 | 0 | - | 1610435 | 1610533 | 0 | 1 | 98 | 0 | -2.0602526 | 0.0007051 | 0.0120915 |
| chr20 | 62852583 | 62854105 | TCFL5 | 0 | - | 62852583 | 62854105 | 0 | 2 | 306,90 | 0,1433 | -1.0862579 | 0.0019640 | 0.0261315 |
| chr6 | 35486699 | 35496934 | TEAD3 | 0 | - | 35486699 | 35496934 | 0 | 2 | 13,37 | 0,10199 | 1.6635436 | 0.0120862 | 0.0955289 |
| chr16 | 69361455 | 69367169 | TERF2 | 0 | - | 69361455 | 69367169 | 0 | 2 | 35,363 | 0,5352 | -0.6930632 | 0.0015699 | 0.0220889 |
| chr7 | 116252390 | 116252439 | TES | 0 | + | 116252390 | 116252439 | 0 | 1 | 49 | 0 | 1.5036089 | 0.0069706 | 0.0657760 |
| chr17 | 64193677 | 64193876 | TEX2 | 0 | - | 64193677 | 64193876 | 0 | 1 | 199 | 0 | -1.2263337 | 0.0098423 | 0.0832823 |
| chr17 | 64212707 | 64213055 | TEX2 | 0 | - | 64212707 | 64213055 | 0 | 1 | 348 | 0 | -1.1402896 | 0.0002752 | 0.0057888 |
| chr17 | 64213403 | 64213601 | TEX2 | 0 | - | 64213403 | 64213601 | 0 | 1 | 198 | 0 | -1.3435379 | 0.0000590 | 0.0017068 |
| chr3 | 30671866 | 30672113 | TGFBR2 | 0 | + | 30671866 | 30672113 | 0 | 1 | 247 | 0 | -2.1177507 | 0.0000150 | 0.0005641 |
| chr18 | 3457612 | 3458109 | TGIF1 | 0 | + | 3457612 | 3458109 | 0 | 1 | 497 | 0 | -0.6925916 | 0.0000099 | 0.0003967 |
| chr20 | 36612013 | 36612309 | TGIF2-C20orf24 | 0 | + | 36612013 | 36612309 | 0 | 1 | 296 | 0 | -0.9065870 | 0.0000024 | 0.0001164 |
| chr2 | 85326871 | 85327069 | TGOLN2 | 0 | - | 85326871 | 85327069 | 0 | 1 | 198 | 0 | -2.1799054 | 0.0000000 | 0.0000021 |
| chr2 | 85327120 | 85327997 | TGOLN2 | 0 | - | 85327120 | 85327997 | 0 | 2 | 566,81 | 0,797 | -1.2142393 | 0.0000005 | 0.0000271 |
| chr2 | 241633414 | 241637014 | THAP4 | 0 | - | 241633414 | 241637014 | 0 | 2 | 666,74 | 0,3527 | -1.0999839 | 0.0000000 | 0.0000000 |
| chr4 | 75527045 | 75527243 | THAP6 | 0 | + | 75527045 | 75527243 | 0 | 1 | 198 | 0 | -3.3732465 | 0.0003061 | 0.0063050 |
| chr19 | 2813352 | 2813401 | THOP1 | 0 | + | 2813352 | 2813401 | 0 | 1 | 49 | 0 | 1.3342262 | 0.0021879 | 0.0281577 |
| chr19 | 2813502 | 2813551 | THOP1 | 0 | + | 2813502 | 2813551 | 0 | 1 | 49 | 0 | 3.2392626 | 0.0000485 | 0.0014626 |
| chr1 | 36289190 | 36289488 | THRAP3 | 0 | + | 36289190 | 36289488 | 0 | 1 | 298 | 0 | -0.7742458 | 0.0044021 | 0.0473052 |
| chr1 | 201955491 | 201957366 | TIMM17A | 0 | + | 201955491 | 201957366 | 0 | 2 | 62,86 | 0,1790 | -0.9534230 | 0.0009988 | 0.0158648 |
| chr19 | 7926968 | 7927167 | TIMM44 | 0 | - | 7926968 | 7927167 | 0 | 1 | 199 | 0 | -1.1052743 | 0.0068985 | 0.0652992 |
| chr3 | 119498521 | 119498769 | TIMMDC1 | 0 | + | 119498521 | 119498769 | 0 | 1 | 248 | 0 | 1.1267875 | 0.0006599 | 0.0115684 |
| chr17 | 78854261 | 78854359 | TIMP2 | 0 | - | 78854261 | 78854359 | 0 | 1 | 98 | 0 | -1.3666711 | 0.0000005 | 0.0000294 |
| chr17 | 78854557 | 78854804 | TIMP2 | 0 | - | 78854557 | 78854804 | 0 | 1 | 247 | 0 | -1.1793328 | 0.0000000 | 0.0000026 |
| chr17 | 78855150 | 78855347 | TIMP2 | 0 | - | 78855150 | 78855347 | 0 | 1 | 197 | 0 | -0.9674042 | 0.0002232 | 0.0048809 |
| chr17 | 78855446 | 78855545 | TIMP2 | 0 | - | 78855446 | 78855545 | 0 | 1 | 99 | 0 | -1.1967944 | 0.0000509 | 0.0015127 |
| chr17 | 78925143 | 78925390 | TIMP2 | 0 | - | 78925143 | 78925390 | 0 | 1 | 247 | 0 | 0.5328691 | 0.0000865 | 0.0023240 |
| chr22 | 32801714 | 32801962 | TIMP3 | 0 | + | 32801714 | 32801962 | 0 | 1 | 248 | 0 | 0.7334162 | 0.0120956 | 0.0955619 |
| chr22 | 32802013 | 32849491 | TIMP3 | 0 | + | 32802013 | 32849491 | 0 | 2 | 110,40 | 0,47439 | 1.0764580 | 0.0001390 | 0.0033783 |
| chr22 | 32859351 | 32859550 | TIMP3 | 0 | + | 32859351 | 32859550 | 0 | 1 | 199 | 0 | -1.0948463 | 0.0000053 | 0.0002309 |
| chr22 | 32859700 | 32859999 | TIMP3 | 0 | + | 32859700 | 32859999 | 0 | 1 | 299 | 0 | -1.6052079 | 0.0000000 | 0.0000000 |
| chr1 | 31577221 | 31577369 | TINAGL1 | 0 | + | 31577221 | 31577369 | 0 | 1 | 148 | 0 | 1.2009888 | 0.0018179 | 0.0246391 |
| chr1 | 31585829 | 31586942 | TINAGL1 | 0 | + | 31585829 | 31586942 | 0 | 3 | 48,46,104 | 0,881,1010 | 0.9212257 | 0.0001778 | 0.0040699 |
| chr1 | 31586992 | 31587289 | TINAGL1 | 0 | + | 31586992 | 31587289 | 0 | 1 | 297 | 0 | 0.5657040 | 0.0122792 | 0.0965896 |
| chr3 | 156675391 | 156694025 | TIPARP | 0 | + | 156675391 | 156694025 | 0 | 4 | 1,31,958,6 | 0,1273,2266,18629 | -0.6677773 | 0.0000817 | 0.0022204 |
| chr3 | 156703647 | 156705026 | TIPARP | 0 | + | 156703647 | 156705026 | 0 | 2 | 56,343 | 0,1037 | -0.6698173 | 0.0123992 | 0.0970706 |
| chr16 | 84479797 | 84479995 | TLDC1 | 0 | - | 84479797 | 84479995 | 0 | 1 | 198 | 0 | 0.9641619 | 0.0048327 | 0.0507899 |
| chr9 | 81689288 | 81689436 | TLE1 | 0 | - | 81689288 | 81689436 | 0 | 1 | 148 | 0 | -2.1490518 | 0.0028875 | 0.0344905 |
| chr9 | 35724252 | 35724301 | TLN1 | 0 | - | 35724252 | 35724301 | 0 | 1 | 49 | 0 | -2.0205319 | 0.0098888 | 0.0835198 |
| chr9 | 35724589 | 35724842 | TLN1 | 0 | - | 35724589 | 35724842 | 0 | 2 | 136,13 | 0,241 | -2.6946420 | 0.0000000 | 0.0000024 |
| chr3 | 149377384 | 149377731 | TM4SF1 | 0 | - | 149377384 | 149377731 | 0 | 1 | 347 | 0 | 0.6154207 | 0.0121684 | 0.0959689 |
| chr14 | 24192726 | 24194729 | TM9SF1 | 0 | - | 24192726 | 24194729 | 0 | 2 | 544,55 | 0,1949 | -1.1841472 | 0.0000000 | 0.0000000 |
| chr2 | 218274644 | 218274693 | TMBIM1 | 0 | - | 218274644 | 218274693 | 0 | 1 | 49 | 0 | -2.7574765 | 0.0000000 | 0.0000001 |
| chr2 | 218275244 | 218275392 | TMBIM1 | 0 | - | 218275244 | 218275392 | 0 | 1 | 148 | 0 | -0.9691706 | 0.0001213 | 0.0030601 |
| chr2 | 218281974 | 218292507 | TMBIM1 | 0 | - | 218281974 | 218292507 | 0 | 2 | 208,42 | 0,10492 | 0.7508486 | 0.0037169 | 0.0417780 |
| chr17 | 78126571 | 78126846 | TMC6 | 0 | - | 78126571 | 78126846 | 0 | 2 | 78,70 | 0,206 | 1.6309191 | 0.0000048 | 0.0002088 |
| chr3 | 50354799 | 50355197 | TMEM115 | 0 | - | 50354799 | 50355197 | 0 | 1 | 398 | 0 | -0.6344520 | 0.0027431 | 0.0332922 |
| chr11 | 102452895 | 102452944 | TMEM123 | 0 | - | 102452895 | 102452944 | 0 | 1 | 49 | 0 | 2.2489986 | 0.0016587 | 0.0230339 |
| chr2 | 96252405 | 96252853 | TMEM127 | 0 | - | 96252405 | 96252853 | 0 | 1 | 448 | 0 | -1.2190825 | 0.0000007 | 0.0000370 |
| chr4 | 55395913 | 55396308 | TMEM165 | 0 | + | 55395913 | 55396308 | 0 | 1 | 395 | 0 | 1.0665083 | 0.0000105 | 0.0004157 |
| chr1 | 203014906 | 203015004 | TMEM183A | 0 | + | 203014906 | 203015004 | 0 | 1 | 98 | 0 | 1.4274597 | 0.0017400 | 0.0238071 |
| chr3 | 149982820 | 149982918 | TMEM183B | 0 | - | 149982820 | 149982918 | 0 | 1 | 98 | 0 | 1.0865461 | 0.0122024 | 0.0961534 |
| chr22 | 38221031 | 38221627 | TMEM184B | 0 | - | 38221031 | 38221627 | 0 | 1 | 596 | 0 | -0.6980179 | 0.0006549 | 0.0114907 |
| chr2 | 120221662 | 120222061 | TMEM185B | 0 | - | 120221662 | 120222061 | 0 | 1 | 399 | 0 | -0.5440031 | 0.0118300 | 0.0942448 |
| chr2 | 120222161 | 120222410 | TMEM185B | 0 | - | 120222161 | 120222410 | 0 | 1 | 249 | 0 | -0.9953302 | 0.0003791 | 0.0075101 |
| chr20 | 50081520 | 50081816 | TMEM189-UBE2V1 | 0 | - | 50081520 | 50081816 | 0 | 1 | 296 | 0 | 1.1991937 | 0.0003733 | 0.0074201 |
| chr9 | 71745148 | 71746266 | TMEM2 | 0 | - | 71745148 | 71746266 | 0 | 2 | 432,66 | 0,1053 | -0.8243753 | 0.0006926 | 0.0119436 |
| chr19 | 11345658 | 11345707 | TMEM205 | 0 | - | 11345658 | 11345707 | 0 | 1 | 49 | 0 | -1.6191819 | 0.0000002 | 0.0000116 |
| chr2 | 27041050 | 27041099 | TMEM214 | 0 | + | 27041050 | 27041099 | 0 | 1 | 49 | 0 | -1.1698071 | 0.0123488 | 0.0968208 |
| chr2 | 27041548 | 27041647 | TMEM214 | 0 | + | 27041548 | 27041647 | 0 | 1 | 99 | 0 | -1.9600969 | 0.0000008 | 0.0000463 |
| chr11 | 61789175 | 61789224 | TMEM258 | 0 | - | 61789175 | 61789224 | 0 | 1 | 49 | 0 | 5.7580200 | 0.0000405 | 0.0012632 |
| chr19 | 1010143 | 1010881 | TMEM259 | 0 | - | 1010143 | 1010881 | 0 | 1 | 738 | 0 | 0.5551681 | 0.0036242 | 0.0409150 |
| chr3 | 14142135 | 14142583 | TMEM43 | 0 | + | 14142135 | 14142583 | 0 | 1 | 448 | 0 | -0.7210619 | 0.0000704 | 0.0019797 |
| chr3 | 194587772 | 194588167 | TMEM44 | 0 | - | 194587772 | 194588167 | 0 | 1 | 395 | 0 | -0.8506419 | 0.0063747 | 0.0619612 |
| chr6 | 44148301 | 44148350 | TMEM63B | 0 | + | 44148301 | 44148350 | 0 | 1 | 49 | 0 | -3.1856712 | 0.0000047 | 0.0002088 |
| chr17 | 28327204 | 28327500 | TMEM97 | 0 | + | 28327204 | 28327500 | 0 | 1 | 296 | 0 | -0.7199214 | 0.0008095 | 0.0134602 |
| chrX | 12976329 | 12976841 | TMSB4X | 0 | + | 12976329 | 12976841 | 0 | 2 | 33,65 | 0,448 | -0.6078357 | 0.0080869 | 0.0726375 |
| chr14 | 103126327 | 103126674 | TNFAIP2 | 0 | + | 103126327 | 103126674 | 0 | 1 | 347 | 0 | -0.7329112 | 0.0030326 | 0.0359387 |
| chr14 | 103135402 | 103135948 | TNFAIP2 | 0 | + | 103135402 | 103135948 | 0 | 1 | 546 | 0 | 0.7491791 | 0.0000157 | 0.0005827 |
| chr6 | 137867238 | 137867486 | TNFAIP3 | 0 | + | 137867238 | 137867486 | 0 | 1 | 248 | 0 | 1.6986454 | 0.0000536 | 0.0015783 |
| chr6 | 137878572 | 137878821 | TNFAIP3 | 0 | + | 137878572 | 137878821 | 0 | 1 | 249 | 0 | -0.9001919 | 0.0052352 | 0.0537712 |
| chr16 | 3021940 | 3022186 | TNFRSF12A | 0 | + | 3021940 | 3022186 | 0 | 1 | 246 | 0 | -1.4986154 | 0.0000000 | 0.0000008 |
| chr6 | 47284070 | 47284369 | TNFRSF21 | 0 | - | 47284070 | 47284369 | 0 | 1 | 299 | 0 | -0.9203744 | 0.0034840 | 0.0398419 |
| chr5 | 151062208 | 151080963 | TNIP1 | 0 | - | 151062208 | 151080963 | 0 | 4 | 5,135,172,84 | 0,1405,2752,18672 | 0.9444618 | 0.0000262 | 0.0008805 |
| chr4 | 2747876 | 2755978 | TNIP2 | 0 | - | 2747876 | 2755978 | 0 | 2 | 70,30 | 0,8073 | 2.5175689 | 0.0000563 | 0.0016421 |
| chr10 | 91798444 | 91798592 | TNKS2 | 0 | + | 91798444 | 91798592 | 0 | 1 | 148 | 0 | 1.4760388 | 0.0011256 | 0.0173989 |
| chr19 | 12719281 | 12722213 | TNPO2 | 0 | - | 12719281 | 12722213 | 0 | 3 | 56,112,80 | 0,1598,2853 | 1.4300323 | 0.0007065 | 0.0121027 |
| chr7 | 129017047 | 129018145 | TNPO3 | 0 | - | 129017047 | 129018145 | 0 | 2 | 10,189 | 0,910 | -1.8191253 | 0.0003286 | 0.0066704 |
| chr22 | 40266054 | 40266253 | TNRC6B | 0 | + | 40266054 | 40266253 | 0 | 1 | 199 | 0 | -1.3844908 | 0.0054854 | 0.0556157 |
| chr2 | 217802982 | 217803031 | TNS1 | 0 | - | 217802982 | 217803031 | 0 | 1 | 49 | 0 | -3.9305012 | 0.0000000 | 0.0000003 |
| chr2 | 217848395 | 217848843 | TNS1 | 0 | - | 217848395 | 217848843 | 0 | 1 | 448 | 0 | -1.1231130 | 0.0003750 | 0.0074459 |
| chr7 | 47368913 | 47369460 | TNS3 | 0 | - | 47368913 | 47369460 | 0 | 1 | 547 | 0 | -1.4191452 | 0.0000087 | 0.0003576 |
| chr17 | 40487147 | 40487345 | TNS4 | 0 | - | 40487147 | 40487345 | 0 | 1 | 198 | 0 | -0.9180398 | 0.0006995 | 0.0120408 |
| chr17 | 50863263 | 50864052 | TOB1 | 0 | - | 50863263 | 50864052 | 0 | 1 | 789 | 0 | -0.7792026 | 0.0000003 | 0.0000172 |
| chr22 | 41436167 | 41436762 | TOB2 | 0 | - | 41436167 | 41436762 | 0 | 1 | 595 | 0 | -0.7483424 | 0.0020900 | 0.0272275 |
| chr22 | 41436812 | 41437407 | TOB2 | 0 | - | 41436812 | 41437407 | 0 | 1 | 595 | 0 | -1.1134800 | 0.0000013 | 0.0000663 |
| chr22 | 38681949 | 38682323 | TOMM22 | 0 | + | 38681949 | 38682323 | 0 | 2 | 147,1 | 0,374 | 1.4413219 | 0.0000044 | 0.0001971 |
| chr9 | 129813921 | 129814166 | TOR1A | 0 | - | 129813921 | 129814166 | 0 | 1 | 245 | 0 | -0.7797160 | 0.0036595 | 0.0412108 |
| chr1 | 3745735 | 3745784 | TP73-AS1 | 0 | - | 3745735 | 3745784 | 0 | 1 | 49 | 0 | -3.9389688 | 0.0096747 | 0.0820944 |
| chr20 | 63865278 | 63865327 | TPD52L2 | 0 | + | 63865278 | 63865327 | 0 | 1 | 49 | 0 | 1.4602665 | 0.0079579 | 0.0717281 |
| chr20 | 63890852 | 63891050 | TPD52L2 | 0 | + | 63890852 | 63891050 | 0 | 1 | 198 | 0 | -1.3399614 | 0.0000738 | 0.0020610 |
| chr12 | 6870751 | 6870800 | TPI1 | 0 | + | 6870751 | 6870800 | 0 | 1 | 49 | 0 | 2.0630088 | 0.0028201 | 0.0338863 |
| chr19 | 16067507 | 16076159 | TPM4 | 0 | + | 16067507 | 16076159 | 0 | 2 | 232,113 | 0,8540 | 0.6137041 | 0.0030669 | 0.0362032 |
| chr11 | 6614358 | 6614556 | TPP1 | 0 | - | 6614358 | 6614556 | 0 | 1 | 198 | 0 | -0.9070923 | 0.0001924 | 0.0043175 |
| chr22 | 26540859 | 26541254 | TPST2 | 0 | - | 26540859 | 26541254 | 0 | 1 | 395 | 0 | -0.7832805 | 0.0086423 | 0.0760810 |
| chr2 | 84870301 | 84870649 | TRABD2A | 0 | - | 84870301 | 84870649 | 0 | 1 | 348 | 0 | -0.9427074 | 0.0000009 | 0.0000515 |
| chr12 | 112140859 | 112142169 | TRAFD1 | 0 | + | 112140859 | 112142169 | 0 | 2 | 366,81 | 0,1230 | -0.9752030 | 0.0009225 | 0.0149011 |
| chr3 | 36831262 | 36831311 | TRANK1 | 0 | - | 36831262 | 36831311 | 0 | 1 | 49 | 0 | -3.5228587 | 0.0014738 | 0.0211135 |
| chr8 | 140450807 | 140451206 | TRAPPC9 | 0 | - | 140450807 | 140451206 | 0 | 1 | 399 | 0 | -1.4103720 | 0.0013989 | 0.0203793 |
| chr6 | 42228330 | 42228579 | TRERF1 | 0 | - | 42228330 | 42228579 | 0 | 1 | 249 | 0 | -1.2499572 | 0.0113817 | 0.0921421 |
| chr3 | 48467151 | 48467200 | TREX1 | 0 | + | 48467151 | 48467200 | 0 | 1 | 49 | 0 | -2.1302361 | 0.0026244 | 0.0323444 |
| chr3 | 48467299 | 48467596 | TREX1 | 0 | + | 48467299 | 48467596 | 0 | 1 | 297 | 0 | -1.0281599 | 0.0000113 | 0.0004441 |
| chr8 | 125436185 | 125437073 | TRIB1 | 0 | + | 125436185 | 125437073 | 0 | 1 | 888 | 0 | -0.5682549 | 0.0007971 | 0.0133234 |
| chr1 | 228394423 | 228397158 | TRIM11 | 0 | - | 228394423 | 228397158 | 0 | 3 | 830,101,16 | 0,2524,2720 | 0.7129783 | 0.0000905 | 0.0024098 |
| chr17 | 15628507 | 15628998 | TRIM16 | 0 | - | 15628507 | 15628998 | 0 | 1 | 491 | 0 | -0.9425558 | 0.0000005 | 0.0000286 |
| chr17 | 18735078 | 18735573 | TRIM16L | 0 | + | 18735078 | 18735573 | 0 | 1 | 495 | 0 | -1.0258993 | 0.0000004 | 0.0000268 |
| chr17 | 56891855 | 56892204 | TRIM25 | 0 | - | 56891855 | 56892204 | 0 | 1 | 349 | 0 | 0.6205687 | 0.0087100 | 0.0764296 |
| chr6 | 25984192 | 25984241 | TRIM38 | 0 | + | 25984192 | 25984241 | 0 | 1 | 49 | 0 | -1.9678483 | 0.0064882 | 0.0628276 |
| chr5 | 14290769 | 14291217 | TRIO | 0 | + | 14290769 | 14291217 | 0 | 1 | 448 | 0 | -0.9302666 | 0.0032592 | 0.0379200 |
| chr2 | 229859278 | 229859527 | TRIP12 | 0 | - | 229859278 | 229859527 | 0 | 1 | 249 | 0 | -1.1429251 | 0.0000611 | 0.0017580 |
| chr5 | 917063 | 917260 | TRIP13 | 0 | + | 917063 | 917260 | 0 | 1 | 197 | 0 | 0.9559012 | 0.0003328 | 0.0067411 |
| chr5 | 917704 | 917753 | TRIP13 | 0 | + | 917704 | 917753 | 0 | 1 | 49 | 0 | -2.1473245 | 0.0012153 | 0.0184160 |
| chr3 | 101565898 | 101566097 | TRMT10C | 0 | + | 101565898 | 101566097 | 0 | 1 | 199 | 0 | -0.9360491 | 0.0052595 | 0.0539292 |
| chr14 | 103534681 | 103534929 | TRMT61A | 0 | + | 103534681 | 103534929 | 0 | 1 | 248 | 0 | -1.0342500 | 0.0081740 | 0.0730578 |
| chr13 | 44434486 | 44434635 | TSC22D1 | 0 | - | 44434486 | 44434635 | 0 | 1 | 149 | 0 | -0.7339594 | 0.0000011 | 0.0000581 |
| chr3 | 150409834 | 150410333 | TSC22D2 | 0 | + | 150409834 | 150410333 | 0 | 1 | 499 | 0 | -1.1134705 | 0.0000091 | 0.0003748 |
| chr12 | 3283153 | 3283449 | TSPAN9 | 0 | + | 3283153 | 3283449 | 0 | 1 | 296 | 0 | 0.9172746 | 0.0101140 | 0.0849276 |
| chr6 | 116278877 | 116279373 | TSPYL1 | 0 | - | 116278877 | 116279373 | 0 | 1 | 496 | 0 | -0.9005300 | 0.0000000 | 0.0000006 |
| chr16 | 1349288 | 1349578 | TSR3 | 0 | - | 1349288 | 1349578 | 0 | 1 | 290 | 0 | -0.5972007 | 0.0000056 | 0.0002421 |
| chr22 | 37011293 | 37018299 | TST | 0 | - | 37011293 | 37018299 | 0 | 2 | 33,162 | 0,6845 | -1.2577974 | 0.0000445 | 0.0013606 |
| chr18 | 24019869 | 24020166 | TTC39C | 0 | + | 24019869 | 24020166 | 0 | 1 | 297 | 0 | -0.6364899 | 0.0004718 | 0.0088575 |
| chr1 | 117075060 | 117075505 | TTF2 | 0 | + | 117075060 | 117075505 | 0 | 1 | 445 | 0 | -1.0587060 | 0.0001152 | 0.0029258 |
| chr1 | 117075556 | 117075803 | TTF2 | 0 | + | 117075556 | 117075803 | 0 | 1 | 247 | 0 | -1.1551200 | 0.0059112 | 0.0587805 |
