Simulation_analysis
scottzijiezhang
2019-10-25
RADAR model
##RADAR simulation model
####covariate_effect_size is set to 0.5, small. For example, the covariate can refer to sex(male/female) in this case. Male:1, female:0. So 1*covariate_effect_size will be added to the replicates testing males.
####gene/site specific intercept: assume to be normally distributed with mean=gene_specific_intercept_mean, sd=gene_specific_intercept_sd. Assume to be the same for all replicates.
####beta: the effect size for groups(conditons). I first ramdomly sample 4000 sites to be DM sites, and then generate uniformly distributed beta (4000) with lower limit=beta_low_limit, and higher limit=beta_high_limit. Finally I multiply those selected sites with those beta. ATTENTION: I randomly sample 4000 sites for each replicates seperately.
## normally distributed random effect parameter (psi) across peaks instead of fixed value for random effect parameter.
RADAR_simulateData2 <- function(n_Sites=20000,replicate=8, percent_diff_exp=0.2, covariate_effect_size=1,
psi=5, gene_specific_intercept_mean=5, gene_specific_intercept_sd=0.6,
beta = 0.5, seed = 1212) {
##condition:control/treatment
control<-matrix(0,nrow=n_Sites, ncol=replicate)
treatment<-matrix(1,nrow=n_Sites, ncol=replicate)
##covariate:sex,female/male
set.seed( seed )
control_female_index<-sort(sample(1:replicate,replicate/2))
####sample four indexes for female in control group
control_male_index<-setdiff(1:replicate,control_female_index)
control_sex<-rep( 0, replicate )
control_sex[control_male_index]<-1
####male set to 1
#set.seed(4)
treatment_female_index<-sort(sample(1:replicate,replicate/2))
####sample four indexs for female in treatment group
treatment_male_index<-setdiff(1:replicate,treatment_female_index)
treatment_sex<-rep(0,replicate)
treatment_sex[treatment_male_index]<-1
####male set to 1
control_sex_all_sites<-matrix(rep(control_sex, n_Sites), nrow = n_Sites, byrow = TRUE)
treatment_sex_all_sites<-matrix(rep(treatment_sex, n_Sites), nrow = n_Sites, byrow = TRUE)
####sample diff_sites: all replicate get the same set of diff sites, 4000(round(n_Sites*percent_diff_exp))
diff_site_all_replicates<- sample(n_Sites, round(n_Sites*percent_diff_exp))
nonDiff_site_all_replicates <- setdiff(1:n_Sites, diff_site_all_replicates)
#set.seed(12)
#beta_fold_change<- runif(round(n_Sites*percent_diff_exp),beta_low_limit,beta_high_limit)
beta_fold_change<-ifelse( diff_site_all_replicates %in% sample(diff_site_all_replicates,round(n_Sites*percent_diff_exp)/2) ,-beta,beta)
####generate 4000 betas (normally distributed centered at 0 )
##gene_specific_effect, gene/site specific intercept
mu<-rnorm(n_Sites,gene_specific_intercept_mean, gene_specific_intercept_sd)
#mu<- gene_specific_intercept_mean
##E: random effects, gamma distributed (can be adjusted)
psiDistri <- pmax( rnorm(n_Sites, mean = psi, sd = 1), 1 )
#sdDistri <- sapply(1:n_Sites, function(x) rnorm(1, mean = 5/(mu[x]), sd = 0.1) )
E<-t( sapply(1:n_Sites, function(x){ rgamma(replicate*2,shape = psiDistri[x], rate = psiDistri[x]) }) )
#E<-t( sapply(1:n_Sites, function(x){ exp(rnorm(replicate*2, mean = 0, sd = sdDistri[x] ) ) }) )
sex_beta <- ifelse( 1:n_Sites %in% sample(1:n_Sites,n_Sites/2) ,-covariate_effect_size,covariate_effect_size)
#sex_beta <- beta
##combine all parameters to generate final lamda, then to generate final data
control_all_replicates_pre<-NULL