| chr20 | 38011585 | 38012030 | TTI1 | 0 | - | 38011585 | 38012030 | 0 | 1 | 445 | 0 | -1.1961354 | 0.0000166 | 0.0006127 |
| chr12 | 49127978 | 49128125 | TUBA1B | 0 | - | 49127978 | 49128125 | 0 | 1 | 147 | 0 | -1.3452807 | 0.0006506 | 0.0114484 |
| chr12 | 49128416 | 49128756 | TUBA1B | 0 | - | 49128416 | 49128756 | 0 | 1 | 340 | 0 | -0.9648354 | 0.0044857 | 0.0478436 |
| chr12 | 49272403 | 49272796 | TUBA1C | 0 | + | 49272403 | 49272796 | 0 | 1 | 393 | 0 | -0.8888334 | 0.0103443 | 0.0862011 |
| chr12 | 49273043 | 49273240 | TUBA1C | 0 | + | 49273043 | 49273240 | 0 | 1 | 197 | 0 | -1.2878053 | 0.0009311 | 0.0150139 |
| chr2 | 219250309 | 219251254 | TUBA4A | 0 | - | 219250309 | 219251254 | 0 | 1 | 945 | 0 | -0.6719836 | 0.0016068 | 0.0224854 |
| chr6 | 3225590 | 3225787 | TUBB2B | 0 | - | 3225590 | 3225787 | 0 | 1 | 197 | 0 | -1.0515478 | 0.0082514 | 0.0735391 |
| chr16 | 89923335 | 89932643 | TUBB3 | 0 | + | 89923335 | 89932643 | 0 | 2 | 124,73 | 0,9236 | 1.3078646 | 0.0031418 | 0.0368475 |
| chr9 | 137241991 | 137242812 | TUBB4B | 0 | + | 137241991 | 137242812 | 0 | 2 | 31,317 | 0,505 | -0.5220941 | 0.0016662 | 0.0231018 |
| chr7 | 19698406 | 19698704 | TWISTNB | 0 | - | 19698406 | 19698704 | 0 | 1 | 298 | 0 | -0.9964494 | 0.0004635 | 0.0087790 |
| chr1 | 32194098 | 32195437 | TXLNA | 0 | + | 32194098 | 32195437 | 0 | 2 | 63,536 | 0,804 | 0.5145151 | 0.0026730 | 0.0327445 |
| chr16 | 11691387 | 11691634 | TXNDC11 | 0 | - | 11691387 | 11691634 | 0 | 1 | 247 | 0 | -1.0247494 | 0.0115699 | 0.0930322 |
| chr1 | 52032285 | 52032834 | TXNDC12 | 0 | - | 52032285 | 52032834 | 0 | 1 | 549 | 0 | -0.9865574 | 0.0000224 | 0.0007821 |
| chr1 | 52033085 | 52033384 | TXNDC12 | 0 | - | 52033085 | 52033384 | 0 | 1 | 299 | 0 | -1.4186750 | 0.0000258 | 0.0008753 |
| chr6 | 7882349 | 7882398 | TXNDC5 | 0 | - | 7882349 | 7882398 | 0 | 1 | 49 | 0 | -2.1318461 | 0.0119782 | 0.0950490 |
| chr6 | 7882648 | 7882897 | TXNDC5 | 0 | - | 7882648 | 7882897 | 0 | 1 | 249 | 0 | -1.0460753 | 0.0020371 | 0.0267307 |
| chr6 | 7883098 | 7884389 | TXNDC5 | 0 | - | 7883098 | 7884389 | 0 | 2 | 169,31 | 0,1261 | -0.9868966 | 0.0074734 | 0.0687297 |
| chr22 | 19880713 | 19883419 | TXNRD2 | 0 | - | 19880713 | 19883419 | 0 | 2 | 5,95 | 0,2612 | -1.7059693 | 0.0027496 | 0.0333261 |
| chr10 | 70139278 | 70140026 | TYSND1 | 0 | - | 70139278 | 70140026 | 0 | 1 | 748 | 0 | -0.5056639 | 0.0037840 | 0.0423494 |
| chr19 | 18571853 | 18573335 | UBA52 | 0 | + | 18571853 | 18573335 | 0 | 2 | 57,43 | 0,1440 | 0.7855348 | 0.0032407 | 0.0377863 |
| chr16 | 4609321 | 4609808 | UBALD1 | 0 | - | 4609321 | 4609808 | 0 | 1 | 487 | 0 | 0.6508100 | 0.0000207 | 0.0007420 |
| chr9 | 34241285 | 34242081 | UBAP1 | 0 | + | 34241285 | 34242081 | 0 | 1 | 796 | 0 | -0.8409042 | 0.0001257 | 0.0031132 |
| chr12 | 124911794 | 124912589 | UBC | 0 | - | 124911794 | 124912589 | 0 | 1 | 795 | 0 | -1.6312024 | 0.0000000 | 0.0000000 |
| chr12 | 124912639 | 124912688 | UBC | 0 | - | 124912639 | 124912688 | 0 | 1 | 49 | 0 | -1.7033312 | 0.0000002 | 0.0000133 |
| chr12 | 124912838 | 124912937 | UBC | 0 | - | 124912838 | 124912937 | 0 | 1 | 99 | 0 | -2.3860133 | 0.0000000 | 0.0000000 |
| chr12 | 124913236 | 124913384 | UBC | 0 | - | 124913236 | 124913384 | 0 | 1 | 148 | 0 | -1.6254487 | 0.0000000 | 0.0000005 |
| chr3 | 23807288 | 23807337 | UBE2E1 | 0 | + | 23807288 | 23807337 | 0 | 1 | 49 | 0 | 1.6434367 | 0.0001688 | 0.0039029 |
| chr21 | 44787948 | 44801379 | UBE2G2 | 0 | - | 44787948 | 44801379 | 0 | 3 | 18,36,96 | 0,112,13336 | 1.1083128 | 0.0062790 | 0.0613935 |
| chr7 | 129833859 | 129833908 | UBE2H | 0 | - | 129833859 | 129833908 | 0 | 1 | 49 | 0 | -1.5748003 | 0.0065840 | 0.0634503 |
| chr7 | 129834058 | 129834305 | UBE2H | 0 | - | 129834058 | 129834305 | 0 | 1 | 247 | 0 | -0.8468585 | 0.0000469 | 0.0014238 |
| chr7 | 129880933 | 129952960 | UBE2H | 0 | - | 129880933 | 129952960 | 0 | 2 | 39,458 | 0,71570 | 0.9409507 | 0.0000000 | 0.0000000 |
| chr17 | 76389681 | 76390275 | UBE2O | 0 | - | 76389681 | 76390275 | 0 | 1 | 594 | 0 | -1.5922091 | 0.0000002 | 0.0000152 |
| chr17 | 76396374 | 76396820 | UBE2O | 0 | - | 76396374 | 76396820 | 0 | 1 | 446 | 0 | -1.1492140 | 0.0004942 | 0.0091317 |
| chr17 | 76452760 | 76453107 | UBE2O | 0 | - | 76452760 | 76453107 | 0 | 1 | 347 | 0 | 1.4678144 | 0.0001245 | 0.0031018 |
| chr1 | 154550112 | 154550458 | UBE2Q1 | 0 | - | 154550112 | 154550458 | 0 | 1 | 346 | 0 | -0.7052772 | 0.0000302 | 0.0009998 |
| chr15 | 25360468 | 25370705 | UBE3A | 0 | - | 25360468 | 25370705 | 0 | 2 | 60,140 | 0,10098 | -1.5331775 | 0.0024071 | 0.0303597 |
| chrX | 56564584 | 56564732 | UBQLN2 | 0 | + | 56564584 | 56564732 | 0 | 1 | 148 | 0 | -2.5516902 | 0.0000000 | 0.0000000 |
| chrX | 56565128 | 56565326 | UBQLN2 | 0 | + | 56565128 | 56565326 | 0 | 1 | 198 | 0 | -0.9368028 | 0.0079454 | 0.0716863 |
| chr6 | 42564022 | 42564370 | UBR2 | 0 | + | 42564022 | 42564370 | 0 | 1 | 348 | 0 | 1.9308619 | 0.0000000 | 0.0000002 |
| chr10 | 97570414 | 97570712 | UBTD1 | 0 | + | 97570414 | 97570712 | 0 | 1 | 298 | 0 | -1.1766939 | 0.0032006 | 0.0374149 |
| chr5 | 172211524 | 172211721 | UBTD2 | 0 | - | 172211524 | 172211721 | 0 | 1 | 197 | 0 | -1.8526254 | 0.0004590 | 0.0087131 |
| chr19 | 4445056 | 4445354 | UBXN6 | 0 | - | 4445056 | 4445354 | 0 | 1 | 298 | 0 | -1.4360056 | 0.0000321 | 0.0010580 |
| chr6 | 34859068 | 34859466 | UHRF1BP1 | 0 | + | 34859068 | 34859466 | 0 | 1 | 398 | 0 | -0.8958241 | 0.0028633 | 0.0342850 |
| chr12 | 131922616 | 131922864 | ULK1 | 0 | + | 131922616 | 131922864 | 0 | 1 | 248 | 0 | -1.3046619 | 0.0004710 | 0.0088575 |
| chr12 | 109098343 | 109098491 | UNG | 0 | + | 109098343 | 109098491 | 0 | 1 | 148 | 0 | 1.0886675 | 0.0091035 | 0.0789490 |
| chr10 | 12014055 | 12014153 | UPF2 | 0 | - | 12014055 | 12014153 | 0 | 1 | 98 | 0 | -2.3776893 | 0.0006984 | 0.0120337 |
| chr10 | 12028763 | 12028862 | UPF2 | 0 | - | 12028763 | 12028862 | 0 | 1 | 99 | 0 | -3.4076997 | 0.0000000 | 0.0000000 |
| chr10 | 12028962 | 12029507 | UPF2 | 0 | - | 12028962 | 12029507 | 0 | 1 | 545 | 0 | -0.9463494 | 0.0007448 | 0.0126289 |
| chr19 | 29207407 | 29207945 | UQCRFS1 | 0 | - | 29207407 | 29207945 | 0 | 1 | 538 | 0 | -1.0289108 | 0.0000810 | 0.0022112 |
| chr21 | 32347430 | 32347629 | URB1 | 0 | - | 32347430 | 32347629 | 0 | 1 | 199 | 0 | -1.2727930 | 0.0004033 | 0.0078545 |
| chr1 | 229636266 | 229636514 | URB2 | 0 | + | 229636266 | 229636514 | 0 | 1 | 248 | 0 | -1.4393273 | 0.0029470 | 0.0350850 |
| chr7 | 43877581 | 43877729 | URGCP | 0 | - | 43877581 | 43877729 | 0 | 1 | 148 | 0 | -1.5700579 | 0.0000031 | 0.0001450 |
| chr7 | 43877928 | 43878175 | URGCP | 0 | - | 43877928 | 43878175 | 0 | 1 | 247 | 0 | -1.3323139 | 0.0000000 | 0.0000016 |
| chr1 | 45014999 | 45015475 | UROD | 0 | + | 45014999 | 45015475 | 0 | 2 | 8,139 | 0,338 | -1.6081157 | 0.0000090 | 0.0003702 |
| chr19 | 35271087 | 35278740 | USF2 | 0 | + | 35271087 | 35278740 | 0 | 2 | 55,43 | 0,7611 | -1.4537314 | 0.0003993 | 0.0078091 |
| chr1 | 62444813 | 62445112 | USP1 | 0 | + | 62444813 | 62445112 | 0 | 1 | 299 | 0 | -0.7431708 | 0.0017506 | 0.0238709 |
| chr1 | 62448627 | 62450405 | USP1 | 0 | + | 62448627 | 62450405 | 0 | 2 | 40,160 | 0,1619 | -1.7001327 | 0.0000231 | 0.0007987 |
| chr1 | 62450456 | 62450755 | USP1 | 0 | + | 62450456 | 62450755 | 0 | 1 | 299 | 0 | -1.1653658 | 0.0006721 | 0.0117016 |
| chr16 | 84744769 | 84745568 | USP10 | 0 | + | 84744769 | 84745568 | 0 | 1 | 799 | 0 | -0.5570937 | 0.0003653 | 0.0073001 |
| chr16 | 84779072 | 84779121 | USP10 | 0 | + | 84779072 | 84779121 | 0 | 1 | 49 | 0 | -1.7125061 | 0.0117487 | 0.0939270 |
| chr22 | 18150190 | 18157623 | USP18 | 0 | + | 18150190 | 18157623 | 0 | 2 | 33,66 | 0,7368 | 1.8631801 | 0.0002617 | 0.0055550 |
| chr22 | 18157673 | 18157722 | USP18 | 0 | + | 18157673 | 18157722 | 0 | 1 | 49 | 0 | 2.7761044 | 0.0000029 | 0.0001368 |
| chr9 | 129880746 | 129881093 | USP20 | 0 | + | 129880746 | 129881093 | 0 | 1 | 347 | 0 | -1.3920053 | 0.0070445 | 0.0662312 |
| chr17 | 21002427 | 21003022 | USP22 | 0 | - | 21002427 | 21003022 | 0 | 1 | 595 | 0 | 0.7256014 | 0.0014143 | 0.0205137 |
| chr1 | 55068699 | 55069046 | USP24 | 0 | - | 55068699 | 55069046 | 0 | 1 | 347 | 0 | -0.9004557 | 0.0072866 | 0.0674910 |
| chr2 | 85616194 | 85616443 | USP39 | 0 | + | 85616194 | 85616443 | 0 | 1 | 249 | 0 | 1.1030728 | 0.0086425 | 0.0760810 |
| chr7 | 6154066 | 6155159 | USP42 | 0 | + | 6154066 | 6155159 | 0 | 1 | 1093 | 0 | -0.7059268 | 0.0011992 | 0.0182178 |
| chr10 | 73516964 | 73517311 | USP54 | 0 | - | 73516964 | 73517311 | 0 | 1 | 347 | 0 | -1.2370746 | 0.0056901 | 0.0572747 |
| chr10 | 11462649 | 11462847 | USP6NL | 0 | - | 11462649 | 11462847 | 0 | 1 | 198 | 0 | -1.0963299 | 0.0060867 | 0.0600321 |
| chr6 | 148747135 | 148747334 | UST | 0 | + | 148747135 | 148747334 | 0 | 1 | 199 | 0 | 1.7384700 | 0.0014192 | 0.0205592 |
| chrX | 129925178 | 129926098 | UTP14A | 0 | + | 129925178 | 129926098 | 0 | 2 | 18,180 | 0,741 | 1.3332357 | 0.0013763 | 0.0201640 |
| chr17 | 51260536 | 51263329 | UTP18 | 0 | + | 51260536 | 51263329 | 0 | 2 | 391,56 | 0,2738 | 1.5869522 | 0.0000000 | 0.0000019 |
| chr1 | 7780550 | 7780648 | VAMP3 | 0 | + | 7780550 | 7780648 | 0 | 1 | 98 | 0 | -2.0650149 | 0.0001987 | 0.0044315 |
| chr1 | 7780844 | 7781138 | VAMP3 | 0 | + | 7780844 | 7781138 | 0 | 1 | 294 | 0 | -1.6541558 | 0.0000240 | 0.0008202 |
| chr1 | 115685509 | 115691797 | VANGL1 | 0 | + | 115685509 | 115691797 | 0 | 2 | 19,679 | 0,5610 | -0.5402352 | 0.0046649 | 0.0493697 |
| chr19 | 45507480 | 45517763 | VASP | 0 | + | 45507480 | 45517763 | 0 | 2 | 297,101 | 0,10183 | 0.9104929 | 0.0000000 | 0.0000000 |
| chr10 | 73998114 | 73998362 | VCL | 0 | + | 73998114 | 73998362 | 0 | 1 | 248 | 0 | 0.8396443 | 0.0025997 | 0.0321548 |
| chr6 | 43770557 | 43771202 | VEGFA | 0 | + | 43770557 | 43771202 | 0 | 1 | 645 | 0 | -0.7227280 | 0.0002396 | 0.0051702 |
| chr6 | 43782045 | 43784746 | VEGFA | 0 | + | 43782045 | 43784746 | 0 | 2 | 43,206 | 0,2496 | -0.6275763 | 0.0036205 | 0.0409150 |
| chr3 | 10141784 | 10150102 | VHL | 0 | + | 10141784 | 10150102 | 0 | 3 | 404,123,316 | 0,4730,8003 | 1.0449091 | 0.0000000 | 0.0000000 |
| chr16 | 31090904 | 31091103 | VKORC1 | 0 | - | 31090904 | 31091103 | 0 | 1 | 199 | 0 | -1.1386881 | 0.0000238 | 0.0008160 |
| chr3 | 51441604 | 51441851 | VPRBP | 0 | - | 51441604 | 51441851 | 0 | 1 | 247 | 0 | -1.6958531 | 0.0000116 | 0.0004520 |
| chr12 | 122867277 | 122867576 | VPS37B | 0 | - | 122867277 | 122867576 | 0 | 1 | 299 | 0 | 0.7762733 | 0.0006774 | 0.0117491 |
| chr15 | 42158900 | 42159199 | VPS39 | 0 | - | 42158900 | 42159199 | 0 | 1 | 299 | 0 | 1.4954489 | 0.0001402 | 0.0034020 |
| chr10 | 28532787 | 28532985 | WAC | 0 | + | 28532787 | 28532985 | 0 | 1 | 198 | 0 | 1.7010455 | 0.0001234 | 0.0030857 |
| chr10 | 28619538 | 28619786 | WAC | 0 | + | 28619538 | 28619786 | 0 | 1 | 248 | 0 | -0.7484081 | 0.0023269 | 0.0295474 |
| chr10 | 86497252 | 86500642 | WAPL | 0 | - | 86497252 | 86500642 | 0 | 2 | 68,925 | 0,2466 | -0.5125007 | 0.0074985 | 0.0688907 |
| chr1 | 27407351 | 27407450 | WASF2 | 0 | - | 27407351 | 27407450 | 0 | 1 | 99 | 0 | -1.6525881 | 0.0004414 | 0.0084309 |
| chr2 | 223918003 | 223945288 | WDFY1 | 0 | - | 223918003 | 223945288 | 0 | 2 | 8,141 | 0,27145 | 1.0803478 | 0.0110492 | 0.0901661 |
| chrX | 48605096 | 48605145 | WDR13 | 0 | + | 48605096 | 48605145 | 0 | 1 | 49 | 0 | 2.1941040 | 0.0000857 | 0.0023199 |
| chr14 | 100381223 | 100381572 | WDR25 | 0 | + | 100381223 | 100381572 | 0 | 1 | 349 | 0 | -0.8993859 | 0.0049154 | 0.0513602 |
| chr1 | 224433752 | 224434249 | WDR26 | 0 | - | 224433752 | 224434249 | 0 | 1 | 497 | 0 | 0.9804899 | 0.0003570 | 0.0071667 |
| chr1 | 109041881 | 109042129 | WDR47 | 0 | - | 109041881 | 109042129 | 0 | 1 | 248 | 0 | 0.9876243 | 0.0103306 | 0.0861465 |
| chr3 | 49007501 | 49012000 | WDR6 | 0 | + | 49007501 | 49012000 | 0 | 2 | 31,366 | 0,4134 | -0.5361343 | 0.0019150 | 0.0256104 |
| chr3 | 49012297 | 49012742 | WDR6 | 0 | + | 49012297 | 49012742 | 0 | 1 | 445 | 0 | -0.8665824 | 0.0000005 | 0.0000290 |
| chr3 | 52278295 | 52278643 | WDR82 | 0 | - | 52278295 | 52278643 | 0 | 1 | 348 | 0 | 0.9282880 | 0.0123690 | 0.0968820 |
| chr19 | 12675735 | 12675784 | WDR83 | 0 | + | 12675735 | 12675784 | 0 | 1 | 49 | 0 | -3.3329627 | 0.0101625 | 0.0851172 |
| chr1 | 68232253 | 68232551 | WLS | 0 | - | 68232253 | 68232551 | 0 | 1 | 298 | 0 | 0.5193135 | 0.0071648 | 0.0667729 |
| chr12 | 753473 | 813739 | WNK1 | 0 | + | 753473 | 813739 | 0 | 2 | 852,98 | 0,60169 | -0.9724534 | 0.0000000 | 0.0000007 |
| chr12 | 896148 | 896547 | WNK1 | 0 | + | 896148 | 896547 | 0 | 1 | 399 | 0 | -0.5517475 | 0.0061886 | 0.0607058 |
| chr12 | 909953 | 910102 | WNK1 | 0 | + | 909953 | 910102 | 0 | 1 | 149 | 0 | -1.9514428 | 0.0020509 | 0.0268527 |
| chr22 | 45921357 | 45921703 | WNT7B | 0 | - | 45921357 | 45921703 | 0 | 1 | 346 | 0 | -1.1088673 | 0.0000002 | 0.0000127 |
| chr3 | 14156488 | 14158597 | XPC | 0 | - | 14156488 | 14158597 | 0 | 2 | 8,587 | 0,1523 | -0.8251558 | 0.0001649 | 0.0038322 |
| chr2 | 61537736 | 61538283 | XPO1 | 0 | - | 61537736 | 61538283 | 0 | 1 | 547 | 0 | 1.1162076 | 0.0000000 | 0.0000002 |
| chr20 | 21303354 | 21326307 | XRN2 | 0 | + | 21303354 | 21326307 | 0 | 2 | 120,29 | 0,22925 | 1.5783877 | 0.0003399 | 0.0068621 |
| chr11 | 102110461 | 102110808 | YAP1 | 0 | + | 102110461 | 102110808 | 0 | 1 | 347 | 0 | 1.3439124 | 0.0000086 | 0.0003566 |
| chr11 | 102227534 | 102230249 | YAP1 | 0 | + | 102227534 | 102230249 | 0 | 2 | 48,548 | 0,2168 | -0.6706428 | 0.0010260 | 0.0162116 |
| chr1 | 32775437 | 32776030 | YARS | 0 | - | 32775437 | 32776030 | 0 | 1 | 593 | 0 | -0.7426675 | 0.0066478 | 0.0638951 |
| chr12 | 32747087 | 32747284 | YARS2 | 0 | - | 32747087 | 32747284 | 0 | 1 | 197 | 0 | -0.9172869 | 0.0110055 | 0.0899598 |
| chr22 | 21628189 | 21628535 | YDJC | 0 | - | 21628189 | 21628535 | 0 | 1 | 346 | 0 | -0.7898041 | 0.0000261 | 0.0008772 |
| chr3 | 183811831 | 183811930 | YEATS2 | 0 | + | 183811831 | 183811930 | 0 | 1 | 99 | 0 | -2.3331860 | 0.0053900 | 0.0549252 |
| chr19 | 38303953 | 38304593 | YIF1B | 0 | - | 38303953 | 38304593 | 0 | 1 | 640 | 0 | 0.7710107 | 0.0012321 | 0.0185819 |
| chr14 | 74798069 | 74798168 | YLPM1 | 0 | + | 74798069 | 74798168 | 0 | 1 | 99 | 0 | -2.4716663 | 0.0091534 | 0.0791545 |
| chr1 | 207049275 | 207049622 | YOD1 | 0 | - | 207049275 | 207049622 | 0 | 1 | 347 | 0 | -0.7126331 | 0.0043309 | 0.0469039 |
| chr17 | 59397151 | 59397299 | YPEL2 | 0 | + | 59397151 | 59397299 | 0 | 1 | 148 | 0 | 1.7705732 | 0.0019024 | 0.0254980 |
| chr2 | 30158806 | 30159003 | YPEL5 | 0 | + | 30158806 | 30159003 | 0 | 1 | 197 | 0 | -0.8928278 | 0.0033389 | 0.0385095 |
| chr20 | 63203100 | 63203396 | YTHDF1 | 0 | - | 63203100 | 63203396 | 0 | 1 | 296 | 0 | -1.1033687 | 0.0000006 | 0.0000340 |
| chr20 | 63203642 | 63203691 | YTHDF1 | 0 | - | 63203642 | 63203691 | 0 | 1 | 49 | 0 | -1.4510965 | 0.0078782 | 0.0711871 |
| chr8 | 63186194 | 63209964 | YTHDF3 | 0 | + | 63186194 | 63209964 | 0 | 2 | 1552,282 | 0,23489 | -0.7925959 | 0.0000000 | 0.0000023 |
| chr20 | 44906848 | 44906946 | YWHAB | 0 | + | 44906848 | 44906946 | 0 | 1 | 98 | 0 | -1.7630349 | 0.0001411 | 0.0034096 |
| chr20 | 44906997 | 44907095 | YWHAB | 0 | + | 44906997 | 44907095 | 0 | 1 | 98 | 0 | -2.0321565 | 0.0002853 | 0.0059549 |
| chr7 | 76329289 | 76330087 | YWHAG | 0 | - | 76329289 | 76330087 | 0 | 1 | 798 | 0 | -0.5775911 | 0.0026777 | 0.0327445 |
| chr22 | 31944543 | 31944791 | YWHAH | 0 | + | 31944543 | 31944791 | 0 | 1 | 248 | 0 | 0.5405578 | 0.0061899 | 0.0607058 |
| chr8 | 100952923 | 100953296 | YWHAZ | 0 | - | 100952923 | 100953296 | 0 | 2 | 71,28 | 0,346 | -2.4583080 | 0.0000027 | 0.0001266 |
| chr14 | 100239851 | 100239900 | YY1 | 0 | + | 100239851 | 100239900 | 0 | 1 | 49 | 0 | -2.0802258 | 0.0027948 | 0.0336719 |
| chr22 | 49886636 | 49886685 | ZBED4 | 0 | + | 49886636 | 49886685 | 0 | 1 | 49 | 0 | -2.7376766 | 0.0050077 | 0.0520851 |
| chr22 | 49886736 | 49886835 | ZBED4 | 0 | + | 49886736 | 49886835 | 0 | 1 | 99 | 0 | -1.7449371 | 0.0117380 | 0.0938835 |
| chr6 | 151365413 | 151366060 | ZBTB2 | 0 | - | 151365413 | 151366060 | 0 | 1 | 647 | 0 | -0.6533257 | 0.0053264 | 0.0544612 |
| chrX | 120254376 | 120254525 | ZBTB33 | 0 | + | 120254376 | 120254525 | 0 | 1 | 149 | 0 | -1.5110693 | 0.0115137 | 0.0926548 |
| chr9 | 126880359 | 126880607 | ZBTB34 | 0 | + | 126880359 | 126880607 | 0 | 1 | 248 | 0 | -1.1723368 | 0.0123199 | 0.0967430 |
| chr12 | 57003724 | 57004272 | ZBTB39 | 0 | - | 57003724 | 57004272 | 0 | 1 | 548 | 0 | -1.0312122 | 0.0000943 | 0.0024864 |
| chr12 | 57004622 | 57004920 | ZBTB39 | 0 | - | 57004622 | 57004920 | 0 | 1 | 298 | 0 | -1.2946096 | 0.0041542 | 0.0453132 |
| chr17 | 7462546 | 7463589 | ZBTB4 | 0 | - | 7462546 | 7463589 | 0 | 1 | 1043 | 0 | -0.5086739 | 0.0030542 | 0.0360999 |
| chr19 | 4047663 | 4048011 | ZBTB7A | 0 | - | 4047663 | 4048011 | 0 | 1 | 348 | 0 | -0.6545634 | 0.0036858 | 0.0414546 |
| chr19 | 4054337 | 4054436 | ZBTB7A | 0 | - | 4054337 | 4054436 | 0 | 1 | 99 | 0 | -1.8907890 | 0.0001613 | 0.0037738 |
| chr19 | 4054487 | 4054685 | ZBTB7A | 0 | - | 4054487 | 4054685 | 0 | 1 | 198 | 0 | -1.4238954 | 0.0013591 | 0.0199912 |
| chr1 | 155015222 | 155015671 | ZBTB7B | 0 | + | 155015222 | 155015671 | 0 | 1 | 449 | 0 | -0.9007816 | 0.0000402 | 0.0012538 |
| chr1 | 203796339 | 203796488 | ZC3H11A | 0 | + | 203796339 | 203796488 | 0 | 1 | 149 | 0 | 1.3831136 | 0.0035961 | 0.0407434 |
| chr13 | 45985450 | 45985747 | ZC3H13 | 0 | - | 45985450 | 45985747 | 0 | 1 | 297 | 0 | -1.1550746 | 0.0095753 | 0.0815185 |
| chr19 | 47067273 | 47067620 | ZC3H4 | 0 | - | 47067273 | 47067620 | 0 | 1 | 347 | 0 | -1.1998796 | 0.0014061 | 0.0204345 |
| chr22 | 41356791 | 41357457 | ZC3H7B | 0 | + | 41356791 | 41357457 | 0 | 2 | 18,281 | 0,386 | 1.5710720 | 0.0000009 | 0.0000510 |
| chr22 | 41357956 | 41358304 | ZC3H7B | 0 | + | 41357956 | 41358304 | 0 | 1 | 348 | 0 | -1.7592103 | 0.0000012 | 0.0000651 |
| chr7 | 139079536 | 139080083 | ZC3HAV1 | 0 | - | 139079536 | 139080083 | 0 | 1 | 547 | 0 | -0.6119022 | 0.0000129 | 0.0005006 |
| chr11 | 57698956 | 57699352 | ZDHHC5 | 0 | + | 57698956 | 57699352 | 0 | 1 | 396 | 0 | -0.9581416 | 0.0007719 | 0.0129641 |
| chr12 | 71657265 | 71663422 | ZFC3H1 | 0 | - | 71657265 | 71663422 | 0 | 2 | 37,410 | 0,5748 | -1.2398939 | 0.0000497 | 0.0014884 |
| chr16 | 72788426 | 72788675 | ZFHX3 | 0 | - | 72788426 | 72788675 | 0 | 1 | 249 | 0 | -1.8450292 | 0.0000062 | 0.0002649 |
| chr16 | 72958280 | 72958429 | ZFHX3 | 0 | - | 72958280 | 72958429 | 0 | 1 | 149 | 0 | -1.8328545 | 0.0012577 | 0.0188530 |
| chr14 | 68788702 | 68788801 | ZFP36L1 | 0 | - | 68788702 | 68788801 | 0 | 1 | 99 | 0 | -1.2732186 | 0.0007218 | 0.0123073 |
| chr14 | 68789500 | 68789849 | ZFP36L1 | 0 | - | 68789500 | 68789849 | 0 | 1 | 349 | 0 | -0.7657291 | 0.0004784 | 0.0089214 |
| chr2 | 43225147 | 43225345 | ZFP36L2 | 0 | - | 43225147 | 43225345 | 0 | 1 | 198 | 0 | -1.9377309 | 0.0000000 | 0.0000023 |
| chr20 | 52152747 | 52153095 | ZFP64 | 0 | - | 52152747 | 52153095 | 0 | 1 | 348 | 0 | -1.1510053 | 0.0001008 | 0.0026298 |
| chrX | 24210568 | 24211416 | ZFX | 0 | + | 24210568 | 24211416 | 0 | 1 | 848 | 0 | -0.8115218 | 0.0005717 | 0.0102710 |
| chr14 | 73024140 | 73024437 | ZFYVE1 | 0 | - | 73024140 | 73024437 | 0 | 1 | 297 | 0 | -1.6751604 | 0.0025924 | 0.0321310 |
| chr1 | 52238315 | 52238963 | ZFYVE9 | 0 | + | 52238315 | 52238963 | 0 | 1 | 648 | 0 | -0.9258917 | 0.0066173 | 0.0637034 |
| chr16 | 2832138 | 2832235 | ZG16B | 0 | + | 2832138 | 2832235 | 0 | 1 | 97 | 0 | -1.2043495 | 0.0048486 | 0.0508419 |
| chr20 | 41202098 | 41202297 | ZHX3 | 0 | - | 41202098 | 41202297 | 0 | 1 | 199 | 0 | -1.2931422 | 0.0125502 | 0.0979564 |
| chr7 | 100035043 | 100035292 | ZKSCAN1 | 0 | + | 100035043 | 100035292 | 0 | 1 | 249 | 0 | -1.8180050 | 0.0004744 | 0.0088765 |
| chr15 | 42448217 | 42448566 | ZNF106 | 0 | - | 42448217 | 42448566 | 0 | 1 | 349 | 0 | -0.9722831 | 0.0035978 | 0.0407434 |
| chr2 | 218642276 | 218642674 | ZNF142 | 0 | - | 218642276 | 218642674 | 0 | 1 | 398 | 0 | -1.3517275 | 0.0000780 | 0.0021554 |
| chr2 | 218642725 | 218642774 | ZNF142 | 0 | - | 218642725 | 218642774 | 0 | 1 | 49 | 0 | -4.1210147 | 0.0044713 | 0.0478055 |
| chr20 | 53575794 | 53576192 | ZNF217 | 0 | - | 53575794 | 53576192 | 0 | 1 | 398 | 0 | -0.6774097 | 0.0036566 | 0.0412033 |
| chr18 | 35337329 | 35337527 | ZNF24 | 0 | - | 35337329 | 35337527 | 0 | 1 | 198 | 0 | -1.1765988 | 0.0070716 | 0.0663830 |
| chr1 | 200407093 | 200407441 | ZNF281 | 0 | - | 200407093 | 200407441 | 0 | 1 | 348 | 0 | -0.6413490 | 0.0046299 | 0.0490570 |
| chr1 | 200408639 | 200408738 | ZNF281 | 0 | - | 200408639 | 200408738 | 0 | 1 | 99 | 0 | -1.9649971 | 0.0067685 | 0.0644397 |
| chr19 | 9161479 | 9161776 | ZNF317 | 0 | + | 9161479 | 9161776 | 0 | 1 | 297 | 0 | -1.1848366 | 0.0009796 | 0.0156699 |
| chr20 | 45971435 | 45972145 | ZNF335 | 0 | - | 45971435 | 45972145 | 0 | 2 | 26,24 | 0,687 | 3.2587160 | 0.0018410 | 0.0248550 |
| chr12 | 6666823 | 6667070 | ZNF384 | 0 | - | 6666823 | 6667070 | 0 | 1 | 247 | 0 | -1.0553635 | 0.0010348 | 0.0163226 |
| chr7 | 99493830 | 99494078 | ZNF394 | 0 | - | 99493830 | 99494078 | 0 | 1 | 248 | 0 | -1.1194224 | 0.0006371 | 0.0112328 |
| chr10 | 75399667 | 75400115 | ZNF503 | 0 | - | 75399667 | 75400115 | 0 | 1 | 448 | 0 | -1.2873881 | 0.0000005 | 0.0000290 |
| chr19 | 42226175 | 42226570 | ZNF526 | 0 | + | 42226175 | 42226570 | 0 | 1 | 395 | 0 | -0.7038448 | 0.0125085 | 0.0977405 |
| chr19 | 58262604 | 58262950 | ZNF544 | 0 | + | 58262604 | 58262950 | 0 | 1 | 346 | 0 | -1.1948422 | 0.0017242 | 0.0236175 |
| chr19 | 58263395 | 58263593 | ZNF544 | 0 | + | 58263395 | 58263593 | 0 | 1 | 198 | 0 | -2.0481844 | 0.0001565 | 0.0036900 |
| chr19 | 9653100 | 9656562 | ZNF562 | 0 | - | 9653100 | 9656562 | 0 | 2 | 782,16 | 0,3447 | -0.6464303 | 0.0116947 | 0.0936613 |
| chr19 | 42079630 | 42079779 | ZNF574 | 0 | + | 42079630 | 42079779 | 0 | 1 | 149 | 0 | -1.1917698 | 0.0016942 | 0.0233831 |
| chr19 | 42079829 | 42079928 | ZNF574 | 0 | + | 42079829 | 42079928 | 0 | 1 | 99 | 0 | -1.4501988 | 0.0076320 | 0.0697627 |
| chr19 | 55645084 | 55645525 | ZNF581 | 0 | + | 55645084 | 55645525 | 0 | 1 | 441 | 0 | -0.8966898 | 0.0000046 | 0.0002051 |
| chr19 | 58417883 | 58418129 | ZNF584 | 0 | + | 58417883 | 58418129 | 0 | 1 | 246 | 0 | -1.2256322 | 0.0041363 | 0.0451750 |
| chr15 | 84784036 | 84784184 | ZNF592 | 0 | + | 84784036 | 84784184 | 0 | 1 | 148 | 0 | -1.8968653 | 0.0000551 | 0.0016133 |
| chr16 | 2009672 | 2009721 | ZNF598 | 0 | - | 2009672 | 2009721 | 0 | 1 | 49 | 0 | 2.2281969 | 0.0028773 | 0.0343912 |
| chr15 | 64499420 | 64500067 | ZNF609 | 0 | + | 64499420 | 64500067 | 0 | 1 | 647 | 0 | -0.5523682 | 0.0067640 | 0.0644309 |
| chr15 | 64674598 | 64674697 | ZNF609 | 0 | + | 64674598 | 64674697 | 0 | 1 | 99 | 0 | -1.8479193 | 0.0124305 | 0.0972735 |
| chr15 | 64675296 | 64675544 | ZNF609 | 0 | + | 64675296 | 64675544 | 0 | 1 | 248 | 0 | -2.1744541 | 0.0000006 | 0.0000368 |
| chr12 | 48343299 | 48343597 | ZNF641 | 0 | - | 48343299 | 48343597 | 0 | 1 | 298 | 0 | -1.7337208 | 0.0000081 | 0.0003380 |
| chr16 | 31080456 | 31080505 | ZNF646 | 0 | + | 31080456 | 31080505 | 0 | 1 | 49 | 0 | -Inf | 0.0048489 | 0.0508419 |
| chr12 | 124011563 | 124012258 | ZNF664 | 0 | + | 124011563 | 124012258 | 0 | 1 | 695 | 0 | 0.6080820 | 0.0056048 | 0.0565410 |
| chr1 | 248847225 | 248847323 | ZNF672 | 0 | + | 248847225 | 248847323 | 0 | 1 | 98 | 0 | -1.5822865 | 0.0050511 | 0.0524753 |
| chr1 | 248847374 | 248847523 | ZNF672 | 0 | + | 248847374 | 248847523 | 0 | 1 | 149 | 0 | -2.0971762 | 0.0000067 | 0.0002852 |
| chr1 | 151286414 | 151286762 | ZNF687 | 0 | + | 151286414 | 151286762 | 0 | 1 | 348 | 0 | -1.0532559 | 0.0008176 | 0.0135576 |
| chr1 | 151287013 | 151287411 | ZNF687 | 0 | + | 151287013 | 151287411 | 0 | 1 | 398 | 0 | -0.9786186 | 0.0001408 | 0.0034086 |
| chr1 | 151287511 | 151288158 | ZNF687 | 0 | + | 151287511 | 151288158 | 0 | 1 | 647 | 0 | -1.5023983 | 0.0000000 | 0.0000001 |
| chr16 | 30604794 | 30605240 | ZNF689 | 0 | - | 30604794 | 30605240 | 0 | 1 | 446 | 0 | -1.2182030 | 0.0017105 | 0.0234833 |
| chr8 | 37698223 | 37698771 | ZNF703 | 0 | + | 37698223 | 37698771 | 0 | 1 | 548 | 0 | -0.6947996 | 0.0086793 | 0.0762565 |
| chr22 | 20406319 | 20406418 | ZNF74 | 0 | + | 20406319 | 20406418 | 0 | 1 | 99 | 0 | -2.8248262 | 0.0019972 | 0.0263599 |
| chr16 | 30525626 | 30525773 | ZNF768 | 0 | - | 30525626 | 30525773 | 0 | 1 | 147 | 0 | -1.6621557 | 0.0000045 | 0.0002015 |
| chr15 | 34982689 | 34983388 | ZNF770 | 0 | - | 34982689 | 34983388 | 0 | 1 | 699 | 0 | 0.8333118 | 0.0074386 | 0.0684970 |
| chr7 | 149455393 | 149456036 | ZNF777 | 0 | - | 149455393 | 149456036 | 0 | 1 | 643 | 0 | -0.8512077 | 0.0006754 | 0.0117376 |
| chr19 | 56087659 | 56088102 | ZNF787 | 0 | - | 56087659 | 56088102 | 0 | 1 | 443 | 0 | -0.5551027 | 0.0085779 | 0.0756598 |
| chr19 | 55615743 | 55616091 | ZNF865 | 0 | + | 55615743 | 55616091 | 0 | 1 | 348 | 0 | -0.8575277 | 0.0111642 | 0.0908186 |
| chr20 | 49246349 | 49246597 | ZNFX1 | 0 | - | 49246349 | 49246597 | 0 | 1 | 248 | 0 | -1.1273096 | 0.0001013 | 0.0026385 |
| chr20 | 49247694 | 49247942 | ZNFX1 | 0 | - | 49247694 | 49247942 | 0 | 1 | 248 | 0 | -1.6136024 | 0.0000001 | 0.0000071 |
| chr20 | 49248690 | 49248988 | ZNFX1 | 0 | - | 49248690 | 49248988 | 0 | 1 | 298 | 0 | -1.1210889 | 0.0000613 | 0.0017634 |
| chr20 | 49249138 | 49249187 | ZNFX1 | 0 | - | 49249138 | 49249187 | 0 | 1 | 49 | 0 | -2.0476562 | 0.0000004 | 0.0000234 |
| chr20 | 49249237 | 49249436 | ZNFX1 | 0 | - | 49249237 | 49249436 | 0 | 1 | 199 | 0 | -1.3401037 | 0.0000000 | 0.0000002 |
| chr20 | 49270017 | 49270514 | ZNFX1 | 0 | - | 49270017 | 49270514 | 0 | 1 | 497 | 0 | -1.0846300 | 0.0000000 | 0.0000015 |
| chr20 | 49270864 | 49271012 | ZNFX1 | 0 | - | 49270864 | 49271012 | 0 | 1 | 148 | 0 | -0.8472732 | 0.0016132 | 0.0225463 |
| chr20 | 49271063 | 49271162 | ZNFX1 | 0 | - | 49271063 | 49271162 | 0 | 1 | 99 | 0 | -0.9219365 | 0.0003682 | 0.0073346 |
| chr20 | 49271212 | 49271660 | ZNFX1 | 0 | - | 49271212 | 49271660 | 0 | 1 | 448 | 0 | -0.7167768 | 0.0006643 | 0.0116109 |
| chr22 | 29049284 | 29049532 | ZNRF3 | 0 | + | 29049284 | 29049532 | 0 | 1 | 248 | 0 | -1.8721172 | 0.0000167 | 0.0006127 |
| chr19 | 13830236 | 13830584 | ZSWIM4 | 0 | + | 13830236 | 13830584 | 0 | 1 | 348 | 0 | -1.2279881 | 0.0002609 | 0.0055456 |
| chrX | 57593034 | 57593533 | ZXDB | 0 | + | 57593034 | 57593533 | 0 | 1 | 499 | 0 | -1.0775748 | 0.0000444 | 0.0013606 |
| chrX | 57593633 | 57594032 | ZXDB | 0 | + | 57593633 | 57594032 | 0 | 1 | 399 | 0 | -1.1747192 | 0.0001687 | 0.0039029 |
| chr17 | 36950370 | 36953148 | AATF | 0 | + | 36950370 | 36953148 | 0 | 2 | 36,263 | 0,2516 | -1.1944395 | 0.0018416 | 0.0248550 |
| chrX | 1601128 | 1601624 | AKAP17A | 0 | + | 1601128 | 1601624 | 0 | 1 | 496 | 0 | -0.6041205 | 0.0075590 | 0.0692704 |
| chr15 | 44717765 | 44717814 | B2M | 0 | + | 44717765 | 44717814 | 0 | 1 | 49 | 0 | -1.0328905 | 0.0004002 | 0.0078158 |
| chr17 | 38672898 | 38673543 | C17orf96 | 0 | - | 38672898 | 38673543 | 0 | 1 | 645 | 0 | -0.5015039 | 0.0093409 | 0.0800496 |
| chr17 | 38673594 | 38673792 | C17orf96 | 0 | - | 38673594 | 38673792 | 0 | 1 | 198 | 0 | -1.1452435 | 0.0070860 | 0.0664239 |
| chr17 | 38674537 | 38674934 | C17orf96 | 0 | - | 38674537 | 38674934 | 0 | 1 | 397 | 0 | -0.8723555 | 0.0035045 | 0.0400380 |
| chr21 | 44145282 | 44145527 | C21orf33 | 0 | + | 44145282 | 44145527 | 0 | 1 | 245 | 0 | -1.0946675 | 0.0015744 | 0.0221169 |
| chr14 | 94060211 | 94060410 | DDX24 | 0 | - | 94060211 | 94060410 | 0 | 1 | 199 | 0 | -1.5041326 | 0.0000072 | 0.0003047 |
| chr14 | 94062390 | 94062589 | DDX24 | 0 | - | 94062390 | 94062589 | 0 | 1 | 199 | 0 | -1.0117202 | 0.0032982 | 0.0381866 |
| chr14 | 94079492 | 94081212 | DDX24 | 0 | - | 94079492 | 94081212 | 0 | 2 | 256,94 | 0,1627 | -0.6311563 | 0.0053988 | 0.0549841 |
| chr10 | 77821512 | 77821910 | DLG5 | 0 | - | 77821512 | 77821910 | 0 | 1 | 398 | 0 | -1.2772959 | 0.0055197 | 0.0558693 |
| chr7 | 76482525 | 76483019 | DTX2 | 0 | + | 76482525 | 76483019 | 0 | 1 | 494 | 0 | -0.8160760 | 0.0011896 | 0.0181026 |
| chr8 | 143725782 | 143726331 | FAM83H | 0 | - | 143725782 | 143726331 | 0 | 1 | 549 | 0 | -0.8995750 | 0.0000384 | 0.0012190 |
| chr8 | 143726981 | 143727530 | FAM83H | 0 | - | 143726981 | 143727530 | 0 | 1 | 549 | 0 | -2.0689506 | 0.0000000 | 0.0000000 |
| chr8 | 143727681 | 143728080 | FAM83H | 0 | - | 143727681 | 143728080 | 0 | 1 | 399 | 0 | -1.7140702 | 0.0000000 | 0.0000001 |
| chr8 | 143728131 | 143728230 | FAM83H | 0 | - | 143728131 | 143728230 | 0 | 1 | 99 | 0 | -1.4513627 | 0.0004525 | 0.0086019 |
| chr21 | 33524767 | 33528181 | GART | 0 | - | 33524767 | 33528181 | 0 | 2 | 234,15 | 0,3400 | 0.9719061 | 0.0064851 | 0.0628276 |
| chr1 | 155234498 | 155235093 | GBA | 0 | - | 155234498 | 155235093 | 0 | 1 | 595 | 0 | -0.6798723 | 0.0101582 | 0.0851172 |
| chr1 | 155213875 | 155214470 | GBAP1 | 0 | - | 155213875 | 155214470 | 0 | 1 | 595 | 0 | -0.7344676 | 0.0043786 | 0.0472594 |
| chr1 | 149851448 | 149851594 | HIST2H2AA3 | 0 | + | 149851448 | 149851594 | 0 | 1 | 146 | 0 | 1.9770504 | 0.0000106 | 0.0004170 |
| chr1 | 149851448 | 149851594 | HIST2H2AA4 | 0 | + | 149851448 | 149851594 | 0 | 1 | 146 | 0 | 1.9770504 | 0.0000106 | 0.0004170 |
| chr17 | 46171429 | 46171478 | KANSL1 | 0 | - | 46171429 | 46171478 | 0 | 1 | 49 | 0 | -4.1525590 | 0.0035658 | 0.0405446 |
| chr19 | 34341619 | 34342018 | KIAA0355 | 0 | + | 34341619 | 34342018 | 0 | 1 | 399 | 0 | -0.9780257 | 0.0081031 | 0.0726408 |
| chr14 | 106494250 | 106494494 | LINC00221 | 0 | + | 106494250 | 106494494 | 0 | 1 | 244 | 0 | -0.9467427 | 0.0023578 | 0.0298776 |
| chr17 | 36501096 | 36501194 | MYO19 | 0 | - | 36501096 | 36501194 | 0 | 1 | 98 | 0 | -2.2733014 | 0.0032626 | 0.0379200 |
| chr16 | 4461726 | 4463770 | NMRAL1 | 0 | - | 4461726 | 4463770 | 0 | 2 | 234,111 | 0,1934 | -0.9419719 | 0.0000288 | 0.0009560 |
| chr10 | 47923375 | 47923524 | NPY4R | 0 | + | 47923375 | 47923524 | 0 | 1 | 149 | 0 | -1.9314252 | 0.0000326 | 0.0010651 |
| chr8 | 143816442 | 143816786 | PUF60 | 0 | - | 143816442 | 143816786 | 0 | 1 | 344 | 0 | -0.6281409 | 0.0012330 | 0.0185819 |
| chr17 | 35809505 | 35817743 | TAF15 | 0 | + | 35809505 | 35817743 | 0 | 2 | 72,28 | 0,8211 | 0.9265505 | 0.0102730 | 0.0858039 |
| chr2 | 3387764 | 3387962 | TRAPPC12 | 0 | + | 3387764 | 3387962 | 0 | 1 | 198 | 0 | -1.2161058 | 0.0049743 | 0.0518269 |
| chr17 | 1728311 | 1728410 | WDR81 | 0 | + | 1728311 | 1728410 | 0 | 1 | 99 | 0 | -2.0239300 | 0.0033755 | 0.0388433 |
| chrX | 2486763 | 2487511 | ZBED1 | 0 | - | 2486763 | 2487511 | 0 | 1 | 748 | 0 | -0.5716358 | 0.0003953 | 0.0077476 |
| chrX | 2488459 | 2488907 | ZBED1 | 0 | - | 2488459 | 2488907 | 0 | 1 | 448 | 0 | -1.0215236 | 0.0000000 | 0.0000030 |
** Note: ** Compare to A549 host cell line, there are more differentially methylated sites on the Hela host cells in this data set.