treat_all_replicates_pre<-NULL
for (i in 1:replicate) {
conditions<-cbind(control[,i], treatment[,i])
conditions[diff_site_all_replicates,]<-conditions[diff_site_all_replicates,]*beta_fold_change
conditions[nonDiff_site_all_replicates,]<-conditions[nonDiff_site_all_replicates,]*0
sex<-cbind(control_sex_all_sites[,i], treatment_sex_all_sites[,i])
sex<-sex*sex_beta
control_all_replicates_pre<-cbind(control_all_replicates_pre, conditions[,1]+sex[,1])
treat_all_replicates_pre<-cbind(treat_all_replicates_pre, conditions[,2]+sex[,2])
}
control_all_replicates_pre<-control_all_replicates_pre + log( E[,1:replicate] )
treat_all_replicates_pre<-treat_all_replicates_pre + log( E[,(replicate+1):(2*replicate)] )
####add error, E
control_all_replicates_pre<-control_all_replicates_pre+mu
treat_all_replicates_pre<-treat_all_replicates_pre+mu
####add gene/site-specific intercept
control_all_replicates<-exp(control_all_replicates_pre)
treat_all_replicates<-exp(treat_all_replicates_pre)
meth1=matrix(0,n_Sites,replicate)
meth2=matrix(0,n_Sites,replicate)
for (i in 1:n_Sites) {
#meth1[i,]=rpois(replicate,lamda=control_all_replicates[i,])
#meth2[i,]=rpois(replicate,lamda=treat_all_replicates[i,])
for (j in 1:replicate) {
meth1[i,j]=max( 1, rpois(1,lambda=control_all_replicates[i,j]) )
meth2[i,j]=max( 1, rpois(1,lambda=treat_all_replicates[i,j]) )
}
}
res=list(4)
res[[paste0("IP_","control")]]=meth1
res[[paste0("IP_","treat")]]=meth2
res[["df_site_index"]]=diff_site_all_replicates
res[["variable"]]=data.frame(predictor = c(rep(0,replicate),rep(1,replicate)), sex = c(control_sex,treatment_sex) )
return (res)
}
load("~/Tools/RADARmannual/data/simulationAnalysis.RData")## fisher's exact test
fisherTest <- function( control_ip, treated_ip, control_input, treated_input ){
testResult <- foreach(i = 1:nrow(control_ip) , .combine = rbind )%do%{
tmpTest <- fisher.test( cbind( c( rowSums(treated_ip)[i], rowSums(treated_input)[i] ), c( rowSums(control_ip)[i], rowSums(control_input)[i] ) ), alternative = "two.sided" )
data.frame( logFC = log(tmpTest$estimate), pvalue = tmpTest$p.value )
}
#rm(list=ls(name=foreach:::.foreachGlobals), pos=foreach:::.foreachGlobals)
testResult$fdr <- p.adjust(testResult$pvalue, method = "fdr")
return(testResult)
}
## wrapper for the MetDiff test
MeTDiffTest <- function( control_ip, treated_ip, control_input, treated_input ){
testResult <- foreach(i = 1:nrow(control_ip) , .combine = rbind )%do%{
x <- t( as.matrix(control_ip[i,]) )
y <- t( as.matrix(control_input[i,]) )
xx <- t( as.matrix(treated_ip[i,]) )
yy <- t( as.matrix(treated_input[i,]) )
xxx = cbind(x,xx)
yyy = cbind(y,yy)
logl1 <- MeTDiff:::.betabinomial.lh(x,y+1)
logl2 <- MeTDiff:::.betabinomial.lh(xx,yy+1)
logl3 <- MeTDiff:::.betabinomial.lh(xxx,yyy+1)
tst <- (logl1$logl+logl2$logl-logl3$logl)*2
pvalues <- 1 - pchisq(tst,2)
log.fc <- log( (sum(xx)+1)/(1+sum(yy)) * (1+sum(y))/(1+sum(x)) )
data.frame( logFC = log.fc, pvalue = pvalues )
}
#rm(list=ls(name=foreach:::.foreachGlobals), pos=foreach:::.foreachGlobals)
testResult$fdr <- p.adjust(testResult$pvalue, method = "fdr")
return(testResult)
}
Bltest <- function(control_ip, treated_ip, control_input, treated_input){
control_ip_total <- sum(colSums(control_ip))
control_input_total <- sum(colSums(control_input))
treated_ip_total <- sum(colSums(treated_ip))
treated_input_total <- sum(colSums(treated_input))
print("start test...")