Metagene of differential peaks
peakDistribution(jointPeak[which(jointPeak$fdr<0.1 & abs(jointPeak$logFC)>0.5 ),],"~/Database/genome/hg19/hg19_UCSC.gtf")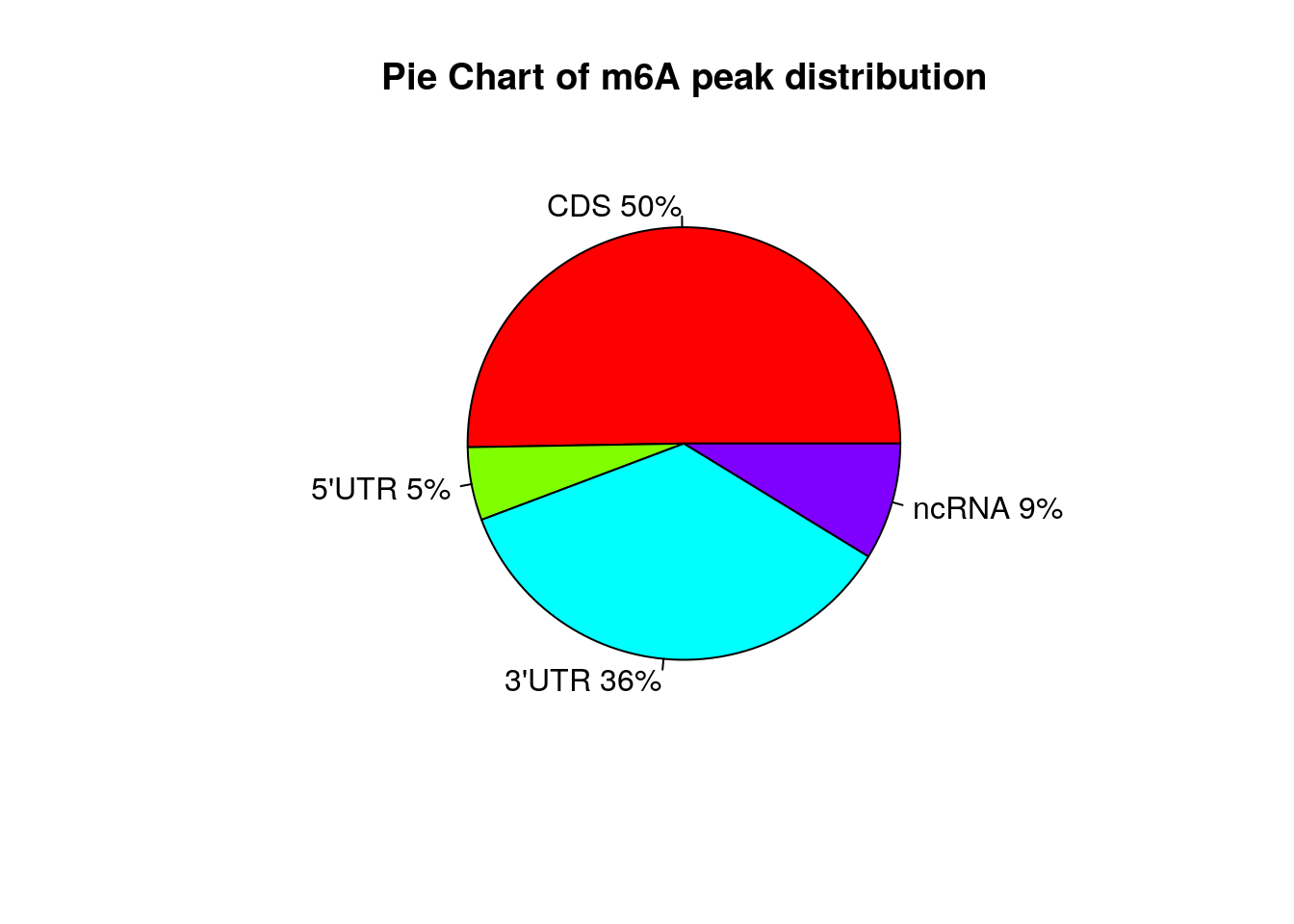
Pathway analysis of the differntial Methylated gene
qnb_res_eg <- bitr(jointPeak[which(jointPeak$fdr<0.1 & abs(jointPeak$logFC)>0.5 ),"name"],fromType = "SYMBOL", toType = "ENTREZID",OrgDb = "org.Hs.eg.db" )
qnb_kegg <- enrichKEGG(qnb_res_eg$ENTREZID,organism = "hsa",pAdjustMethod = "none", pvalueCutoff = 0.05,minGSSize = 3)
emapplot(qnb_kegg )
Differential expression analysis
deseq2_res_hela <- RADAR::DESeq2(RSV.Hela$geneSum, matrix(c("Ctl","Ctl","Infected","Infected"),ncol = 1) )[1] "without pc ..."
Using input matrix without additional normalization...#write.table(deseq2_res_hela[which(deseq2_res_hela$p.adjust<0.05),], file = "~/RSV/2017/Host_diff_expression.xls",sep = "\t", quote = F,row.names = F)
knitr::kable(deseq2_res_hela[order(deseq2_res_hela$p.adjust,decreasing = F)[1:2000],], format = "pandoc", row.names = F)| GeneID | log2FC | p.adjust | pvalue |
|---|---|---|---|
| ARNT2 | 6.14 | 0 | 0 |
| CYR61 | 3.54 | 0 | 0 |
| DDX58 | 7.17 | 0 | 0 |
| GPRC5A | 3.67 | 0 | 0 |
| HPD | -3.55 | 0 | 0 |
| HSPA8 | -3.37 | 0 | 0 |
| ICAM1 | 4.93 | 0 | 0 |
| IFI6 | 7.16 | 0 | 0 |
| IFIT1 | 5.06 | 0 | 0 |
| IFIT2 | 6.31 | 0 | 0 |
| IFITM1 | 3.21 | 0 | 0 |
| ISG15 | 7.41 | 0 | 0 |
| ITGA5 | 4.54 | 0 | 0 |
| NT5E | 4.98 | 0 | 0 |
| OASL | 8.10 | 0 | 0 |
| PLAUR | 4.44 | 0 | 0 |
| SAT1 | 5.22 | 0 | 0 |
| SERPINE2 | 4.94 | 0 | 0 |
| SERPING1 | 5.07 | 0 | 0 |
| SOD2 | 4.12 | 0 | 0 |
| SPOCK1 | 3.87 | 0 | 0 |
| TXNIP | 4.22 | 0 | 0 |
| ZC3HAV1 | 3.57 | 0 | 0 |
| GDF15 | 6.87 | 0 | 0 |
| TIMP1 | 2.86 | 0 | 0 |
| LAMB3 | 3.56 | 0 | 0 |
| H1F0 | 3.34 | 0 | 0 |
| CTSL | 2.79 | 0 | 0 |
| TINAGL1 | 3.78 | 0 | 0 |
| ZNFX1 | 3.09 | 0 | 0 |
| ECM1 | 6.33 | 0 | 0 |
| CCT5 | -2.58 | 0 | 0 |
| SLC7A5 | -2.71 | 0 | 0 |
| SRSF2 | -2.88 | 0 | 0 |
| TRIP13 | -3.80 | 0 | 0 |
| RBM3 | -3.58 | 0 | 0 |
| HERC5 | 4.13 | 0 | 0 |
| DUSP5 | 4.36 | 0 | 0 |
| ODC1 | -3.22 | 0 | 0 |
| MUC16 | 3.56 | 0 | 0 |
| RRM2 | -3.91 | 0 | 0 |
| CD68 | 5.48 | 0 | 0 |
| ISG20 | 3.44 | 0 | 0 |
| C3 | 4.91 | 0 | 0 |
| FOLR1 | 3.37 | 0 | 0 |
| HELZ2 | 4.37 | 0 | 0 |
| ADAMTSL4 | 3.84 | 0 | 0 |
| BIRC5 | -3.97 | 0 | 0 |
| MYBL2 | -3.04 | 0 | 0 |
| OPTN | 3.31 | 0 | 0 |
| TNIP1 | 2.89 | 0 | 0 |
| PABPC4 | -2.98 | 0 | 0 |
| ANXA1 | 3.11 | 0 | 0 |
| DHX9 | -3.22 | 0 | 0 |
| LAMA1 | -2.59 | 0 | 0 |
| CTSB | 2.48 | 0 | 0 |
| EIF4B | -2.64 | 0 | 0 |
| SRSF1 | -3.26 | 0 | 0 |
| TIMP3 | 2.69 | 0 | 0 |
| SLC3A2 | -2.78 | 0 | 0 |
| IL32 | 5.17 | 0 | 0 |
| MFGE8 | 3.94 | 0 | 0 |
| ZNF385A | 3.76 | 0 | 0 |
| ITGA2 | 4.47 | 0 | 0 |
| NME1 | -2.65 | 0 | 0 |
| CEMIP | 5.57 | 0 | 0 |
| SDC4 | 3.08 | 0 | 0 |
| CDC20 | -3.64 | 0 | 0 |
| HLA-A | 3.53 | 0 | 0 |
| TRANK1 | 4.59 | 0 | 0 |
| CD276 | 2.66 | 0 | 0 |
| TEP1 | 3.66 | 0 | 0 |
| HSP90AA1 | -2.39 | 0 | 0 |
| SRSF3 | -2.90 | 0 | 0 |
| BCL6 | 3.26 | 0 | 0 |
| EIF3CL | -2.96 | 0 | 0 |
| MAFF | 4.74 | 0 | 0 |
| JUN | 4.21 | 0 | 0 |
| TFRC | -2.80 | 0 | 0 |
| IER5 | 3.06 | 0 | 0 |
| CCNB1 | -2.94 | 0 | 0 |
| ALYREF | -3.31 | 0 | 0 |
| EXO1 | -3.92 | 0 | 0 |
| GRN | 2.78 | 0 | 0 |
| GRB10 | 3.01 | 0 | 0 |
| EIF3C | -2.92 | 0 | 0 |
| DDX39A | -3.32 | 0 | 0 |
| DHX15 | -2.86 | 0 | 0 |
| SRSF7 | -3.25 | 0 | 0 |
| CD44 | 2.53 | 0 | 0 |
| APOL6 | 3.57 | 0 | 0 |
| DDX60 | 5.13 | 0 | 0 |
| TRAP1 | -2.59 | 0 | 0 |
| ATF3 | 2.83 | 0 | 0 |
| TMEM132A | 3.16 | 0 | 0 |
| EIF3A | -2.34 | 0 | 0 |
| UBE2H | 2.64 | 0 | 0 |
| SMC4 | -3.54 | 0 | 0 |
| GADD45A | 3.28 | 0 | 0 |
| EIF5A | -2.10 | 0 | 0 |
| MCM7 | -2.36 | 0 | 0 |
| C1S | 2.83 | 0 | 0 |
| KIF20A | -3.18 | 0 | 0 |
| TUBA1B | -1.94 | 0 | 0 |
| MYADM | 2.74 | 0 | 0 |
| PCDH1 | 5.15 | 0 | 0 |
| GSN | 3.06 | 0 | 0 |
| CSAG3 | 5.86 | 0 | 0 |
| MCM4 | -2.42 | 0 | 0 |
| RRM1 | -2.69 | 0 | 0 |
| FOSL1 | 4.49 | 0 | 0 |
| LGALS3BP | 2.29 | 0 | 0 |
| CCL5 | 11.76 | 0 | 0 |
| OSMR | 2.49 | 0 | 0 |
| ITGA3 | 2.94 | 0 | 0 |
| CASP4 | 4.18 | 0 | 0 |
| LAMB2 | 2.87 | 0 | 0 |
| IPO4 | -3.30 | 0 | 0 |
| SSRP1 | -2.20 | 0 | 0 |
| C1RL | 3.26 | 0 | 0 |
| PODXL | 2.39 | 0 | 0 |
| LDLR | 2.24 | 0 | 0 |
| PML | 3.41 | 0 | 0 |
| PHLDA1 | 4.10 | 0 | 0 |
| PLK1 | -3.58 | 0 | 0 |
| TRA2B | -2.53 | 0 | 0 |
| EIF3L | -2.40 | 0 | 0 |
| MCM5 | -2.53 | 0 | 0 |
| CSE1L | -2.60 | 0 | 0 |
| YBX1 | -2.09 | 0 | 0 |
| ACTL8 | -3.95 | 0 | 0 |
| NCL | -1.90 | 0 | 0 |
| ATP5F1 | -2.52 | 0 | 0 |
| ATP2B4 | 2.57 | 0 | 0 |
| B2M | 2.53 | 0 | 0 |
| MKI67 | -2.94 | 0 | 0 |
| CENPN | -2.79 | 0 | 0 |
| GNS | 2.31 | 0 | 0 |
| PDLIM7 | 2.67 | 0 | 0 |
| MCM2 | -2.50 | 0 | 0 |
| OAS3 | 2.45 | 0 | 0 |
| TOP2A | -2.67 | 0 | 0 |
| IPO5 | -2.32 | 0 | 0 |
| OXCT1 | -2.60 | 0 | 0 |
| PBXIP1 | 2.99 | 0 | 0 |
| SQRDL | 3.29 | 0 | 0 |
| CCNA2 | -3.45 | 0 | 0 |
| S100A6 | 2.20 | 0 | 0 |
| ZSWIM4 | 3.64 | 0 | 0 |
| TMEM173 | 3.39 | 0 | 0 |
| TNFRSF21 | 3.64 | 0 | 0 |
| EIF3B | -2.05 | 0 | 0 |
| AHNAK2 | 2.18 | 0 | 0 |
| RBMX | -2.67 | 0 | 0 |
| TGM2 | 6.50 | 0 | 0 |
| MGLL | 2.67 | 0 | 0 |
| IRF1 | 3.30 | 0 | 0 |
| TNFAIP2 | 2.34 | 0 | 0 |
| COL17A1 | 6.59 | 0 | 0 |
| CCT3 | -1.86 | 0 | 0 |
| MCM3 | -2.14 | 0 | 0 |
| ASPM | -3.97 | 0 | 0 |
| SLPI | 2.56 | 0 | 0 |
| ADSL | -2.66 | 0 | 0 |
| MT2A | 2.50 | 0 | 0 |
| QSOX1 | 2.48 | 0 | 0 |
| TIMP2 | 2.01 | 0 | 0 |
| ANLN | -2.70 | 0 | 0 |
| BHLHE40 | 2.39 | 0 | 0 |
| CALCOCO1 | 2.55 | 0 | 0 |
| EPHA2 | 2.69 | 0 | 0 |
| CDKN1A | 4.97 | 0 | 0 |
| U2AF2 | -2.18 | 0 | 0 |
| HSD17B10 | -2.37 | 0 | 0 |
| EPRS | -2.16 | 0 | 0 |
| PARK7 | -2.42 | 0 | 0 |
| ETHE1 | 2.76 | 0 | 0 |
| BUB1B | -4.04 | 0 | 0 |
| OAS1 | 2.33 | 0 | 0 |
| LMO7 | 2.06 | 0 | 0 |
| PABPC1 | -1.72 | 0 | 0 |
| CTGF | 3.68 | 0 | 0 |
| PRC1 | -2.40 | 0 | 0 |
| IL7R | 9.06 | 0 | 0 |
| FOXM1 | -2.92 | 0 | 0 |
| STIP1 | -2.08 | 0 | 0 |
| PHF21A | 2.81 | 0 | 0 |
| SAMD9 | 4.40 | 0 | 0 |
| SERBP1 | -1.71 | 0 | 0 |
| ADM | 3.15 | 0 | 0 |
| TUBB4B | -1.85 | 0 | 0 |
| NPM1 | -2.17 | 0 | 0 |
| ZWINT | -3.40 | 0 | 0 |
| GRB7 | 4.33 | 0 | 0 |
| BCAR3 | 2.09 | 0 | 0 |
| IFIH1 | 4.15 | 0 | 0 |
| RAN | -2.08 | 0 | 0 |
| CDC45 | -4.49 | 0 | 0 |
| PIK3IP1 | 5.44 | 0 | 0 |
| CPS1 | -1.74 | 0 | 0 |
| XPO1 | -2.45 | 0 | 0 |
| RPL4 | -1.73 | 0 | 0 |
| IGSF8 | 2.45 | 0 | 0 |
| SLC41A1 | 2.32 | 0 | 0 |
| HNRNPM | -2.04 | 0 | 0 |
| DDX46 | -2.59 | 0 | 0 |
| AHNAK | 1.89 | 0 | 0 |
| SERPINE1 | 5.76 | 0 | 0 |
| ACSS2 | 3.07 | 0 | 0 |
| CUL4A | -2.03 | 0 | 0 |
| SRGN | 2.15 | 0 | 0 |
| PHB2 | -1.93 | 0 | 0 |
| ASS1 | -1.84 | 0 | 0 |
| MXD1 | 4.13 | 0 | 0 |
| LRG1 | 3.29 | 0 | 0 |
| AIMP2 | -2.89 | 0 | 0 |
| GFPT2 | 3.44 | 0 | 0 |
| ENO1 | -1.61 | 0 | 0 |
| ZFP36L1 | 2.55 | 0 | 0 |
| NQO1 | -2.08 | 0 | 0 |
| PRSS23 | 1.71 | 0 | 0 |
| SLC25A10 | -2.65 | 0 | 0 |
| IFI16 | 3.51 | 0 | 0 |
| ITGB8 | 3.65 | 0 | 0 |
| NCF2 | 9.14 | 0 | 0 |
| XRCC6 | -1.68 | 0 | 0 |
| NT5DC2 | -3.06 | 0 | 0 |
| FAM83D | -2.81 | 0 | 0 |
| PES1 | -2.21 | 0 | 0 |
| TPP1 | 2.50 | 0 | 0 |
| OLR1 | 2.19 | 0 | 0 |
| NCBP1 | -2.97 | 0 | 0 |
| CCT6A | -1.84 | 0 | 0 |
| CCT2 | -2.00 | 0 | 0 |
| NOP58 | -2.95 | 0 | 0 |
| RAB11FIP5 | 2.46 | 0 | 0 |
| TUBA1C | -1.64 | 0 | 0 |
| ORC1 | -3.72 | 0 | 0 |
| KHNYN | 2.12 | 0 | 0 |
| LGMN | 2.22 | 0 | 0 |
| TMBIM1 | 2.13 | 0 | 0 |
| MARC1 | -2.27 | 0 | 0 |
| LYPD3 | 2.24 | 0 | 0 |
| TOMM40 | -2.08 | 0 | 0 |
| TNFAIP3 | 2.60 | 0 | 0 |
| TACC3 | -2.69 | 0 | 0 |
| CEBPB | 2.24 | 0 | 0 |
| WBP2 | 2.06 | 0 | 0 |
| G6PD | -2.39 | 0 | 0 |
| PRR13 | 1.81 | 0 | 0 |
| EFNB1 | 2.83 | 0 | 0 |
| SRSF6 | -2.18 | 0 | 0 |
| NPTX1 | -1.76 | 0 | 0 |
| PRMT1 | -2.37 | 0 | 0 |
| CYP2S1 | 3.47 | 0 | 0 |
| ETV5 | 3.01 | 0 | 0 |
| RPL5 | -1.70 | 0 | 0 |
| LRP10 | 2.52 | 0 | 0 |
| CENPF | -2.53 | 0 | 0 |
| CLU | 1.89 | 0 | 0 |
| TCP1 | -1.88 | 0 | 0 |
| FKBP10 | 1.88 | 0 | 0 |
| NOP16 | -2.53 | 0 | 0 |
| ATAD2 | -2.70 | 0 | 0 |
| PSMD3 | -1.99 | 0 | 0 |
| CDKN2B | 3.00 | 0 | 0 |
| SPAG5 | -2.30 | 0 | 0 |
| KIF18B | -3.55 | 0 | 0 |
| NOP56 | -2.09 | 0 | 0 |
| SHCBP1 | -3.70 | 0 | 0 |
| PHGDH | -1.94 | 0 | 0 |
| NCAPG | -3.76 | 0 | 0 |
| CDC6 | -2.47 | 0 | 0 |
| SNRNP25 | -3.25 | 0 | 0 |
| RPA1 | -2.02 | 0 | 0 |
| TRIM16L | -2.63 | 0 | 0 |
| KLHL13 | -2.52 | 0 | 0 |
| KIF2C | -2.17 | 0 | 0 |
| DNAJC11 | -2.89 | 0 | 0 |
| IFI44 | 8.46 | 0 | 0 |
| RETSAT | 2.03 | 0 | 0 |
| WDR77 | -2.13 | 0 | 0 |
| BMP6 | 1.93 | 0 | 0 |
| FUS | -1.73 | 0 | 0 |
| BOP1 | -2.41 | 0 | 0 |
| SFPQ | -1.75 | 0 | 0 |
| NFKBIA | 2.24 | 0 | 0 |
| NUP88 | -2.47 | 0 | 0 |
| PLEKHA6 | 4.36 | 0 | 0 |
| LDHB | -1.66 | 0 | 0 |
| FBL | -1.92 | 0 | 0 |
| TDRD7 | 4.57 | 0 | 0 |
| CPM | 2.34 | 0 | 0 |
| HNRNPA2B1 | -1.69 | 0 | 0 |
| ILF3 | -1.75 | 0 | 0 |
| HLA-B | 4.26 | 0 | 0 |
| DAZAP1 | -1.81 | 0 | 0 |
| C6orf223 | 2.94 | 0 | 0 |
| SERINC2 | 3.22 | 0 | 0 |
| KRT17 | 1.73 | 0 | 0 |
| PFAS | -2.97 | 0 | 0 |
| SUPT16H | -2.14 | 0 | 0 |
| FBXO32 | 2.58 | 0 | 0 |
| JUNB | 2.10 | 0 | 0 |
| NPR3 | -2.35 | 0 | 0 |
| DHX58 | 7.97 | 0 | 0 |
| IRAK2 | 6.30 | 0 | 0 |
| GINS2 | -3.21 | 0 | 0 |
| GDI2 | -1.96 | 0 | 0 |
| RUVBL2 | -1.86 | 0 | 0 |
| PRSS56 | -1.89 | 0 | 0 |
| HSPD1 | -1.87 | 0 | 0 |
| TROAP | -2.37 | 0 | 0 |
| NDRG1 | 1.65 | 0 | 0 |
| IFIT3 | 6.48 | 0 | 0 |
| BAG1 | -1.99 | 0 | 0 |
| FAM83A | 3.74 | 0 | 0 |
| XRCC5 | -1.70 | 0 | 0 |
| GTPBP1 | 1.99 | 0 | 0 |
| DANCR | -3.72 | 0 | 0 |
| DHFR | -3.25 | 0 | 0 |
| SAPCD2 | -2.44 | 0 | 0 |
| PA2G4 | -1.64 | 0 | 0 |
| S100A10 | 1.63 | 0 | 0 |
| DDB1 | -1.57 | 0 | 0 |
| TBC1D2 | 2.53 | 0 | 0 |
| HNRNPDL | -2.02 | 0 | 0 |
| TLDC1 | 2.77 | 0 | 0 |
| UHRF1 | -2.74 | 0 | 0 |
| GTSE1 | -3.33 | 0 | 0 |
| POLD2 | -1.87 | 0 | 0 |
| C1R | 3.31 | 0 | 0 |
| SLC12A4 | 2.36 | 0 | 0 |
| MRPL51 | -2.15 | 0 | 0 |
| TK1 | -2.24 | 0 | 0 |
| ID3 | -2.38 | 0 | 0 |
| MRPS34 | -1.90 | 0 | 0 |
| BAZ2A | 1.71 | 0 | 0 |
| TNFSF9 | 1.99 | 0 | 0 |
| SNHG16 | -2.06 | 0 | 0 |
| ELF3 | 1.91 | 0 | 0 |
| MAZ | -2.14 | 0 | 0 |
| RUVBL1 | -2.25 | 0 | 0 |
| ALDOC | 2.70 | 0 | 0 |
| MRPL37 | -1.99 | 0 | 0 |
| S100A9 | 7.73 | 0 | 0 |
| SLFN5 | 2.87 | 0 | 0 |
| CSDE1 | -1.75 | 0 | 0 |
| HSP90AB1 | -1.41 | 0 | 0 |
| ZNF646 | 2.43 | 0 | 0 |
| KPNA2 | -1.62 | 0 | 0 |
| PI4K2A | 2.05 | 0 | 0 |
| MSLN | 2.13 | 0 | 0 |
| TNS4 | 2.23 | 0 | 0 |
| KIF11 | -2.66 | 0 | 0 |
| PSAP | 1.49 | 0 | 0 |
| KPNB1 | -1.54 | 0 | 0 |
| GBP3 | 6.71 | 0 | 0 |
| RAB43 | 2.88 | 0 | 0 |
| MTCH2 | -1.98 | 0 | 0 |
| SEMA4B | 2.34 | 0 | 0 |
| IARS | -1.65 | 0 | 0 |
| PLSCR1 | 2.75 | 0 | 0 |
| ETV4 | 2.91 | 0 | 0 |
| FARSB | -2.89 | 0 | 0 |
| RELB | 3.31 | 0 | 0 |
| POLD4 | 2.89 | 0 | 0 |
| DNHD1 | 2.85 | 0 | 0 |
| MCM10 | -3.01 | 0 | 0 |
| MRPL24 | -2.32 | 0 | 0 |
| PRDX3 | -1.84 | 0 | 0 |
| SLC25A13 | -3.00 | 0 | 0 |
| FGF2 | 4.13 | 0 | 0 |
| SRRT | -2.20 | 0 | 0 |
| H2AFZ | -2.07 | 0 | 0 |
| NUSAP1 | -2.38 | 0 | 0 |
| NCAPH | -2.97 | 0 | 0 |
| SGK1 | -1.71 | 0 | 0 |
| NONO | -1.57 | 0 | 0 |
| HEG1 | 2.00 | 0 | 0 |
| TUBA1A | -1.52 | 0 | 0 |
| H2AFJ | 2.40 | 0 | 0 |
| GRWD1 | -2.26 | 0 | 0 |
| APEX1 | -1.73 | 0 | 0 |
| HLA-C | 3.32 | 0 | 0 |
| C1QTNF1 | 4.37 | 0 | 0 |
| TRAF1 | 4.53 | 0 | 0 |
| WARS | 1.51 | 0 | 0 |
| RACGAP1 | -1.97 | 0 | 0 |
| NOC2L | -1.93 | 0 | 0 |
| TMC6 | 2.19 | 0 | 0 |
| C1QBP | -1.78 | 0 | 0 |
| SORBS2 | -2.28 | 0 | 0 |
| SELM | 2.08 | 0 | 0 |
| ANKRD33B | 2.16 | 0 | 0 |
| CARS2 | -2.44 | 0 | 0 |
| CCT7 | -1.46 | 0 | 0 |
| NEFH | -1.87 | 0 | 0 |
| CEP55 | -2.45 | 0 | 0 |
| UTP20 | -2.96 | 0 | 0 |
| HCP5 | 3.93 | 0 | 0 |
| RRBP1 | 2.07 | 0 | 0 |
| TUFM | -1.65 | 0 | 0 |
| ALAS1 | 1.78 | 0 | 0 |
| THSD4 | 3.44 | 0 | 0 |
| TRIM16 | -2.03 | 0 | 0 |
| DTYMK | -2.85 | 0 | 0 |
| TIMM13 | -1.97 | 0 | 0 |
| ETF1 | -1.66 | 0 | 0 |
| MRPL12 | -1.82 | 0 | 0 |
| LINC00473 | -3.32 | 0 | 0 |
| CDCA3 | -3.27 | 0 | 0 |
| PNRC1 | 2.64 | 0 | 0 |
| NUP188 | -1.87 | 0 | 0 |
| ARMC6 | -2.71 | 0 | 0 |
| SECTM1 | 3.38 | 0 | 0 |
| B4GALT5 | 1.92 | 0 | 0 |
| TP53INP2 | 3.73 | 0 | 0 |
| PTPRH | 3.48 | 0 | 0 |
| CDK1 | -2.46 | 0 | 0 |
| FXYD5 | 2.03 | 0 | 0 |
| C8orf33 | -1.79 | 0 | 0 |
| QPRT | 1.84 | 0 | 0 |
| CD63 | 1.51 | 0 | 0 |
| GRINA | 1.77 | 0 | 0 |
| PSMC5 | -2.04 | 0 | 0 |
| ENOSF1 | -2.74 | 0 | 0 |
| PCNA | -1.96 | 0 | 0 |
| OTUD1 | 3.57 | 0 | 0 |
| FAM120A | -1.79 | 0 | 0 |
| HSPA4 | -1.63 | 0 | 0 |
| DLGAP5 | -3.22 | 0 | 0 |
| DUSP4 | 2.53 | 0 | 0 |
| HNRNPUL1 | -1.63 | 0 | 0 |
| MAPKAP1 | -2.27 | 0 | 0 |
| HYAL1 | 1.78 | 0 | 0 |
| PGK1 | -1.36 | 0 | 0 |
| KIF23 | -2.51 | 0 | 0 |
| PHLDA3 | 2.14 | 0 | 0 |
| LRPPRC | -1.82 | 0 | 0 |
| RPL6 | -1.59 | 0 | 0 |
| ATP5B | -1.43 | 0 | 0 |
| SH3TC2 | 4.41 | 0 | 0 |
| NOV | 3.09 | 0 | 0 |
| NDC1 | -2.52 | 0 | 0 |
| STAT2 | 2.62 | 0 | 0 |
| RANBP1 | -1.91 | 0 | 0 |
| DDX49 | -2.20 | 0 | 0 |
| IFRD2 | -2.21 | 0 | 0 |
| SLC38A1 | -1.72 | 0 | 0 |
| MCM6 | -2.01 | 0 | 0 |
| NELFCD | -1.85 | 0 | 0 |
| NDUFA9 | -2.10 | 0 | 0 |
| MALAT1 | 1.79 | 0 | 0 |
| SEPHS1 | -2.63 | 0 | 0 |
| SH3BGRL3 | 1.92 | 0 | 0 |
| MAST4 | 3.39 | 0 | 0 |
| YPEL3 | 4.31 | 0 | 0 |
| TKT | -1.42 | 0 | 0 |
| SDR16C5 | 2.82 | 0 | 0 |
| RPSA | -1.55 | 0 | 0 |
| OPA1 | -2.30 | 0 | 0 |
| GABARAPL1 | 1.42 | 0 | 0 |
| MAEA | -2.78 | 0 | 0 |
| RRAS | 2.14 | 0 | 0 |
| ABCD3 | -2.18 | 0 | 0 |
| ITPR1 | -1.77 | 0 | 0 |
| TIPARP | 2.57 | 0 | 0 |
| MTMR11 | 4.39 | 0 | 0 |
| RAB5B | 1.59 | 0 | 0 |
| PDP1 | 2.54 | 0 | 0 |
| CHAF1A | -2.30 | 0 | 0 |
| EIF5AL1 | -1.92 | 0 | 0 |
| DNAJA3 | -2.08 | 0 | 0 |
| HOTAIRM1 | 2.91 | 0 | 0 |
| C11orf68 | 2.44 | 0 | 0 |
| GJB3 | 3.45 | 0 | 0 |
| SPRY4 | 7.34 | 0 | 0 |
| TYMS | -1.83 | 0 | 0 |
| PBK | -3.16 | 0 | 0 |
| CMSS1 | -2.46 | 0 | 0 |
| DROSHA | -1.97 | 0 | 0 |
| SP6 | 4.08 | 0 | 0 |
| BLM | -4.44 | 0 | 0 |
| HNRNPD | -1.70 | 0 | 0 |
| KIF22 | -2.01 | 0 | 0 |
| ORAI3 | 4.00 | 0 | 0 |
| DNAAF5 | -2.26 | 0 | 0 |
| FZD4 | 3.42 | 0 | 0 |
| STK17A | 2.22 | 0 | 0 |
| CERCAM | 1.88 | 0 | 0 |
| GALNT2 | -1.46 | 0 | 0 |
| FARP1 | -2.26 | 0 | 0 |
| ECT2 | -2.06 | 0 | 0 |
| EIF4G1 | -1.48 | 0 | 0 |
| PLCG2 | 2.93 | 0 | 0 |
| PGAM1 | -1.57 | 0 | 0 |
| BUB1 | -2.07 | 0 | 0 |
| UGCG | 2.51 | 0 | 0 |
| CCAR1 | -2.21 | 0 | 0 |
| KLF10 | 2.05 | 0 | 0 |
| NCSTN | 1.64 | 0 | 0 |
| NCAPD2 | -1.73 | 0 | 0 |
| CKS2 | -1.84 | 0 | 0 |
| RPSAP58 | -1.54 | 0 | 0 |
| NUP160 | -1.96 | 0 | 0 |
| GNL3L | -2.26 | 0 | 0 |
| SEPT2 | -1.78 | 0 | 0 |
| RBMS2 | 1.87 | 0 | 0 |
| GAA | 2.00 | 0 | 0 |
| YWHAE | -1.44 | 0 | 0 |
| ZFP36 | 3.08 | 0 | 0 |
| CITED2 | 1.62 | 0 | 0 |
| PRKDC | -1.64 | 0 | 0 |
| UTP14A | -2.05 | 0 | 0 |
| LINC00707 | 3.21 | 0 | 0 |
| FANCI | -2.04 | 0 | 0 |
| MRPL3 | -2.02 | 0 | 0 |
| HEATR5A | -2.06 | 0 | 0 |
| TNC | 4.67 | 0 | 0 |
| CLPTM1L | -1.58 | 0 | 0 |
| RARRES3 | 6.65 | 0 | 0 |
| CBS | -1.79 | 0 | 0 |
| LONP1 | -1.84 | 0 | 0 |
| TRIM21 | 2.38 | 0 | 0 |
| LMNB1 | -2.21 | 0 | 0 |
| TRIB1 | 2.36 | 0 | 0 |
| AGRN | 1.79 | 0 | 0 |
| SLC25A6 | -1.79 | 0 | 0 |
| PARP10 | 2.51 | 0 | 0 |
| SOX13 | 2.10 | 0 | 0 |
| TFDP1 | -1.86 | 0 | 0 |
| CORO6 | 3.28 | 0 | 0 |
| FEN1 | -1.76 | 0 | 0 |
| SKIV2L2 | -2.42 | 0 | 0 |
| PLIN3 | 1.53 | 0 | 0 |
| YPEL5 | 2.31 | 0 | 0 |
| EBNA1BP2 | -1.58 | 0 | 0 |
| IL6 | 7.09 | 0 | 0 |
| RCC1 | -1.75 | 0 | 0 |
| KIF4A | -3.05 | 0 | 0 |
| ARID3B | 3.15 | 0 | 0 |
| NOP2 | -1.49 | 0 | 0 |
| RHOC | 1.48 | 0 | 0 |
| NCOA5 | -2.70 | 0 | 0 |
| S100A11 | 1.32 | 0 | 0 |
| DYNLL1 | -1.57 | 0 | 0 |
| CST3 | 1.74 | 0 | 0 |
| PEA15 | 1.46 | 0 | 0 |
| CCNB2 | -2.51 | 0 | 0 |
| SERPINB1 | 1.77 | 0 | 0 |
| ARPC4 | -1.60 | 0 | 0 |
| PMPCA | -1.82 | 0 | 0 |
| TIMELESS | -1.92 | 0 | 0 |
| ERBB3 | 2.61 | 0 | 0 |
| NCAPD3 | -2.16 | 0 | 0 |
| PLEKHA4 | 3.45 | 0 | 0 |
| OCEL1 | 2.37 | 0 | 0 |
| NME1-NME2 | -1.31 | 0 | 0 |
| NLN | -2.18 | 0 | 0 |
| IRF9 | 3.10 | 0 | 0 |
| NFE2 | 4.32 | 0 | 0 |
| PRMT7 | -1.94 | 0 | 0 |
| TPX2 | -1.65 | 0 | 0 |
| PIM1 | 2.22 | 0 | 0 |
| LARS | -1.89 | 0 | 0 |
| MRTO4 | -2.16 | 0 | 0 |
| KLF6 | 1.92 | 0 | 0 |
| TAF15 | -2.43 | 0 | 0 |
| MTHFD1 | -1.58 | 0 | 0 |
| NCAPG2 | -2.58 | 0 | 0 |
| NCLN | -2.17 | 0 | 0 |
| NFAT5 | 2.05 | 0 | 0 |
| ZNF217 | 2.26 | 0 | 0 |
| HSPA9 | -1.35 | 0 | 0 |
| CDCA5 | -2.54 | 0 | 0 |
| TOR4A | 2.11 | 0 | 0 |
| ASUN | -2.37 | 0 | 0 |
| ADCY6 | 1.83 | 0 | 0 |
| KDM6B | 2.66 | 0 | 0 |
| SDHA | -1.28 | 0 | 0 |
| LDHA | -1.58 | 0 | 0 |
| CITED4 | 1.81 | 0 | 0 |
| ATP5D | -1.78 | 0 | 0 |
| LSM4 | -1.51 | 0 | 0 |
| TUBB6 | -1.44 | 0 | 0 |
| MCRS1 | -1.85 | 0 | 0 |
| PSME3 | -1.62 | 0 | 0 |
| OAS2 | 9.33 | 0 | 0 |
| PSMD1 | -1.73 | 0 | 0 |
| PGP | -1.59 | 0 | 0 |
| KARS | -1.51 | 0 | 0 |
| FANCD2 | -3.30 | 0 | 0 |
| DMAP1 | -2.38 | 0 | 0 |
| UBC | 1.25 | 0 | 0 |
| CACNB2 | -3.94 | 0 | 0 |
| TRAFD1 | 1.66 | 0 | 0 |
| CYCS | -1.66 | 0 | 0 |
| AMOTL2 | 1.68 | 0 | 0 |
| DHX37 | -2.11 | 0 | 0 |
| MED15 | 1.92 | 0 | 0 |
| INSIG1 | 1.78 | 0 | 0 |
| ABCF2 | -1.67 | 0 | 0 |
| MYLK3 | -2.93 | 0 | 0 |
| PSMB5 | -1.58 | 0 | 0 |
| ACTG1 | -1.19 | 0 | 0 |
| LTBP3 | 2.05 | 0 | 0 |
| TIMM10 | -2.61 | 0 | 0 |
| SYNCRIP | -1.82 | 0 | 0 |
| MLST8 | -1.78 | 0 | 0 |
| PPP2R5C | -2.36 | 0 | 0 |
| DIO2 | -3.79 | 0 | 0 |
| VAT1 | 1.48 | 0 | 0 |
| MR1 | 3.09 | 0 | 0 |
| RNF44 | 2.02 | 0 | 0 |
| NEK2 | -2.61 | 0 | 0 |
| PTTG1 | -1.76 | 0 | 0 |
| PLK3 | 3.43 | 0 | 0 |
| TGFB1 | 2.57 | 0 | 0 |
| AURKA | -1.79 | 0 | 0 |
| MSMO1 | 2.21 | 0 | 0 |
| KCNK1 | 1.97 | 0 | 0 |
| TALDO1 | -1.56 | 0 | 0 |
| CLIP2 | 3.00 | 0 | 0 |
| RPL10A | -1.28 | 0 | 0 |
| NRD1 | -1.59 | 0 | 0 |
| TRUB2 | -1.95 | 0 | 0 |
| WDR3 | -1.82 | 0 | 0 |
| CDCA8 | -2.39 | 0 | 0 |
| HDAC5 | 1.91 | 0 | 0 |
| AFG3L2 | -1.82 | 0 | 0 |
| HNRNPR | -1.50 | 0 | 0 |
| MRRF | -2.67 | 0 | 0 |
| NOLC1 | -1.32 | 0 | 0 |
| NEDD9 | 1.81 | 0 | 0 |
| SLC38A2 | 1.61 | 0 | 0 |
| RPL23 | -1.60 | 0 | 0 |
| CYC1 | -1.50 | 0 | 0 |
| VCP | -1.28 | 0 | 0 |
| PTGER4 | 3.68 | 0 | 0 |
| PRSS8 | 5.92 | 0 | 0 |
| TIMM50 | -1.73 | 0 | 0 |
| COTL1 | -1.44 | 0 | 0 |
| IDE | -2.38 | 0 | 0 |
| HNRNPAB | -1.37 | 0 | 0 |
| DKC1 | -1.54 | 0 | 0 |
| ANKH | -1.71 | 0 | 0 |
| ARSA | 2.90 | 0 | 0 |
| PHLDB2 | 1.53 | 0 | 0 |
| NPC1 | 2.01 | 0 | 0 |
| ADGRD1 | 2.00 | 0 | 0 |
| BTG2 | 2.06 | 0 | 0 |
| NATD1 | 2.83 | 0 | 0 |
| CLEC2D | -1.94 | 0 | 0 |
| GAB2 | 2.46 | 0 | 0 |
| NDC80 | -3.27 | 0 | 0 |
| C15orf39 | 1.95 | 0 | 0 |
| ZMYM6NB | 4.08 | 0 | 0 |
| RPLP0 | -1.19 | 0 | 0 |
| IFI27 | 9.02 | 0 | 0 |
| CDC123 | -1.77 | 0 | 0 |
| SAMM50 | -2.03 | 0 | 0 |
| DEPDC1 | -2.88 | 0 | 0 |
| TPBG | 2.44 | 0 | 0 |
| POLE | -1.84 | 0 | 0 |
| SF3B1 | -1.50 | 0 | 0 |
| NUPR1 | 1.82 | 0 | 0 |
| PRR11 | -2.67 | 0 | 0 |
| NEAT1 | 1.50 | 0 | 0 |
| RRM2B | 2.30 | 0 | 0 |
| RPS2 | -1.29 | 0 | 0 |
| RAB8A | -1.80 | 0 | 0 |
| SKA3 | -3.84 | 0 | 0 |
| LIG1 | -2.25 | 0 | 0 |
| BTN3A1 | 2.33 | 0 | 0 |
| SRSF10 | -2.34 | 0 | 0 |
| NARS | -1.65 | 0 | 0 |
| RRP1B | -1.90 | 0 | 0 |
| MUC13 | 4.61 | 0 | 0 |
| BCLAF1 | -2.27 | 0 | 0 |
| NFKBIE | 2.67 | 0 | 0 |
| PALM2-AKAP2 | 1.70 | 0 | 0 |
| MBNL3 | -3.57 | 0 | 0 |
| SLC7A5P2 | -2.47 | 0 | 0 |
| SYNPO | 1.63 | 0 | 0 |
| HES4 | 2.28 | 0 | 0 |
| GTF3C2 | -1.52 | 0 | 0 |
| RBM14 | -1.69 | 0 | 0 |
| LAS1L | -1.68 | 0 | 0 |
| SSH2 | 2.07 | 0 | 0 |
| UNC13A | 3.19 | 0 | 0 |
| C7orf50 | -2.19 | 0 | 0 |
| EEF1B2 | -1.52 | 0 | 0 |
| GNL2 | -1.71 | 0 | 0 |
| C1orf43 | -1.29 | 0 | 0 |
| MYEOV | 5.72 | 0 | 0 |
| AKAP2 | 1.72 | 0 | 0 |
| RBBP7 | -1.34 | 0 | 0 |
| PPP4R3B | -2.22 | 0 | 0 |
| SMC1A | -1.51 | 0 | 0 |
| IREB2 | -2.28 | 0 | 0 |
| THBS1 | 2.22 | 0 | 0 |
| STRA13 | -1.74 | 0 | 0 |
| LOC100130476 | 4.16 | 0 | 0 |
| PAICS | -1.67 | 0 | 0 |
| PPIF | -1.51 | 0 | 0 |
| PSMC3 | -1.38 | 0 | 0 |
| SMG7 | 1.51 | 0 | 0 |
| CDK2AP1 | -2.28 | 0 | 0 |
| FAM214B | 2.54 | 0 | 0 |
| RHEBL1 | 4.12 | 0 | 0 |
| AMD1 | -1.68 | 0 | 0 |
| IARS2 | -1.69 | 0 | 0 |
| PARP9 | 1.95 | 0 | 0 |
| ZWILCH | -2.25 | 0 | 0 |
| P4HA2 | 2.04 | 0 | 0 |
| FOSL2 | 1.61 | 0 | 0 |
| HSPE1 | -2.03 | 0 | 0 |
| MRPS27 | -1.59 | 0 | 0 |
| CKAP2L | -2.77 | 0 | 0 |
| PSMB2 | -1.57 | 0 | 0 |
| AEN | 1.51 | 0 | 0 |
| EIF1AX | -1.77 | 0 | 0 |
| RRP7A | -1.52 | 0 | 0 |
| IRF2BPL | 1.96 | 0 | 0 |
| CCT4 | -1.24 | 0 | 0 |
| WBSCR22 | -1.64 | 0 | 0 |
| PLAC8 | 1.49 | 0 | 0 |
| BCL3 | 2.38 | 0 | 0 |
| JHDM1D-AS1 | 3.17 | 0 | 0 |
| FH | -1.58 | 0 | 0 |
| PCSK6 | -2.91 | 0 | 0 |
| GRAMD1A | 1.88 | 0 | 0 |
| PRDM1 | 4.20 | 0 | 0 |
| AKT1S1 | -1.73 | 0 | 0 |
| LAMC2 | 5.94 | 0 | 0 |
| IMPDH2 | -1.38 | 0 | 0 |
| ANAPC7 | -2.00 | 0 | 0 |
| ATP5A1 | -1.33 | 0 | 0 |
| TSC22D4 | 1.91 | 0 | 0 |
| PSMG1 | -1.98 | 0 | 0 |
| RMRP | -1.50 | 0 | 0 |
| CACYBP | -1.68 | 0 | 0 |
| LANCL2 | -3.34 | 0 | 0 |
| HBS1L | -2.17 | 0 | 0 |
| NOL11 | -1.91 | 0 | 0 |
| MTDH | -1.53 | 0 | 0 |
| SPDL1 | -2.22 | 0 | 0 |
| PTGES | 1.60 | 0 | 0 |
| PKMYT1 | -2.22 | 0 | 0 |
| SLC6A6 | 1.78 | 0 | 0 |
| IMPA2 | -1.63 | 0 | 0 |
| KDM7A | 2.96 | 0 | 0 |
| KIF3C | 2.58 | 0 | 0 |
| C19orf33 | 1.67 | 0 | 0 |
| IGFBP3 | 1.90 | 0 | 0 |
| ST3GAL1 | 1.82 | 0 | 0 |
| RBM8A | -1.64 | 0 | 0 |
| COQ2 | -3.21 | 0 | 0 |
| ANG | 4.56 | 0 | 0 |
| RBM39 | -1.64 | 0 | 0 |
| IGFBP7 | 1.65 | 0 | 0 |
| C10orf54 | 2.65 | 0 | 0 |
| PLD3 | 1.59 | 0 | 0 |
| HDGF | -1.14 | 0 | 0 |
| BRCA1 | -2.54 | 0 | 0 |
| UBALD2 | 1.63 | 0 | 0 |
| RASIP1 | -1.57 | 0 | 0 |
| AXL | 1.75 | 0 | 0 |
| KIAA0040 | 1.53 | 0 | 0 |
| NUP205 | -1.60 | 0 | 0 |
| PRPF38A | -2.03 | 0 | 0 |
| HENMT1 | -2.51 | 0 | 0 |
| LIPA | 1.86 | 0 | 0 |
| NCAPH2 | -2.11 | 0 | 0 |
| CLN6 | -2.19 | 0 | 0 |
| HSPBP1 | -2.06 | 0 | 0 |
| MRPS7 | -1.45 | 0 | 0 |
| PAPPA | 2.05 | 0 | 0 |
| POLR2E | -1.44 | 0 | 0 |
| LINC-PINT | 3.54 | 0 | 0 |
| PRPF8 | -1.47 | 0 | 0 |
| POLA1 | -2.19 | 0 | 0 |
| ITPRIPL2 | 1.56 | 0 | 0 |
| PDZK1IP1 | 5.60 | 0 | 0 |
| NOTCH2NL | 1.97 | 0 | 0 |
| HIST1H4E | -3.92 | 0 | 0 |
| TARS | -1.38 | 0 | 0 |
| ERH | -1.75 | 0 | 0 |
| MIR4435-2HG | 2.03 | 0 | 0 |
| HDAC1 | -1.68 | 0 | 0 |
| TRMT61A | -2.68 | 0 | 0 |
| HMGN1 | -1.68 | 0 | 0 |
| SFXN4 | -2.64 | 0 | 0 |
| HJURP | -2.23 | 0 | 0 |
| NFKB2 | 1.56 | 0 | 0 |
| ATP5O | -1.64 | 0 | 0 |
| SND1 | -1.35 | 0 | 0 |
| AP3D1 | -1.50 | 0 | 0 |
| ATP5G1 | -1.68 | 0 | 0 |
| MRPL4 | -1.57 | 0 | 0 |
| PPP1CC | -1.57 | 0 | 0 |
| BMF | 6.39 | 0 | 0 |
| HEATR1 | -1.82 | 0 | 0 |
| USP7 | -1.65 | 0 | 0 |
| CTSD | 1.58 | 0 | 0 |
| TSPAN1 | 5.51 | 0 | 0 |
| ACTB | -1.22 | 0 | 0 |
| DDX60L | 2.77 | 0 | 0 |
| GM2A | 1.85 | 0 | 0 |
| NFE2L1 | 1.39 | 0 | 0 |
| IL4R | 2.31 | 0 | 0 |
| POLD1 | -1.85 | 0 | 0 |
| TTLL12 | -1.61 | 0 | 0 |
| FAM65A | 1.49 | 0 | 0 |
| TXNRD2 | -1.84 | 0 | 0 |
| SLC2A3 | 1.40 | 0 | 0 |
| HMGB1 | -1.71 | 0 | 0 |
| CAPN5 | 5.04 | 0 | 0 |
| ATP1B1 | 1.60 | 0 | 0 |
| PDCD6 | -1.24 | 0 | 0 |
| THOP1 | -1.65 | 0 | 0 |
| ELAC2 | -1.35 | 0 | 0 |
| PDAP1 | -1.38 | 0 | 0 |
| CCDC80 | 2.86 | 0 | 0 |
| PSMA3 | -1.99 | 0 | 0 |
| NUP107 | -2.12 | 0 | 0 |
| RNASEH2A | -1.83 | 0 | 0 |
| HAT1 | -2.55 | 0 | 0 |
| KIAA1191 | 1.43 | 0 | 0 |
| RPS7 | -1.27 | 0 | 0 |
| ABTB1 | 2.47 | 0 | 0 |
| PDHA1 | -1.23 | 0 | 0 |
| GPX1 | -1.28 | 0 | 0 |
| ARL8A | 1.86 | 0 | 0 |
| PIEZO1 | -1.51 | 0 | 0 |
| COL4A2 | 1.33 | 0 | 0 |
| KRT15 | 4.46 | 0 | 0 |
| TYMP | 1.74 | 0 | 0 |
| MAP1S | 1.96 | 0 | 0 |
| GNB4 | -1.85 | 0 | 0 |
| DTX3L | 1.82 | 0 | 0 |
| CHST11 | 2.68 | 0 | 0 |
| MRPL20 | -2.10 | 0 | 0 |
| GTF3A | -1.70 | 0 | 0 |