tmpResult <- do.call(cbind.data.frame, exomePeak::bltest(rowSums(control_ip), rowSums(control_input),rowSums(treated_ip),rowSums(treated_input),control_ip_total, control_input_total, treated_ip_total, treated_input_total) )
return( data.frame(logFC = tmpResult[,"log.fc"], pvalue = exp(tmpResult[,"log.p"]), fdr = exp(tmpResult$log.fdr) ) )
}Simple case
We first test the simple scenario where there are 8 cases and 8 controls without covariates.
#### beta = 0.5
library(RADAR)
##
load("~/Tools/RADARmannual/data/Input.RData")
registerDoParallel(10)
simuTest_beta0.5 <- foreach(i = 1:10, .combine = rbind)%dopar%{
iteration <- RADAR_simulateData2(n_Sites = 26324,beta = 0.5, replicate = 8, psi = 5 , gene_specific_intercept_mean = 5, covariate_effect_size = 0 , seed = i*3)
simu.radar <- MeRIP.RADAR()
variable(simu.radar) <- data.frame( iteration$variable$predictor )
simu.radar@Peak_adjExpr <- cbind(iteration$IP_control, iteration$IP_treat)
rownames(simu.radar@Peak_adjExpr) <- paste0("peak",1:26324)
samplenames(simu.radar) <- paste0("s",1:16)
simu.radar@Peak_input <- cbind(CtlInput, TreatInput)
colnames(simu.radar@Peak_input ) <- paste0("s",1:16)
enrichRaio <- t( apply(simu.radar@Peak_adjExpr, 1, function(x){x/mean(x)}*max(1,rnorm(1,5.6,2) ) ) )
simu.radar@Peak_ip <- round( simu.radar@Peak_input * enrichRaio )
simu.radar <- diffIP( simu.radar )
simu.radar@test.est[,"fdr"] <- p.adjust(simu.radar@test.est[,"p_value"],method = "fdr")
simu.Metdiff <- MeTDiffTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.qnb <- QNB::qnbtest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16],plot.dispersion = FALSE)
simu.fisher <- fisherTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.exomePeak <- Bltest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
result <- NULL
for(q in c(0.01, 0.05, 0.1, 0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9,0.95,0.99)){
tmpPower <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" =simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), iteration$df_site_index ) )/length(iteration$df_site_index)})
tmpFDR <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length(which( x < q ) )})
tmpFalsePosRate <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length( setdiff( 1:26324, iteration$df_site_index ) ) })
result <- rbind(result, data.frame( power = tmpPower, fdr = tmpFDR, fpr = tmpFalsePosRate, method = c("RADAR","MeTDiff","exomePeak","Fisher","QNB") , fdr_cutoff = q ) )
}
result
}
save.image("~/Tools/RADARmannual/data/simulationAnalysis.RData")beta = 0.75
registerDoParallel(10)
simuTest_beta0.75 <- foreach(i = 1:10, .combine = rbind)%dopar%{
iteration <- RADAR_simulateData2(n_Sites = 26324,beta = 0.75, replicate = 8, psi = 5 , gene_specific_intercept_mean = 5, covariate_effect_size = 0 , seed = i*3)
simu.radar <- MeRIP.RADAR()
variable(simu.radar) <- data.frame( iteration$variable$predictor )