| CNOT1 | -1.34 | 0 | 0 |
| ISY1-RAB43 | 1.78 | 0 | 0 |
| RPL26L1 | -2.48 | 0 | 0 |
| POLR1A | -2.07 | 0 | 0 |
| NOL10 | -2.51 | 0 | 0 |
| DLD | -1.60 | 0 | 0 |
| TRA2A | -2.18 | 0 | 0 |
| TCOF1 | -1.59 | 0 | 0 |
| OAZ1 | -1.37 | 0 | 0 |
| LMNB2 | -1.38 | 0 | 0 |
| VWA5A | 2.20 | 0 | 0 |
| BTN2A2 | 3.46 | 0 | 0 |
| MVP | 1.65 | 0 | 0 |
| MAP4K4 | 1.26 | 0 | 0 |
| CBX7 | 2.52 | 0 | 0 |
| AK2 | -1.25 | 0 | 0 |
| PTGES3 | -1.29 | 0 | 0 |
| ATAD3A | -2.07 | 0 | 0 |
| MLXIPL | 3.91 | 0 | 0 |
| GPATCH4 | -1.87 | 0 | 0 |
| CASC5 | -3.06 | 0 | 0 |
| CRELD1 | 2.29 | 0 | 0 |
| SLC12A3 | -2.22 | 0 | 0 |
| RSL1D1 | -1.37 | 0 | 0 |
| C11orf86 | -2.28 | 0 | 0 |
| EML2 | 1.53 | 0 | 0 |
| CDKN3 | -3.10 | 0 | 0 |
| BACH1 | 2.13 | 0 | 0 |
| CD9 | 1.34 | 0 | 0 |
| GPRC5B | 7.06 | 0 | 0 |
| WDR18 | -2.19 | 0 | 0 |
| RFC3 | -2.61 | 0 | 0 |
| VEGFA | 1.64 | 0 | 0 |
| CYB5D1 | -2.66 | 0 | 0 |
| ITPRIP | 1.42 | 0 | 0 |
| RELA | 1.39 | 0 | 0 |
| DNAJB5 | 2.52 | 0 | 0 |
| RPL28 | 1.08 | 0 | 0 |
| TOPBP1 | -2.03 | 0 | 0 |
| MPDU1 | -1.80 | 0 | 0 |
| MXI1 | 1.81 | 0 | 0 |
| ITM2B | 1.55 | 0 | 0 |
| CSRNP1 | 2.07 | 0 | 0 |
| ATP5G3 | -1.47 | 0 | 0 |
| POLR1B | -1.71 | 0 | 0 |
| GTPBP2 | 1.59 | 0 | 0 |
| DNMT1 | -1.42 | 0 | 0 |
| CLSPN | -2.52 | 0 | 0 |
| OBFC1 | 2.41 | 0 | 0 |
| PSMA7 | -1.30 | 0 | 0 |
| PCBP1 | -1.22 | 0 | 0 |
| PRELID1 | -1.35 | 0 | 0 |
| SLC35A4 | -1.74 | 0 | 0 |
| SNRNP200 | -1.16 | 0 | 0 |
| APOL1 | 7.15 | 0 | 0 |
| SLF1 | -2.26 | 0 | 0 |
| TIMM44 | -1.66 | 0 | 0 |
| POP1 | -2.24 | 0 | 0 |
| RBM4 | -1.64 | 0 | 0 |
| WDR43 | -1.61 | 0 | 0 |
| POC1A | -2.53 | 0 | 0 |
| JMJD8 | -2.29 | 0 | 0 |
| ANAPC5 | -1.39 | 0 | 0 |
| KIF5B | -1.45 | 0 | 0 |
| CD82 | 2.65 | 0 | 0 |
| DRAM1 | 2.12 | 0 | 0 |
| DDX54 | -1.32 | 0 | 0 |
| CD81 | 1.41 | 0 | 0 |
| PPAP2B | 2.24 | 0 | 0 |
| C19orf66 | 2.44 | 0 | 0 |
| JUP | 1.26 | 0 | 0 |
| UBE2C | -1.82 | 0 | 0 |
| GLP2R | -1.76 | 0 | 0 |
| ELK3 | 2.64 | 0 | 0 |
| PPM1G | -1.24 | 0 | 0 |
| TMEM159 | 2.71 | 0 | 0 |
| FGD6 | 2.08 | 0 | 0 |
| ABL2 | 1.45 | 0 | 0 |
| TCF25 | -1.84 | 0 | 0 |
| SF3B3 | -1.25 | 0 | 0 |
| ATAD3B | -2.21 | 0 | 0 |
| RAB4A | -2.67 | 0 | 0 |
| PSAT1 | -1.81 | 0 | 0 |
| H6PD | 2.15 | 0 | 0 |
| PRMT6 | -2.52 | 0 | 0 |
| PLXNA2 | 1.97 | 0 | 0 |
| BIRC3 | 3.55 | 0 | 0 |
| SCPEP1 | 2.21 | 0 | 0 |
| PEBP1 | -1.20 | 0 | 0 |
| PHB | -1.27 | 0 | 0 |
| PRDX1 | -1.08 | 0 | 0 |
| ECSIT | -1.94 | 0 | 0 |
| TMEM40 | 4.89 | 0 | 0 |
| GFM1 | -1.73 | 0 | 0 |
| RDH10 | 1.86 | 0 | 0 |
| CRTC2 | 1.63 | 0 | 0 |
| SUV39H1 | -2.47 | 0 | 0 |
| LBR | -1.80 | 0 | 0 |
| FOXRED1 | -2.26 | 0 | 0 |
| NSUN2 | -1.15 | 0 | 0 |
| POLR3H | -1.94 | 0 | 0 |
| GLRX | 1.88 | 0 | 0 |
| TIMM21 | -3.45 | 0 | 0 |
| RBM14-RBM4 | -1.68 | 0 | 0 |
| PRNP | 1.56 | 0 | 0 |
| ELAVL1 | -1.41 | 0 | 0 |
| CDC25A | -2.07 | 0 | 0 |
| CENPO | -2.09 | 0 | 0 |
| MED24 | -1.75 | 0 | 0 |
| ACP1 | -1.51 | 0 | 0 |
| SLMO2 | -1.60 | 0 | 0 |
| STARD7 | -1.40 | 0 | 0 |
| COPS3 | -1.59 | 0 | 0 |
| PSMD2 | -1.11 | 0 | 0 |
| APOL2 | 1.51 | 0 | 0 |
| SNRPA | -1.59 | 0 | 0 |
| CCT8 | -1.44 | 0 | 0 |
| SLC4A2 | -1.39 | 0 | 0 |
| PMAIP1 | 3.92 | 0 | 0 |
| POLR1E | -2.23 | 0 | 0 |
| ULK1 | 1.57 | 0 | 0 |
| WLS | 1.21 | 0 | 0 |
| UNG | -1.71 | 0 | 0 |
| PHF1 | 2.93 | 0 | 0 |
| SRM | -1.57 | 0 | 0 |
| SH2B3 | 2.41 | 0 | 0 |
| AMOTL1 | 1.21 | 0 | 0 |
| FAM168B | -1.77 | 0 | 0 |
| HNRNPA0 | -1.23 | 0 | 0 |
| MCL1 | 1.18 | 0 | 0 |
| CD70 | 1.94 | 0 | 0 |
| SIGMAR1 | -1.25 | 0 | 0 |
| TNFRSF12A | 1.45 | 0 | 0 |
| AUP1 | -1.38 | 0 | 0 |
| RARG | 1.61 | 0 | 0 |
| THRAP3 | -1.37 | 0 | 0 |
| TEX19 | 2.82 | 0 | 0 |
| BTN2A1 | 1.92 | 0 | 0 |
| ZFYVE1 | 2.69 | 0 | 0 |
| PPIH | -2.42 | 0 | 0 |
| ETV3 | 2.41 | 0 | 0 |
| PPP1R15A | 1.93 | 0 | 0 |
| HADH | -2.38 | 0 | 0 |
| WDR4 | -2.43 | 0 | 0 |
| BUB3 | -1.33 | 0 | 0 |
| DTL | -1.93 | 0 | 0 |
| WDR1 | -1.19 | 0 | 0 |
| CPT1C | 3.04 | 0 | 0 |
| SNCG | 1.24 | 0 | 0 |
| ZBTB4 | 1.47 | 0 | 0 |
| ZYX | 1.28 | 0 | 0 |
| LINC00482 | 4.52 | 0 | 0 |
| PAQR5 | 2.81 | 0 | 0 |
| MAPKBP1 | 2.17 | 0 | 0 |
| KDM6A | 2.04 | 0 | 0 |
| SHROOM3 | 1.51 | 0 | 0 |
| PTPN11 | -1.56 | 0 | 0 |
| PGAM4 | -1.58 | 0 | 0 |
| WDR62 | -2.26 | 0 | 0 |
| PSMB4 | -1.16 | 0 | 0 |
| STARD10 | 2.49 | 0 | 0 |
| PTGES2 | -1.60 | 0 | 0 |
| MRPS12 | -1.51 | 0 | 0 |
| CFD | 2.01 | 0 | 0 |
| CCNF | -2.12 | 0 | 0 |
| BCL2L1 | 1.44 | 0 | 0 |
| PI3 | 4.98 | 0 | 0 |
| ABCC3 | 2.81 | 0 | 0 |
| PDGFB | 1.96 | 0 | 0 |
| FAM102B | -1.77 | 0 | 0 |
| ZNF687 | 1.60 | 0 | 0 |
| NUF2 | -3.76 | 0 | 0 |
| PVRL2 | 1.56 | 0 | 0 |
| FN1 | 5.63 | 0 | 0 |
| CTDSP2 | 1.27 | 0 | 0 |
| SPEG | 2.36 | 0 | 0 |
| SMAD3 | 1.48 | 0 | 0 |
| PARP1 | -1.16 | 0 | 0 |
| SNPH | 2.82 | 0 | 0 |
| JTB | -1.25 | 0 | 0 |
| PRPF4B | -1.56 | 0 | 0 |
| MSTO2P | -1.74 | 0 | 0 |
| SETD7 | -1.54 | 0 | 0 |
| PUS7 | -1.99 | 0 | 0 |
| SERTAD4-AS1 | 3.09 | 0 | 0 |
| NTN4 | 2.24 | 0 | 0 |
| TMPO | -1.33 | 0 | 0 |
| SRRM2 | -1.23 | 0 | 0 |
| LPCAT4 | 2.28 | 0 | 0 |
| LIF | 3.01 | 0 | 0 |
| BRIX1 | -1.50 | 0 | 0 |
| HNRNPC | -1.05 | 0 | 0 |
| PSMB6 | -1.35 | 0 | 0 |
| CAPG | 1.14 | 0 | 0 |
| DUSP2 | -1.98 | 0 | 0 |
| GNL3 | -1.26 | 0 | 0 |
| MET | -1.28 | 0 | 0 |
| PAPOLA | -1.28 | 0 | 0 |
| KIAA0101 | -2.13 | 0 | 0 |
| NECAP2 | 1.73 | 0 | 0 |
| ZFR | -1.31 | 0 | 0 |
| TRIM28 | -1.25 | 0 | 0 |
| UBE2Z | 1.20 | 0 | 0 |
| DIS3L | -1.98 | 0 | 0 |
| QPCTL | 1.94 | 0 | 0 |
| GAPDH | -1.02 | 0 | 0 |
| AMPD3 | 6.93 | 0 | 0 |
| TOMM22 | -1.53 | 0 | 0 |
| TCERG1 | -1.56 | 0 | 0 |
| GTPBP4 | -1.42 | 0 | 0 |
| NINJ1 | 1.70 | 0 | 0 |
| CDA | 1.75 | 0 | 0 |
| CIRH1A | -1.71 | 0 | 0 |
| PFKP | -1.13 | 0 | 0 |
| GCNT3 | 3.15 | 0 | 0 |
| H2AFX | -1.60 | 0 | 0 |
| HEATR3 | -2.51 | 0 | 0 |
| NUP85 | -1.41 | 0 | 0 |
| TPPP | 2.34 | 0 | 0 |
| ABHD8 | 2.05 | 0 | 0 |
| KIF20B | -3.00 | 0 | 0 |
| PTBP1 | -1.09 | 0 | 0 |
| EMD | -1.46 | 0 | 0 |
| TP53INP1 | 4.17 | 0 | 0 |
| CLUH | -1.39 | 0 | 0 |
| ABCC2 | -1.54 | 0 | 0 |
| TNS2 | 1.78 | 0 | 0 |
| CTSA | 1.20 | 0 | 0 |
| WARS2 | -2.17 | 0 | 0 |
| MTIF2 | -1.85 | 0 | 0 |
| POR | -1.26 | 0 | 0 |
| ALS2CL | 2.07 | 0 | 0 |
| TNFSF10 | 6.16 | 0 | 0 |
| PGAM5 | -1.34 | 0 | 0 |
| F11R | 1.60 | 0 | 0 |
| PSMD13 | -1.47 | 0 | 0 |
| ABCF3 | -1.46 | 0 | 0 |
| TMX2 | -1.54 | 0 | 0 |
| C17orf89 | -2.00 | 0 | 0 |
| MAP3K12 | 3.18 | 0 | 0 |
| CKB | -1.20 | 0 | 0 |
| PCSK9 | 1.31 | 0 | 0 |
| CSPG4 | 1.56 | 0 | 0 |
| SLC25A32 | -2.17 | 0 | 0 |
| SLC50A1 | 1.49 | 0 | 0 |
| NIFK | -1.91 | 0 | 0 |
| RNASE4 | 4.41 | 0 | 0 |
| RUSC2 | 1.60 | 0 | 0 |
| CHORDC1 | -2.13 | 0 | 0 |
| WTAP | 1.56 | 0 | 0 |
| EARS2 | -2.37 | 0 | 0 |
| SCARB2 | 1.44 | 0 | 0 |
| PRKAR1B | -2.34 | 0 | 0 |
| CKS1B | -2.16 | 0 | 0 |
| IMPDH1 | -1.50 | 0 | 0 |
| NOL8 | -1.88 | 0 | 0 |
| ICAM5 | 2.29 | 0 | 0 |
| SNX17 | -1.63 | 0 | 0 |
| RRP9 | -1.70 | 0 | 0 |
| RAB11FIP1 | 1.78 | 0 | 0 |
| ADGRB2 | 2.53 | 0 | 0 |
| KIAA1524 | -2.75 | 0 | 0 |
| CCDC64 | 2.46 | 0 | 0 |
| EIF4H | -1.05 | 0 | 0 |
| MRPL11 | -1.61 | 0 | 0 |
| PMM2 | -1.67 | 0 | 0 |
| TMC8 | 4.67 | 0 | 0 |
| HEXB | 1.30 | 0 | 0 |
| TOMM5 | -1.42 | 0 | 0 |
| NDUFB4 | -1.32 | 0 | 0 |
| DUS3L | -1.97 | 0 | 0 |
| PSMD14 | -1.57 | 0 | 0 |
| VPS9D1-AS1 | -3.07 | 0 | 0 |
| KRT14 | 2.70 | 0 | 0 |
| XPO7 | -1.51 | 0 | 0 |
| NME2 | -1.05 | 0 | 0 |
| SNRPB | -1.26 | 0 | 0 |
| STOM | 1.21 | 0 | 0 |
| KNTC1 | -2.03 | 0 | 0 |
| GINS4 | -2.67 | 0 | 0 |
| DIAPH3 | -2.17 | 0 | 0 |
| SMARCA4 | -1.31 | 0 | 0 |
| TP53I11 | 2.81 | 0 | 0 |
| POLR2F | -1.49 | 0 | 0 |
| TFPI | 3.40 | 0 | 0 |
| ITGB1BP1 | -1.63 | 0 | 0 |
| SLC2A4RG | -1.54 | 0 | 0 |
| HSPB7 | -1.88 | 0 | 0 |
| RGS2 | 1.82 | 0 | 0 |
| RDH11 | 1.33 | 0 | 0 |
| IMP4 | -1.68 | 0 | 0 |
| XRCC3 | -2.56 | 0 | 0 |
| MRFAP1 | -1.13 | 0 | 0 |
| STRAP | -1.20 | 0 | 0 |
| STAT5A | 1.57 | 0 | 0 |
| NDUFC2 | -1.54 | 0 | 0 |
| N4BP1 | 1.88 | 0 | 0 |
| PIEZO2 | -1.49 | 0 | 0 |
| IFIT5 | 3.60 | 0 | 0 |
| ANP32B | -1.18 | 0 | 0 |
| NSA2 | -1.61 | 0 | 0 |
| SEC14L2 | 2.71 | 0 | 0 |
| RAD21 | -1.23 | 0 | 0 |
| TMEM183A | -1.50 | 0 | 0 |
| FRMD6 | 2.61 | 0 | 0 |
| PSMB3 | -1.40 | 0 | 0 |
| EIF4G2 | -1.03 | 0 | 0 |
| NOCT | 2.44 | 0 | 0 |
| FBXO2 | 2.89 | 0 | 0 |
| RBM28 | -2.12 | 0 | 0 |
| PSME1 | 1.40 | 0 | 0 |
| SRSF11 | -1.27 | 0 | 0 |
| ABCE1 | -2.41 | 0 | 0 |
| POLDIP2 | -1.13 | 0 | 0 |
| TXNL4A | -1.71 | 0 | 0 |
| ATG2A | 1.72 | 0 | 0 |
| HGH1 | -1.65 | 0 | 0 |
| EXOSC2 | -1.70 | 0 | 0 |
| LETM1 | -1.59 | 0 | 0 |
| LRP1 | 1.53 | 0 | 0 |
| SEC24D | 1.98 | 0 | 0 |
| NUP155 | -1.36 | 0 | 0 |
| TOE1 | -2.13 | 0 | 0 |
| C15orf52 | 2.30 | 0 | 0 |
| TMEM184B | 1.51 | 0 | 0 |
| HERC6 | 3.05 | 0 | 0 |
| RAD50 | -2.11 | 0 | 0 |
| GART | -1.18 | 0 | 0 |
| RTN2 | 2.02 | 0 | 0 |
| BCS1L | -1.88 | 0 | 0 |
| PSMC2 | -1.31 | 0 | 0 |
| STAG1 | -2.52 | 0 | 0 |
| TELO2 | -1.92 | 0 | 0 |
| TBCD | -2.24 | 0 | 0 |
| USP18 | 2.01 | 0 | 0 |
| SDCCAG3 | -1.60 | 0 | 0 |
| BANF1 | -1.32 | 0 | 0 |
| NQO2 | -1.38 | 0 | 0 |
| DDX56 | -1.36 | 0 | 0 |
| MACF1 | 1.21 | 0 | 0 |
| TNFRSF10B | 1.19 | 0 | 0 |
| ELL2 | 1.27 | 0 | 0 |
| RAD23A | -1.25 | 0 | 0 |
| CENPU | -2.88 | 0 | 0 |
| PDIA5 | 2.03 | 0 | 0 |
| TXN2 | -1.44 | 0 | 0 |
| FANCG | -1.81 | 0 | 0 |
| GBP1 | 5.65 | 0 | 0 |
| IGFBP6 | 8.55 | 0 | 0 |
| HES2 | 2.20 | 0 | 0 |
| OMA1 | -2.88 | 0 | 0 |
| EIF3E | -1.42 | 0 | 0 |
| SARS2 | -2.32 | 0 | 0 |
| HPDL | -1.82 | 0 | 0 |
| PNPT1 | -1.73 | 0 | 0 |
| TNKS1BP1 | 1.40 | 0 | 0 |
| TIMM23 | -1.35 | 0 | 0 |
| NOL12 | -2.00 | 0 | 0 |
| MTO1 | -2.09 | 0 | 0 |
| AHSA1 | -1.23 | 0 | 0 |
| BAG2 | -1.80 | 0 | 0 |
| DNAJC2 | -1.96 | 0 | 0 |
| SLTM | -1.67 | 0 | 0 |
| ZNF581 | 2.02 | 0 | 0 |
| TIMMDC1 | -1.52 | 0 | 0 |
| HSPA14 | -1.48 | 0 | 0 |
| PRPF4 | -1.47 | 0 | 0 |
| CSAG1 | 1.36 | 0 | 0 |
| TBC1D8 | 1.70 | 0 | 0 |
| ANP32A | -1.29 | 0 | 0 |
| CENPA | -2.82 | 0 | 0 |
| MATR3 | -1.14 | 0 | 0 |
| METTL16 | -1.75 | 0 | 0 |
| IL2RG | 7.80 | 0 | 0 |
| HNRNPK | -1.04 | 0 | 0 |
| ASNS | -2.01 | 0 | 0 |
| RICTOR | 1.52 | 0 | 0 |
| LY6K | 1.11 | 0 | 0 |
| NDUFV1 | -1.38 | 0 | 0 |
| HDAC7 | 1.44 | 0 | 0 |
| NAT8L | 1.69 | 0 | 0 |
| POLR2B | -1.46 | 0 | 0 |
| ARSI | 4.90 | 0 | 0 |
| C19orf48 | -1.46 | 0 | 0 |
| PCGF2 | 2.30 | 0 | 0 |
| MORF4L1 | -1.48 | 0 | 0 |
| HTRA1 | -3.02 | 0 | 0 |
| EIF4A1 | -0.94 | 0 | 0 |
| MUC3A | 6.12 | 0 | 0 |
| TBKBP1 | 3.83 | 0 | 0 |
| NDUFS6 | -1.16 | 0 | 0 |
| STK26 | -1.41 | 0 | 0 |
| COL1A1 | -1.71 | 0 | 0 |
| TONSL | -1.77 | 0 | 0 |
| HAUS7 | -2.27 | 0 | 0 |
| RFC5 | -1.92 | 0 | 0 |
| MDM2 | 1.60 | 0 | 0 |
| TMSB10 | 0.94 | 0 | 0 |
| SLC25A3 | -1.04 | 0 | 0 |
| PEG10 | -1.49 | 0 | 0 |
| FAM35A | -2.34 | 0 | 0 |
| E2F1 | -1.73 | 0 | 0 |
| MOGS | 1.32 | 0 | 0 |
| ELF4 | 1.35 | 0 | 0 |
| HNRNPA3 | -1.58 | 0 | 0 |
| DCAF6 | -1.55 | 0 | 0 |
| TMEM59 | 1.39 | 0 | 0 |
| SMTN | 1.60 | 0 | 0 |
| RNF19B | 1.79 | 0 | 0 |
| U2SURP | -1.38 | 0 | 0 |
| ITPR3 | 1.15 | 0 | 0 |
| B3GNT3 | 4.55 | 0 | 0 |
| ILF2 | -1.05 | 0 | 0 |
| KIAA0368 | -1.34 | 0 | 0 |
| MCF2L | -2.18 | 0 | 0 |
| KDM5B | 1.31 | 0 | 0 |
| MRPL30 | -1.64 | 0 | 0 |
| MSN | 0.93 | 0 | 0 |
| GMIP | 2.38 | 0 | 0 |
| ACO2 | -1.19 | 0 | 0 |
| H3F3AP4 | -1.61 | 0 | 0 |
| TRIM56 | 1.76 | 0 | 0 |
| HNRNPL | -1.08 | 0 | 0 |
| H3F3A | -1.62 | 0 | 0 |
| ARNTL2 | 2.62 | 0 | 0 |
| SREBF1 | -1.43 | 0 | 0 |
| IGSF3 | 1.32 | 0 | 0 |
| JAK3 | 3.71 | 0 | 0 |
| CA11 | 2.00 | 0 | 0 |
| EIF2S3 | -1.14 | 0 | 0 |
| PKP4 | -1.84 | 0 | 0 |
| RNASEH1 | -2.09 | 0 | 0 |
| ADGRE5 | 1.33 | 0 | 0 |
| CDCA2 | -2.07 | 0 | 0 |
| LARP6 | 3.56 | 0 | 0 |
| CAD | -1.56 | 0 | 0 |
| RECQL4 | -2.09 | 0 | 0 |
| ZFP36L2 | 1.39 | 0 | 0 |
| DCAF13 | -2.11 | 0 | 0 |
| MKNK2 | 1.24 | 0 | 0 |
| LUC7L2 | -1.48 | 0 | 0 |
| C5orf15 | 1.50 | 0 | 0 |
| NDUFAB1 | -1.65 | 0 | 0 |
| ARFGAP3 | 1.72 | 0 | 0 |
| SLC45A3 | 2.66 | 0 | 0 |
| CDK18 | 1.84 | 0 | 0 |
| TPRA1 | 1.45 | 0 | 0 |
| CHAF1B | -2.13 | 0 | 0 |
| PRPF38B | -1.65 | 0 | 0 |
| MFI2 | 1.51 | 0 | 0 |
| ME2 | -1.99 | 0 | 0 |
| PALM | 1.40 | 0 | 0 |
| ACTL6A | -1.36 | 0 | 0 |
| RDX | -1.46 | 0 | 0 |
| USP5 | -1.18 | 0 | 0 |
| C19orf24 | -1.38 | 0 | 0 |
| HDLBP | -1.00 | 0 | 0 |
| SNAI1 | 2.34 | 0 | 0 |
| RAET1L | 4.29 | 0 | 0 |
| TARDBP | -1.49 | 0 | 0 |
| PPP5C | -1.20 | 0 | 0 |
| KRT16 | 2.45 | 0 | 0 |
| ENC1 | 2.62 | 0 | 0 |
| CCDC71L | 3.19 | 0 | 0 |
| IMMT | -1.15 | 0 | 0 |
| HNRNPH1 | -1.21 | 0 | 0 |
| ALG8 | -2.07 | 0 | 0 |
| CD3EAP | -1.81 | 0 | 0 |
| SENP3-EIF4A1 | -0.91 | 0 | 0 |
| MAD2L1 | -2.28 | 0 | 0 |
| SP100 | 1.63 | 0 | 0 |
| MAFG | 1.24 | 0 | 0 |
| ST6GALNAC6 | 1.97 | 0 | 0 |
| UPP1 | 1.58 | 0 | 0 |
| CFAP57 | 4.59 | 0 | 0 |
| COL13A1 | 4.23 | 0 | 0 |
| BBC3 | 2.68 | 0 | 0 |
| TMEM158 | 2.48 | 0 | 0 |
| WBSCR16 | -1.46 | 0 | 0 |
| PPP1CA | -1.21 | 0 | 0 |
| EIF3D | -1.00 | 0 | 0 |
| SMARCB1 | -1.53 | 0 | 0 |
| HIVEP2 | 2.16 | 0 | 0 |
| C1QTNF1-AS1 | 5.98 | 0 | 0 |
| RPS5 | -0.96 | 0 | 0 |
| ULBP2 | 3.41 | 0 | 0 |
| RPTOR | -1.49 | 0 | 0 |
| MIF | -1.33 | 0 | 0 |
| RNF169 | 1.48 | 0 | 0 |
| INCENP | -1.49 | 0 | 0 |
| TRIM8 | 1.48 | 0 | 0 |
| DDX18 | -1.39 | 0 | 0 |
| SLC19A1 | -2.68 | 0 | 0 |
| KIF14 | -2.97 | 0 | 0 |
| NDUFA11 | -1.21 | 0 | 0 |
| NDUFA10 | -1.59 | 0 | 0 |
| SAE1 | -1.07 | 0 | 0 |
| ANAPC15 | -2.24 | 0 | 0 |
| BCAM | 1.67 | 0 | 0 |
| RIN1 | 1.71 | 0 | 0 |
| TEX261 | -1.46 | 0 | 0 |
| DRAP1 | 1.13 | 0 | 0 |
| USP1 | -1.41 | 0 | 0 |
| ALPP | -2.18 | 0 | 0 |
| EPG5 | 1.99 | 0 | 0 |
| NHP2 | -1.27 | 0 | 0 |
| TXNL1 | -1.75 | 0 | 0 |
| ARL6IP5 | -1.79 | 0 | 0 |
| NASP | -1.23 | 0 | 0 |
| CLTC | -1.08 | 0 | 0 |
| RBM25 | -1.51 | 0 | 0 |
| SLC16A5 | 2.28 | 0 | 0 |
| S100A16 | 1.21 | 0 | 0 |
| OS9 | 1.16 | 0 | 0 |
| GPI | -1.06 | 0 | 0 |
| TRIM38 | 1.95 | 0 | 0 |
| ZNF467 | 5.45 | 0 | 0 |
| NBR1 | 1.05 | 0 | 0 |
| HSCB | 2.03 | 0 | 0 |
| MSH2 | -1.94 | 0 | 0 |
| AP2M1 | -0.98 | 0 | 0 |
| MICAL2 | 1.58 | 0 | 0 |
| ARHGEF40 | 2.40 | 0 | 0 |
| OCIAD2 | 2.01 | 0 | 0 |
| DHX34 | -1.81 | 0 | 0 |
| PLEKHF1 | 1.99 | 0 | 0 |
| YAP1 | 1.18 | 0 | 0 |
| ERI3 | -1.50 | 0 | 0 |
| BAMBI | 1.50 | 0 | 0 |
| ZFAS1 | 1.36 | 0 | 0 |
| KLHDC3 | -1.26 | 0 | 0 |
| GGCT | -1.61 | 0 | 0 |
| TMEM140 | 5.08 | 0 | 0 |
| CLIC3 | 3.28 | 0 | 0 |
| MYO18A | 1.29 | 0 | 0 |
| BRCC3 | -2.21 | 0 | 0 |
| RFFL | 1.98 | 0 | 0 |
| PHF19 | -1.46 | 0 | 0 |
| GAS5 | 1.24 | 0 | 0 |
| AGMAT | -2.67 | 0 | 0 |
| OAF | 1.53 | 0 | 0 |
| APRT | -1.33 | 0 | 0 |
| CERK | -1.99 | 0 | 0 |
| C19orf43 | -1.22 | 0 | 0 |
| RRP1 | -1.52 | 0 | 0 |
| CHD3 | -1.17 | 0 | 0 |
| KLHL24 | 2.00 | 0 | 0 |
| EIF2B5 | -1.53 | 0 | 0 |
| PIR | -1.71 | 0 | 0 |
| GRIN2D | 1.84 | 0 | 0 |
| ZBTB10 | 1.76 | 0 | 0 |
| FAIM | -2.47 | 0 | 0 |
| WHSC1 | -1.43 | 0 | 0 |
| TKFC | -2.28 | 0 | 0 |
| CCNG1 | 1.43 | 0 | 0 |
| GPAA1 | -1.37 | 0 | 0 |
| AGAP1 | -1.80 | 0 | 0 |
| TTL | -1.32 | 0 | 0 |
| BCYRN1 | -1.68 | 0 | 0 |
| HMMR | -2.44 | 0 | 0 |
| EFTUD2 | -1.07 | 0 | 0 |
| PDS5A | -1.37 | 0 | 0 |
| WDHD1 | -2.16 | 0 | 0 |
| FBLIM1 | 3.23 | 0 | 0 |
| PRPF19 | -1.04 | 0 | 0 |
| SMCR8 | 1.22 | 0 | 0 |
| TUBA4A | -1.13 | 0 | 0 |
| YWHAB | -1.07 | 0 | 0 |
| IKBKAP | -1.34 | 0 | 0 |
| GEMIN5 | -1.59 | 0 | 0 |
| ZNF76 | -2.39 | 0 | 0 |
| SSBP1 | -1.66 | 0 | 0 |
| PXDN | -1.12 | 0 | 0 |
| PDSS1 | -3.63 | 0 | 0 |
| POLR3B | -2.09 | 0 | 0 |
| ERP44 | 1.53 | 0 | 0 |
| ST14 | -1.86 | 0 | 0 |
| CD55 | 1.01 | 0 | 0 |
| VDAC1 | -1.05 | 0 | 0 |
| DHX29 | -1.54 | 0 | 0 |
| TIMM8B | -1.81 | 0 | 0 |
| TXNDC5 | -1.03 | 0 | 0 |
| NOP14 | -1.60 | 0 | 0 |
| GMDS | -1.77 | 0 | 0 |
| DNASE2 | 1.20 | 0 | 0 |
| BACE2 | -1.16 | 0 | 0 |
| METTL3 | -1.71 | 0 | 0 |
| RASGEF1A | 1.65 | 0 | 0 |
| AURKB | -1.71 | 0 | 0 |
| MAGOHB | -2.85 | 0 | 0 |
| MAGED2 | 1.29 | 0 | 0 |
| DDX23 | -1.18 | 0 | 0 |
| TEAD3 | 1.64 | 0 | 0 |
| RBM15B | -1.50 | 0 | 0 |
| MSTO1 | -1.78 | 0 | 0 |
| ITPKC | 1.92 | 0 | 0 |
| MPZL2 | 2.38 | 0 | 0 |
| CRAT | 1.76 | 0 | 0 |
| SH3PXD2A | -1.25 | 0 | 0 |
| MEMO1 | -2.03 | 0 | 0 |
| ACVRL1 | 5.11 | 0 | 0 |
| DPP9 | -1.25 | 0 | 0 |
| MT1X | 2.60 | 0 | 0 |
| GEMIN4 | -1.35 | 0 | 0 |
| PNN | -1.30 | 0 | 0 |
| PERP | 1.14 | 0 | 0 |
| NKRF | -1.86 | 0 | 0 |
| DUSP9 | -2.27 | 0 | 0 |
| HIST1H4B | -5.81 | 0 | 0 |
| UBAC1 | -1.55 | 0 | 0 |
| PKM | -0.92 | 0 | 0 |
| ZNF326 | -1.66 | 0 | 0 |
| LOC100288152 | 2.14 | 0 | 0 |
| MAP3K10 | 1.94 | 0 | 0 |
| HNRNPH2 | -1.37 | 0 | 0 |
| MAP2K6 | -3.72 | 0 | 0 |
| PHKB | -1.60 | 0 | 0 |
| GADD45B | 1.53 | 0 | 0 |
| FAM64A | -2.47 | 0 | 0 |
| WDR76 | -2.32 | 0 | 0 |
| SEL1L3 | 1.40 | 0 | 0 |
| PTP4A1 | -1.18 | 0 | 0 |
| SERPINH1 | 1.02 | 0 | 0 |
| ARHGAP26 | 2.42 | 0 | 0 |
| PAGE1 | -1.21 | 0 | 0 |
| SLC2A6 | 2.20 | 0 | 0 |
| LCN2 | 3.72 | 0 | 0 |
| WDR26 | 1.31 | 0 | 0 |
| SNF8 | -1.48 | 0 | 0 |
| DNAJC7 | -1.20 | 0 | 0 |
| TUBGCP2 | -1.60 | 0 | 0 |
| VCL | 1.00 | 0 | 0 |
| FAM179A | -4.59 | 0 | 0 |
| SAV1 | 1.57 | 0 | 0 |
| NPTXR | 1.93 | 0 | 0 |
| NF2 | -1.42 | 0 | 0 |
| RRS1 | -1.60 | 0 | 0 |
| IFITM2 | 1.10 | 0 | 0 |
| MRPL47 | -1.79 | 0 | 0 |
| GMNN | -1.43 | 0 | 0 |
| UMPS | -1.51 | 0 | 0 |
| CHRD | 2.98 | 0 | 0 |
| VAMP2 | 2.18 | 0 | 0 |
| AIFM1 | -1.20 | 0 | 0 |
| DAPK2 | 2.95 | 0 | 0 |
| ADAP1 | 4.97 | 0 | 0 |
| DUSP16 | 1.07 | 0 | 0 |
| PDE4D | -1.07 | 0 | 0 |
| ARL6IP1 | -1.29 | 0 | 0 |
| NRP2 | 2.24 | 0 | 0 |
| SSB | -1.74 | 0 | 0 |
| AAAS | -1.38 | 0 | 0 |
| MME | 4.63 | 0 | 0 |
| ADI1 | -1.28 | 0 | 0 |
| MCCC2 | -1.36 | 0 | 0 |
| TGFBI | 1.16 | 0 | 0 |
| PPAN | -1.43 | 0 | 0 |
| PARP12 | 1.29 | 0 | 0 |
| SCLY | -2.67 | 0 | 0 |
| HRH1 | 3.26 | 0 | 0 |
| CACNA1H | 3.30 | 0 | 0 |
| HMGB2 | -1.56 | 0 | 0 |
| GRK6 | -1.60 | 0 | 0 |
| LRRFIP2 | 1.36 | 0 | 0 |
| CTSS | 7.81 | 0 | 0 |
| MBTPS1 | -1.24 | 0 | 0 |
| DLGAP4 | 1.58 | 0 | 0 |
| ZNF609 | 1.23 | 0 | 0 |
| ATF5 | -1.62 | 0 | 0 |
| HCFC1 | -1.22 | 0 | 0 |
| MAPK4 | -1.23 | 0 | 0 |
| CAPRIN1 | -0.94 | 0 | 0 |
| QARS | -1.14 | 0 | 0 |
| FCHSD1 | 2.47 | 0 | 0 |
| CDC7 | -1.90 | 0 | 0 |
| ADAMTS4 | 3.48 | 0 | 0 |
| NUCKS1 | -1.10 | 0 | 0 |
| TMEM38A | 3.69 | 0 | 0 |
| BRPF3 | 1.41 | 0 | 0 |
| GPRC5C | 1.56 | 0 | 0 |
| TXLNG | -1.33 | 0 | 0 |
| FBXL16 | 1.73 | 0 | 0 |
| VDAC3 | -1.41 | 0 | 0 |
| DNAJC9 | -1.68 | 0 | 0 |
| KRT9 | 3.71 | 0 | 0 |
| NUDT1 | -2.08 | 0 | 0 |
| CRYAB | 2.27 | 0 | 0 |
| HSH2D | 3.63 | 0 | 0 |
| TOP2B | -1.44 | 0 | 0 |
| LEMD1 | 4.91 | 0 | 0 |
| KBTBD6 | -2.08 | 0 | 0 |
| PTMA | -0.94 | 0 | 0 |
| CEBPZ | -1.34 | 0 | 0 |
| ZNF746 | 1.58 | 0 | 0 |
| ABCB8 | -2.13 | 0 | 0 |
| MARS | -1.08 | 0 | 0 |
| CDCA7 | -2.14 | 0 | 0 |
| DNA2 | -3.39 | 0 | 0 |
| ADGRA2 | 3.01 | 0 | 0 |
| HOMER3 | 1.44 | 0 | 0 |
| GTPBP6 | -2.42 | 0 | 0 |
| TYSND1 | 1.26 | 0 | 0 |
| DEK | -1.34 | 0 | 0 |
| NAE1 | -1.60 | 0 | 0 |
| BCL9L | 1.21 | 0 | 0 |
| SHISA5 | 1.08 | 0 | 0 |
| AP2B1 | -1.01 | 0 | 0 |
| MRPS23 | -1.48 | 0 | 0 |
| GNE | 1.38 | 0 | 0 |
| BLOC1S5-TXNDC5 | -0.99 | 0 | 0 |
| NUP93 | -1.28 | 0 | 0 |
| UFD1L | -1.29 | 0 | 0 |
| SEMA4C | 1.50 | 0 | 0 |
| RPUSD1 | -1.51 | 0 | 0 |
| RAD51AP1 | -2.48 | 0 | 0 |
| ATIC | -1.28 | 0 | 0 |
| VASP | 1.14 | 0 | 0 |
| OSGEP | -2.66 | 0 | 0 |
| RSAD2 | 7.69 | 0 | 0 |
| RPP40 | -2.22 | 0 | 0 |
| WDR34 | -1.20 | 0 | 0 |
| FBN1 | 2.14 | 0 | 0 |
| EAF1 | 1.56 | 0 | 0 |
| NPC2 | 1.31 | 0 | 0 |
| OIP5 | -3.01 | 0 | 0 |
| NFS1 | -1.75 | 0 | 0 |
| FLNB | 1.03 | 0 | 0 |
| GORASP2 | -1.38 | 0 | 0 |
| HBEGF | 2.70 | 0 | 0 |
| CEACAM1 | 9.63 | 0 | 0 |
| TSR1 | -1.05 | 0 | 0 |
| UQCRFS1 | -1.17 | 0 | 0 |
| SLC46A1 | 1.23 | 0 | 0 |
| CREBRF | 2.93 | 0 | 0 |
| FOXRED2 | -1.43 | 0 | 0 |
| CHML | -1.49 | 0 | 0 |
| PYCRL | -2.74 | 0 | 0 |
| APOL3 | 5.64 | 0 | 0 |
| KLF4 | 1.04 | 0 | 0 |
| FGFBP1 | 3.21 | 0 | 0 |
| BAK1 | 1.41 | 0 | 0 |
| FUBP1 | -1.12 | 0 | 0 |
| IRF2BP2 | 1.21 | 0 | 0 |
| PCCB | -1.20 | 0 | 0 |
| COX4I1 | -0.91 | 0 | 0 |
| PUF60 | -1.19 | 0 | 0 |
| MEIS3 | 1.99 | 0 | 0 |
| URM1 | -1.57 | 0 | 0 |
| UBA7 | 4.88 | 0 | 0 |
| STIL | -1.95 | 0 | 0 |
| FAM13B | -1.86 | 0 | 0 |
| TTC27 | -1.74 | 0 | 0 |
| RCN3 | 1.32 | 0 | 0 |
| PPP2R5B | 2.69 | 0 | 0 |
| UQCRH | -0.99 | 0 | 0 |
| HRASLS2 | 6.64 | 0 | 0 |
| LSG1 | -1.27 | 0 | 0 |
| PFN1 | -0.92 | 0 | 0 |
| C14orf2 | -1.29 | 0 | 0 |
| NFKBIZ | 2.68 | 0 | 0 |
| PMF1 | -1.36 | 0 | 0 |
| ASAP3 | 2.54 | 0 | 0 |
| SLC25A39 | -1.00 | 0 | 0 |
| PHPT1 | -1.62 | 0 | 0 |
| SWAP70 | 1.41 | 0 | 0 |
| LOC100507412 | -2.74 | 0 | 0 |
| ATP11C | -1.98 | 0 | 0 |
| UBE3C | -1.34 | 0 | 0 |
| TSC2 | -1.57 | 0 | 0 |
| TEAD1 | -1.28 | 0 | 0 |
| FTH1 | 1.01 | 0 | 0 |
| CUL3 | -1.50 | 0 | 0 |
| LAMA5 | 1.18 | 0 | 0 |
| SAMD9L | 3.38 | 0 | 0 |
| LUC7L3 | -1.37 | 0 | 0 |
| EIF4EBP2 | -1.52 | 0 | 0 |
| XYLT2 | 1.59 | 0 | 0 |
| SLC30A9 | -1.92 | 0 | 0 |
| MTG1 | -2.38 | 0 | 0 |
| WDR90 | -2.19 | 0 | 0 |
| SGTA | -1.11 | 0 | 0 |
| LDHD | 2.50 | 0 | 0 |
| L3MBTL2 | -1.54 | 0 | 0 |
| SIL1 | 1.43 | 0 | 0 |
| AFF4 | 1.14 | 0 | 0 |
| CNOT7 | -1.74 | 0 | 0 |
| CYB561A3 | -1.54 | 0 | 0 |
| KCNJ12 | 3.57 | 0 | 0 |
| HSPE1-MOB4 | -1.52 | 0 | 0 |
| MRPL54 | -1.61 | 0 | 0 |
| CAPN2 | -0.98 | 0 | 0 |
| NDUFB2 | -1.24 | 0 | 0 |
| PRKD3 | -1.46 | 0 | 0 |
| TNRC6C-AS1 | 3.36 | 0 | 0 |
| ACSL4 | 1.30 | 0 | 0 |
| FIGNL1 | -2.23 | 0 | 0 |
| ERRFI1 | 1.71 | 0 | 0 |
| KIAA1522 | 1.13 | 0 | 0 |
| NUP50 | -1.41 | 0 | 0 |
| MELK | -1.76 | 0 | 0 |
| SH3BP5 | -1.23 | 0 | 0 |
| ADRM1 | -1.11 | 0 | 0 |
| SNRPD3 | -1.08 | 0 | 0 |
| AGL | -1.65 | 0 | 0 |
| LAPTM4B | -0.97 | 0 | 0 |
| NPAS1 | 4.63 | 0 | 0 |
| TRIM22 | 9.45 | 0 | 0 |
| EXOSC10 | -1.28 | 0 | 0 |
| DBF4B | -2.20 | 0 | 0 |
| SEZ6L2 | 1.87 | 0 | 0 |
| GCSH | -1.65 | 0 | 0 |
| WDR5 | -1.33 | 0 | 0 |
| KLF5 | 1.74 | 0 | 0 |
| PSRC1 | -1.63 | 0 | 0 |
| ZNF367 | -2.36 | 0 | 0 |
| RABEPK | -1.63 | 0 | 0 |
| ANAPC1 | -1.26 | 0 | 0 |
| TUFT1 | 1.32 | 0 | 0 |
| PCSK1 | -2.64 | 0 | 0 |
| SLX1A | 1.55 | 0 | 0 |
| SLX1B | 1.55 | 0 | 0 |
| NUBP2 | -1.44 | 0 | 0 |
| EFTUD1 | 1.29 | 0 | 0 |
| NEMF | -1.85 | 0 | 0 |
| CMIP | 1.31 | 0 | 0 |
| GALNT6 | 1.19 | 0 | 0 |
| SGOL2 | -2.85 | 0 | 0 |
| DPH2 | -1.58 | 0 | 0 |
| IL27RA | 1.24 | 0 | 0 |
| ZNF865 | 1.80 | 0 | 0 |
| F2RL1 | 5.10 | 0 | 0 |
| HNRNPU | -0.85 | 0 | 0 |
| TRMU | -1.68 | 0 | 0 |
| PVRL4 | 5.94 | 0 | 0 |
| PXN | 1.04 | 0 | 0 |
| GMFB | -1.94 | 0 | 0 |
| ACTA2 | 4.26 | 0 | 0 |
| IDH3B | -1.34 | 0 | 0 |
| WIPI1 | 2.02 | 0 | 0 |
| PSMD11 | -1.03 | 0 | 0 |
| MARCH2 | 1.98 | 0 | 0 |
| CBX3 | -1.19 | 0 | 0 |
| MRPS15 | -1.16 | 0 | 0 |
| ACTN1 | 1.02 | 0 | 0 |
| PDZD2 | 5.39 | 0 | 0 |
| NUMB | 1.27 | 0 | 0 |
| ALG14 | -4.20 | 0 | 0 |
| C16orf59 | -2.81 | 0 | 0 |
| PLEKHA7 | 2.15 | 0 | 0 |
| RBBP4 | -1.26 | 0 | 0 |
| FNDC3A | 1.56 | 0 | 0 |
| WDR61 | -1.47 | 0 | 0 |
| ABCG2 | -1.34 | 0 | 0 |
| DCTPP1 | -1.49 | 0 | 0 |
| MTMR2 | -1.51 | 0 | 0 |
| GCAT | -1.85 | 0 | 0 |
| WDR12 | -1.74 | 0 | 0 |
| SPTBN1 | -1.00 | 0 | 0 |
| AARSD1 | -1.88 | 0 | 0 |
| ACACB | -1.73 | 0 | 0 |
| FAM208A | -1.57 | 0 | 0 |
| UTP11L | -1.61 | 0 | 0 |
| GLYR1 | -1.16 | 0 | 0 |
| ATRIP | -2.50 | 0 | 0 |
| TBC1D22B | 1.74 | 0 | 0 |
| EFNA1 | 1.15 | 0 | 0 |
| KIFC1 | -2.23 | 0 | 0 |
| RRP7BP | -1.56 | 0 | 0 |
| HMGA1 | 0.94 | 0 | 0 |
| CSRNP2 | 1.48 | 0 | 0 |
| TARBP2 | -1.69 | 0 | 0 |
| EIF3K | -0.93 | 0 | 0 |
| LCMT2 | -3.22 | 0 | 0 |
| IQGAP1 | -0.89 | 0 | 0 |
| C6orf89 | 1.20 | 0 | 0 |
| PTPRM | 1.33 | 0 | 0 |
| IGF2BP2 | 1.59 | 0 | 0 |
| ZNF281 | 1.29 | 0 | 0 |
| LOC100507002 | 4.66 | 0 | 0 |
| PLAT | 4.87 | 0 | 0 |