simu.radar@Peak_adjExpr <- cbind(iteration$IP_control, iteration$IP_treat)
rownames(simu.radar@Peak_adjExpr) <- paste0("peak",1:26324)
samplenames(simu.radar) <- paste0("s",1:16)
simu.radar@Peak_input <- cbind(CtlInput, TreatInput)
colnames(simu.radar@Peak_input ) <- paste0("s",1:16)
enrichRaio <- t( apply(simu.radar@Peak_adjExpr, 1, function(x){x/mean(x)}*max(1,rnorm(1,5.6,2) ) ) )
simu.radar@Peak_ip <- round( simu.radar@Peak_input * enrichRaio )
simu.radar <- diffIP( simu.radar )
simu.radar@test.est[,"fdr"] <- p.adjust(simu.radar@test.est[,"p_value"],method = "fdr")
simu.Metdiff <- MeTDiffTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.qnb <- QNB::qnbtest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16],plot.dispersion = FALSE)
simu.fisher <- fisherTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.exomePeak <- Bltest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
result <- NULL
for(q in c(0.01, 0.05, 0.1, 0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9,0.95,0.99)){
tmpPower <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" =simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), iteration$df_site_index ) )/length(iteration$df_site_index)})
tmpFDR <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length(which( x < q ) )})
tmpFalsePosRate <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length( setdiff( 1:26324, iteration$df_site_index ) ) })
result <- rbind(result, data.frame( power = tmpPower, fdr = tmpFDR, fpr = tmpFalsePosRate, method = c("RADAR","MeTDiff","exomePeak","Fisher","QNB") , fdr_cutoff = q ) )
}
result
}
save.image("~/Tools/RADARmannual/data/simulationAnalysis.RData")beta = 1
registerDoParallel(10)
simuTest_beta1 <- foreach(i = 1:10, .combine = rbind)%dopar%{
iteration <- RADAR_simulateData2(n_Sites = 26324,beta = 1, replicate = 8, gene_specific_intercept_mean = 5, covariate_effect_size = 0 , seed = i*3)
simu.radar <- MeRIP.RADAR()
variable(simu.radar) <- data.frame( iteration$variable$predictor )
simu.radar@Peak_adjExpr <- cbind(iteration$IP_control, iteration$IP_treat)
rownames(simu.radar@Peak_adjExpr) <- paste0("peak",1:26324)
samplenames(simu.radar) <- paste0("s",1:16)
simu.radar@Peak_input <- cbind(CtlInput, TreatInput)
colnames(simu.radar@Peak_input ) <- paste0("s",1:16)
enrichRaio <- t( apply(simu.radar@Peak_adjExpr, 1, function(x){x/mean(x)}*max(1,rnorm(1,5.6,2) ) ) )
simu.radar@Peak_ip <- round( simu.radar@Peak_input * enrichRaio )
simu.radar <- diffIP( simu.radar )
simu.radar@test.est[,"fdr"] <- p.adjust(simu.radar@test.est[,"p_value"],method = "fdr")
simu.Metdiff <- MeTDiffTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.qnb <- QNB::qnbtest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16],plot.dispersion = FALSE)
simu.fisher <- fisherTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.exomePeak <- Bltest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
result <- NULL