| PSMA1 | -1.27 | 0 | 0 |
| TMEM63B | 1.35 | 0 | 0 |
| PEPD | -1.52 | 0 | 0 |
| GARS | -1.04 | 0 | 0 |
| RAB10 | -1.17 | 0 | 0 |
| TDRD9 | -1.84 | 0 | 0 |
| ZG16B | 2.62 | 0 | 0 |
| DUSP10 | 3.94 | 0 | 0 |
| SMURF1 | 1.40 | 0 | 0 |
| CASP9 | -1.78 | 0 | 0 |
| CLIC4 | 1.17 | 0 | 0 |
| EMP1 | 1.40 | 0 | 0 |
| MEF2D | 1.15 | 0 | 0 |
| POTEI | -1.27 | 0 | 0 |
| RND1 | 4.06 | 0 | 0 |
| ADGRA3 | -2.17 | 0 | 0 |
| CDKN2C | -1.88 | 0 | 0 |
| POLR3K | -1.59 | 0 | 0 |
| PRCP | 1.39 | 0 | 0 |
| INTS10 | -1.24 | 0 | 0 |
| TM4SF1-AS1 | 3.23 | 0 | 0 |
| TTC3 | -1.53 | 0 | 0 |
| TBL1X | -1.38 | 0 | 0 |
| KRT81 | 3.16 | 0 | 0 |
| PARPBP | -3.01 | 0 | 0 |
| EEF2 | -0.84 | 0 | 0 |
| GDI1 | 1.07 | 0 | 0 |
| EEF1A1 | -0.83 | 0 | 0 |
| CLK3 | 1.29 | 0 | 0 |
| TSTA3 | -1.59 | 0 | 0 |
| CTTN | -1.14 | 0 | 0 |
| ARHGDIA | -1.05 | 0 | 0 |
| DDX21 | -0.95 | 0 | 0 |
| ASNA1 | -1.09 | 0 | 0 |
| TARS2 | -1.26 | 0 | 0 |
| EI24 | -1.18 | 0 | 0 |
| PPIL1 | -1.45 | 0 | 0 |
| BSDC1 | 1.21 | 0 | 0 |
| NDUFC2-KCTD14 | -1.33 | 0 | 0 |
| DSCC1 | -3.37 | 0 | 0 |
| MRPS22 | -1.41 | 0 | 0 |
| CTAG2 | -1.00 | 0 | 0 |
| FAM195A | -1.52 | 0 | 0 |
| YWHAG | -0.93 | 0 | 0 |
| ICMT | -1.29 | 0 | 0 |
| LTBP2 | 3.55 | 0 | 0 |
| ATP1B3 | -0.92 | 0 | 0 |
| KIF15 | -2.79 | 0 | 0 |
| ME1 | -1.75 | 0 | 0 |
| ATP5J | -1.18 | 0 | 0 |
| CSAG4 | 2.28 | 0 | 0 |
| TMEM183B | -1.48 | 0 | 0 |
| CSTF1 | -1.53 | 0 | 0 |
| EIF3J | -1.19 | 0 | 0 |
| SCRN2 | 1.56 | 0 | 0 |
| PPP1R3B | 3.09 | 0 | 0 |
| GLOD4 | -1.58 | 0 | 0 |
| FAM111B | -3.38 | 0 | 0 |
| SLC26A6 | 1.31 | 0 | 0 |
| PTPN12 | 1.45 | 0 | 0 |
| LINC01003 | 3.10 | 0 | 0 |
| MIDN | 1.16 | 0 | 0 |
| CFLAR | 1.70 | 0 | 0 |
| SNRNP70 | -0.99 | 0 | 0 |
| KIAA1551 | 1.76 | 0 | 0 |
| DAD1 | -1.44 | 0 | 0 |
| TMEM205 | 1.23 | 0 | 0 |
| KLHDC4 | -1.61 | 0 | 0 |
| FOXD2-AS1 | 2.69 | 0 | 0 |
| CENPM | -2.42 | 0 | 0 |
| TMBIM6 | 0.84 | 0 | 0 |
| LTBR | 0.98 | 0 | 0 |
| KCTD15 | -1.55 | 0 | 0 |
| DDIAS | -2.92 | 0 | 0 |
| IPO11 | -1.50 | 0 | 0 |
| ARHGAP11A | -2.58 | 0 | 0 |
| NUP98 | -0.97 | 0 | 0 |
| ICT1 | -1.37 | 0 | 0 |
| DPF2 | 1.24 | 0 | 0 |
| ZMYND11 | -1.82 | 0 | 0 |
| ZSWIM8 | 1.35 | 0 | 0 |
| MRPL15 | -1.26 | 0 | 0 |
| BTN3A2 | 1.38 | 0 | 0 |
| MTA1 | -1.18 | 0 | 0 |
| SPRY2 | 2.37 | 0 | 0 |
| LZTS3 | 2.28 | 0 | 0 |
| FIBCD1 | 1.93 | 0 | 0 |
| DES | 3.04 | 0 | 0 |
| GGT1 | 3.27 | 0 | 0 |
| WDR33 | -1.23 | 0 | 0 |
| PARVB | -1.41 | 0 | 0 |
| GADD45GIP1 | -1.12 | 0 | 0 |
| UAP1 | 1.14 | 0 | 0 |
| DLEU1 | -3.79 | 0 | 0 |
| ACIN1 | -0.97 | 0 | 0 |
| HN1L | -0.97 | 0 | 0 |
| ZFYVE19 | 1.35 | 0 | 0 |
| RREB1 | 1.18 | 0 | 0 |
| HSPG2 | 1.28 | 0 | 0 |
| SOWAHC | 1.10 | 0 | 0 |
| ITPA | -1.45 | 0 | 0 |
| MRPL35 | -1.52 | 0 | 0 |
| COPZ1 | -0.94 | 0 | 0 |
| HIST1H4H | -5.36 | 0 | 0 |
| FKBP5 | -1.48 | 0 | 0 |
| PGPEP1 | 2.96 | 0 | 0 |
| DHX38 | -1.20 | 0 | 0 |
| ZNF697 | 1.95 | 0 | 0 |
| FAM171A2 | 2.45 | 0 | 0 |
| CKAP2 | -1.44 | 0 | 0 |
| MLLT1 | -1.33 | 0 | 0 |
| NSMCE1 | -2.21 | 0 | 0 |
| CCNY | -1.63 | 0 | 0 |
| IDH1 | -1.28 | 0 | 0 |
| IDH3G | -1.24 | 0 | 0 |
| CBX5 | -1.19 | 0 | 0 |
| TMEM43 | 1.09 | 0 | 0 |
| NAA20 | -1.58 | 0 | 0 |
| LUC7L | -1.93 | 0 | 0 |
| STMN1 | -1.07 | 0 | 0 |
| LMBR1L | 1.82 | 0 | 0 |
| SFN | 0.99 | 0 | 0 |
| AKAP13 | 1.03 | 0 | 0 |
| JUND | 1.29 | 0 | 0 |
| SEMA3F | 1.23 | 0 | 0 |
| BEAN1 | 4.51 | 0 | 0 |
| GAL | -1.59 | 0 | 0 |
| CPSF2 | -1.35 | 0 | 0 |
| MDH2 | -0.88 | 0 | 0 |
| CAPZA2 | -1.18 | 0 | 0 |
| TSPAN31 | 2.20 | 0 | 0 |
| RNF213 | 0.94 | 0 | 0 |
| NUDT5 | -1.23 | 0 | 0 |
| HIST1H2BG | -3.54 | 0 | 0 |
| ADGRG1 | 1.63 | 0 | 0 |
| YTHDC2 | -1.67 | 0 | 0 |
| ACTR3B | -3.20 | 0 | 0 |
| NUP133 | -1.38 | 0 | 0 |
| KCNN4 | 1.51 | 0 | 0 |
| PRSS16 | 2.62 | 0 | 0 |
| HHAT | 3.32 | 0 | 0 |
| NMT1 | -0.99 | 0 | 0 |
| VTN | 1.39 | 0 | 0 |
| MALSU1 | -1.48 | 0 | 0 |
| HECTD1 | -1.27 | 0 | 0 |
| ANXA6 | -0.91 | 0 | 0 |
| PSG4 | 3.24 | 0 | 0 |
| MBD3 | -1.20 | 0 | 0 |
| MAP7D1 | 1.19 | 0 | 0 |
| FAM81A | -2.35 | 0 | 0 |
| CPSF3L | -1.48 | 0 | 0 |
| ZBTB39 | 1.43 | 0 | 0 |
| HIVEP1 | 1.54 | 0 | 0 |
| WDR74 | -1.35 | 0 | 0 |
| ACAD9 | -1.27 | 0 | 0 |
| NFE2L3 | 6.59 | 0 | 0 |
| SMYD2 | -1.47 | 0 | 0 |
| NOL4L | 2.15 | 0 | 0 |
| SPG7 | -1.34 | 0 | 0 |
| FASTKD2 | -1.80 | 0 | 0 |
| PROCR | 1.85 | 0 | 0 |
| KRTCAP2 | 1.08 | 0 | 0 |
| P4HA1 | 1.34 | 0 | 0 |
| MB | 2.84 | 0 | 0 |
| PTCD3 | -1.46 | 0 | 0 |
| GEN1 | -2.52 | 0 | 0 |
| ATP5J2 | -0.89 | 0 | 0 |
| C6orf1 | 1.96 | 0 | 0 |
| HIST2H2BF | -3.30 | 0 | 0 |
| H2AFY | -0.91 | 0 | 0 |
| SAR1A | 1.09 | 0 | 0 |
| STAT3 | 0.91 | 0 | 0 |
| SRPRB | -1.17 | 0 | 0 |
| ABCC4 | -1.56 | 0 | 0 |
| DDX51 | -1.72 | 0 | 0 |
| BCCIP | -1.40 | 0 | 0 |
| PSG5 | 6.18 | 0 | 0 |
| CUL1 | -1.24 | 0 | 0 |
| CCDC50 | 1.24 | 0 | 0 |
| DDX20 | -1.20 | 0 | 0 |
| TNNT1 | 1.56 | 0 | 0 |
| YPEL2 | 2.45 | 0 | 0 |
| PRAME | 1.64 | 0 | 0 |
| HMGCL | 2.06 | 0 | 0 |
| UBE2D1 | 1.83 | 0 | 0 |
| SLC25A19 | -1.62 | 0 | 0 |
| LIMS2 | 2.30 | 0 | 0 |
| MARK4 | 1.74 | 0 | 0 |
| FIBP | -1.35 | 0 | 0 |
| HARS | -1.11 | 0 | 0 |
| PELI3 | 1.83 | 0 | 0 |
| LOX | 2.37 | 0 | 0 |
| IL15RA | 2.22 | 0 | 0 |
| ARHGEF37 | 2.47 | 0 | 0 |
| PPID | -1.51 | 0 | 0 |
| PRMT3 | -1.94 | 0 | 0 |
| FNDC3B | 1.14 | 0 | 0 |
| ADARB1 | -1.14 | 0 | 0 |
| CCDC68 | 2.76 | 0 | 0 |
| FOXJ2 | 1.31 | 0 | 0 |
| C20orf27 | -1.66 | 0 | 0 |
| UQCRC1 | -0.89 | 0 | 0 |
| INTS3 | -1.66 | 0 | 0 |
| EIF3F | -1.00 | 0 | 0 |
| NDUFA13 | -1.08 | 0 | 0 |
| UBA2 | -1.06 | 0 | 0 |
| IVD | -1.41 | 0 | 0 |
| XPOT | -1.12 | 0 | 0 |
| HMBS | -1.58 | 0 | 0 |
| SLC7A6 | -1.17 | 0 | 0 |
| EMC6 | -1.32 | 0 | 0 |
| RBM10 | -1.22 | 0 | 0 |
| SSSCA1 | -1.50 | 0 | 0 |
| PMF1-BGLAP | -1.44 | 0 | 0 |
| SIPA1 | 1.40 | 0 | 0 |
| TRMT10C | -1.39 | 0 | 0 |
| COL8A1 | 3.33 | 0 | 0 |
| DUT | -1.45 | 0 | 0 |
| MMP19 | 3.65 | 0 | 0 |
| LSM3 | -1.70 | 0 | 0 |
| ANGPTL4 | 6.14 | 0 | 0 |
| TTC38 | -1.33 | 0 | 0 |
| RORC | 1.59 | 0 | 0 |
| KHDRBS1 | -0.93 | 0 | 0 |
| IQSEC2 | 1.89 | 0 | 0 |
| GLMP | 1.29 | 0 | 0 |
| SHMT1 | -1.18 | 0 | 0 |
| GATS | 2.33 | 0 | 0 |
| RABAC1 | 1.48 | 0 | 0 |
| DDIT3 | 1.86 | 0 | 0 |
| SNX5 | -1.47 | 0 | 0 |
| SKA2 | -1.96 | 0 | 0 |
| PHF20L1 | -1.64 | 0 | 0 |
| CCL28 | 3.93 | 0 | 0 |
| COA7 | -1.44 | 0 | 0 |
| PAK1IP1 | -1.37 | 0 | 0 |
| MAP1LC3B | 1.25 | 0 | 0 |
| SNX33 | 1.41 | 0 | 0 |
| GLUL | 0.93 | 0 | 0 |
| CLEC3A | -3.43 | 0 | 0 |
| MDFI | 2.16 | 0 | 0 |
| CCDC120 | 1.92 | 0 | 0 |
| UQCRQ | -1.03 | 0 | 0 |
| GBA | 1.33 | 0 | 0 |
| SLC17A5 | 1.88 | 0 | 0 |
| SMC3 | -1.51 | 0 | 0 |
| G3BP1 | -1.01 | 0 | 0 |
| NT5C2 | -0.99 | 0 | 0 |
| CD40 | 3.50 | 0 | 0 |
| TTLL3 | 1.79 | 0 | 0 |
| INPP5A | -2.74 | 0 | 0 |
| PSMB1 | -1.04 | 0 | 0 |
| ACAT2 | 1.17 | 0 | 0 |
| HLA-E | 3.84 | 0 | 0 |
| ARMC1 | -1.66 | 0 | 0 |
| INF2 | 1.10 | 0 | 0 |
| C1orf131 | -2.01 | 0 | 0 |
| GNAO1 | 1.83 | 0 | 0 |
| FTL | -0.83 | 0 | 0 |
| CLDN4 | 5.55 | 0 | 0 |
| GATB | -1.78 | 0 | 0 |
| A4GALT | 1.95 | 0 | 0 |
| SIN3B | 1.10 | 0 | 0 |
| ERCC6L | -3.36 | 0 | 0 |
| CCDC18 | -3.74 | 0 | 0 |
| SESN3 | -1.88 | 0 | 0 |
| B4GALT4 | 1.46 | 0 | 0 |
| ECHS1 | -1.10 | 0 | 0 |
| ACACA | -1.47 | 0 | 0 |
| ARSB | -2.23 | 0 | 0 |
| RPS6 | -0.87 | 0 | 0 |
| QSER1 | -1.37 | 0 | 0 |
| LACTB | 1.57 | 0 | 0 |
| GNPNAT1 | -1.22 | 0 | 0 |
| PIR-FIGF | -1.66 | 0 | 0 |
Pathway analysis of the differntial expressed gene
deseq_res_hela_eg <- bitr(deseq2_res_hela[order(deseq2_res_hela$p.adjust,decreasing = F)[1:2000],"GeneID"],fromType = "SYMBOL", toType = "ENTREZID",OrgDb = "org.Hs.eg.db" )
eKEGG_hela <- enrichKEGG(deseq_res_hela_eg$ENTREZID,organism = "hsa",pAdjustMethod = "none", pvalueCutoff = 0.05,minGSSize = 3)
#enrichMap(eKEGG,vertex.label.font = 2,n = 25)
emapplot(eKEGG_hela)
Joint pathway analysis in A549 and Hela
This is to address the reviewer’s question “comment on the pathway in common in A549 and Hela cell line”
library(ggplot2)
DEG_all <- list("A549 DEGs"=deseq_res_eg$ENTREZID ,"Hela DEGs"=deseq_res_hela_eg$ENTREZID)
ck <- compareCluster(geneCluster = DEG_all, fun = "enrichKEGG")
dotplot(ck,showCategory=10)+theme(
panel.grid.minor = element_blank(), axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=22, face="bold", hjust=0.5,family = "arial"),
axis.title.y=element_text(size=22, face="bold", vjust=0.4, angle=90, family = "arial"),
legend.title=element_text(size=15, face="bold"),legend.text = element_text(size = 20, face = "bold",family = "arial"),
axis.text.x = element_text(size = 15,face = "bold",family = "arial") ,axis.text.y = element_text(size = 16,face = "bold",family = "arial"))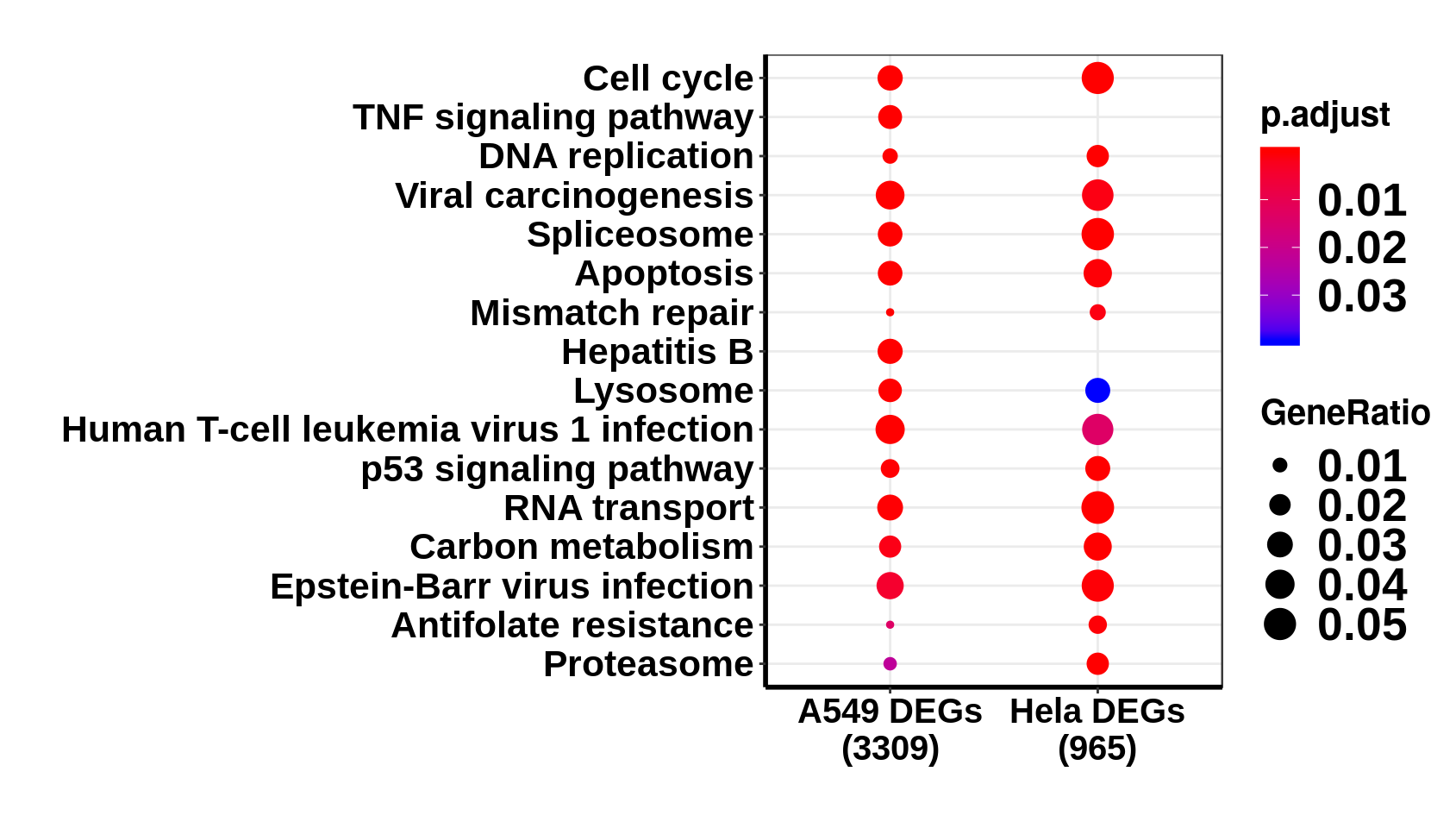
Session information
sessionInfo()R version 3.5.0 (2018-04-23)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 17.10
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/libopenblasp-r0.2.20.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] grid stats4 parallel stats graphics grDevices utils
[8] datasets methods base
other attached packages:
[1] org.Hs.eg.db_3.7.0 enrichplot_1.2.0
[3] clusterProfiler_3.10.0 DESeq2_1.22.1
[5] m6Amonster_0.1.5 RcppArmadillo_0.9.200.5.0
[7] Rcpp_1.0.0 reshape2_1.4.3
[9] GenomicAlignments_1.18.0 SummarizedExperiment_1.12.0
[11] DelayedArray_0.8.0 BiocParallel_1.16.1
[13] matrixStats_0.54.0 rtracklayer_1.42.1
[15] doParallel_1.0.14 iterators_1.0.10
[17] foreach_1.4.4 ggplot2_3.1.0
[19] Rsamtools_1.34.0 Biostrings_2.50.1
[21] XVector_0.22.0 GenomicFeatures_1.34.1
[23] AnnotationDbi_1.44.0 Biobase_2.42.0
[25] GenomicRanges_1.34.0 GenomeInfoDb_1.18.1
[27] IRanges_2.16.0 S4Vectors_0.20.1
[29] BiocGenerics_0.28.0
loaded via a namespace (and not attached):
[1] backports_1.1.2 Hmisc_4.1-1 fastmatch_1.1-0
[4] workflowr_1.1.1 plyr_1.8.4 igraph_1.2.2
[7] lazyeval_0.2.1 splines_3.5.0 urltools_1.7.1
[10] digest_0.6.18 htmltools_0.3.6 GOSemSim_2.8.0
[13] viridis_0.5.1 GO.db_3.7.0 magrittr_1.5
[16] checkmate_1.8.5 memoise_1.1.0 cluster_2.0.7-1
[19] annotate_1.60.0 R.utils_2.7.0 prettyunits_1.0.2
[22] colorspace_1.4-0 blob_1.1.1 ggrepel_0.8.0
[25] dplyr_0.7.8 RADAR_0.1.5 crayon_1.3.4
[28] RCurl_1.95-4.11 jsonlite_1.5 genefilter_1.64.0
[31] bindr_0.1.1 survival_2.43-1 ape_5.2
[34] glue_1.3.0 gtable_0.2.0 zlibbioc_1.28.0
[37] UpSetR_1.3.3 scales_1.0.0 DOSE_3.8.0
[40] DBI_1.0.0 viridisLite_0.3.0 xtable_1.8-3
[43] progress_1.2.0 htmlTable_1.12 units_0.6-1
[46] gridGraphics_0.3-0 foreign_0.8-71 bit_1.1-14
[49] europepmc_0.3 Formula_1.2-3 htmlwidgets_1.3
[52] httr_1.3.1 fgsea_1.8.0 RColorBrewer_1.1-2
[55] acepack_1.4.1 pkgconfig_2.0.2 XML_3.98-1.16
[58] R.methodsS3_1.7.1 farver_1.1.0 nnet_7.3-12
[61] locfit_1.5-9.1 ggplotify_0.0.3 tidyselect_0.2.5
[64] labeling_0.3 rlang_0.3.1 munsell_0.5.0
[67] tools_3.5.0 RSQLite_2.1.1 ggridges_0.5.1
[70] evaluate_0.12 stringr_1.3.1 yaml_2.2.0
[73] knitr_1.20 bit64_0.9-7 purrr_0.2.5
[76] ggraph_1.0.2 bindrcpp_0.2.2 nlme_3.1-137
[79] whisker_0.3-2 R.oo_1.22.0 xml2_1.2.0
[82] DO.db_2.9 biomaRt_2.38.0 compiler_3.5.0
[85] rstudioapi_0.8 Guitar_1.20.0 tibble_2.0.1
[88] tweenr_1.0.0 geneplotter_1.60.0 stringi_1.2.4
[91] highr_0.7 lattice_0.20-38 Matrix_1.2-15
[94] vegan_2.5-3 permute_0.9-4 pillar_1.3.1
[97] triebeard_0.3.0 data.table_1.11.8 cowplot_0.9.3
[100] bitops_1.0-6 qvalue_2.14.0 R6_2.3.0
[103] latticeExtra_0.6-28 vcfR_1.8.0 gridExtra_2.3
[106] codetools_0.2-15 MASS_7.3-51.1 assertthat_0.2.0
[109] rprojroot_1.3-2 withr_2.1.2 pinfsc50_1.1.0
[112] GenomeInfoDbData_1.2.0 mgcv_1.8-26 hms_0.4.2
[115] rpart_4.1-13 tidyr_0.8.2 rvcheck_0.1.1
[118] rmarkdown_1.10 git2r_0.23.0 ggforce_0.1.3
[121] base64enc_0.1-3 This reproducible R Markdown analysis was created with workflowr 1.1.1