for(q in c(0.01, 0.05, 0.1, 0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9,0.95,0.99)){
tmpPower <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" =simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), iteration$df_site_index ) )/length(iteration$df_site_index)})
tmpFDR <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length(which( x < q ) )})
tmpFalsePosRate <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length( setdiff( 1:26324, iteration$df_site_index ) ) })
result <- rbind(result, data.frame( power = tmpPower, fdr = tmpFDR, fpr = tmpFalsePosRate, method = c("RADAR","MeTDiff","exomePeak","Fisher","QNB") , fdr_cutoff = q ) )
}
result
}
save.image("~/Tools/RADARmannual/data/simulationAnalysis.RData")library(RADAR)
g_legend <- function(a.gplot){
tmp <- ggplot_gtable(ggplot_build(a.gplot))
leg <- which(sapply(tmp$grobs, function(x) x$name) == "guide-box")
legend <- tmp$grobs[[leg]]
return(legend)}
p1 <- ggplot(simuTest_beta0.5, aes(x = fdr, y = power, colour = method))+geom_point()+theme_bw()+ggtitle("beta = 0.5")+theme(legend.position = "none", text = element_text(face = "bold"))
p2 <- ggplot(simuTest_beta0.75, aes(x = fdr, y = power, colour = method))+geom_point()+theme_bw()+ggtitle("beta = 0.75")+theme(legend.position = "none", text = element_text(face = "bold"))
p3 <- ggplot(simuTest_beta1, aes(x = fdr, y = power, colour = method))+geom_point()+theme_bw()+ggtitle("beta = 1")+theme(legend.position = "none", text = element_text(face = "bold"))
len <- ggplot(simuTest_beta1, aes(x = fdr, y = power, colour = method))+geom_point()+theme(plot.title = element_text(face = "bold", size = 10,hjust = 0.5), legend.title = element_text(face = "bold", size = 15),legend.text = element_text(face = "bold", size = 15) )
gridExtra::grid.arrange(p1,p2,p3,g_legend(len), ncol = 4)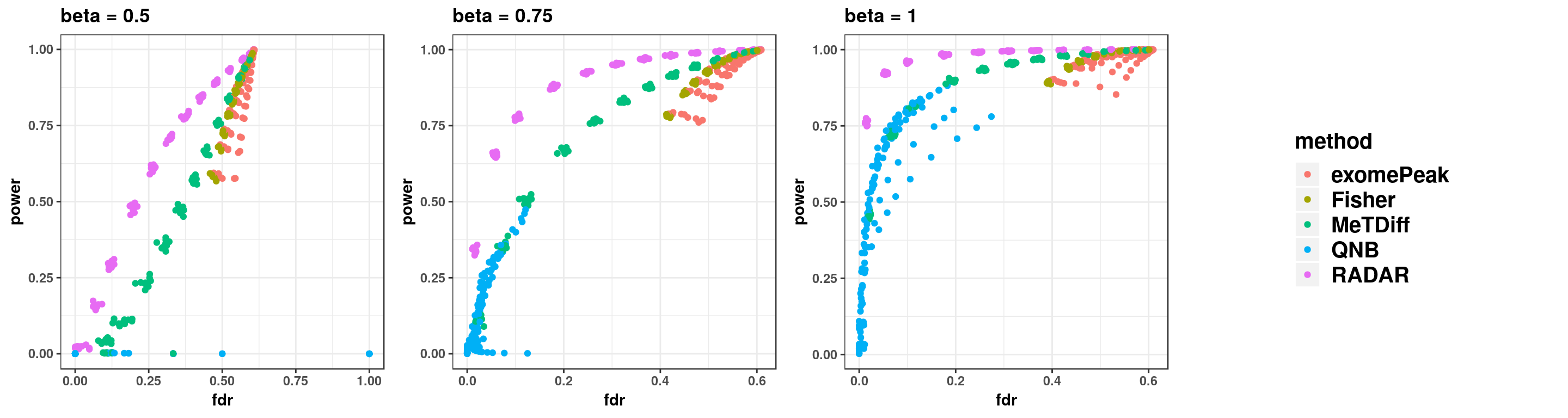
simuTest_beta0.5_mean <- data.frame(FDR = tapply(simuTest_beta0.5$fdr, paste0(simuTest_beta0.5$method,simuTest_beta0.5$fdr_cutoff), mean, na.rm=T),
Power = tapply(simuTest_beta0.5$power, paste0(simuTest_beta0.5$method,simuTest_beta0.5$fdr_cutoff), mean, na.rm=T),
method = tapply(simuTest_beta0.5$method, paste0(simuTest_beta0.5$method,simuTest_beta0.5$fdr_cutoff), function(x) unique(as.character(x)) ),
fdr_cutoff = as.factor(tapply(simuTest_beta0.5$fdr_cutoff, paste0(simuTest_beta0.5$method,simuTest_beta0.5$fdr_cutoff),unique) )
)
simuTest_beta0.75_mean <- data.frame(FDR = tapply(simuTest_beta0.75$fdr, paste0(simuTest_beta0.75$method,simuTest_beta0.75$fdr_cutoff), mean, na.rm=T),
Power = tapply(simuTest_beta0.75$power, paste0(simuTest_beta0.75$method,simuTest_beta0.75$fdr_cutoff), mean, na.rm=T),
method = tapply(simuTest_beta0.75$method, paste0(simuTest_beta0.75$method,simuTest_beta0.75$fdr_cutoff), function(x) unique(as.character(x)) ),
fdr_cutoff = as.factor(tapply(simuTest_beta0.75$fdr_cutoff, paste0(simuTest_beta0.75$method,simuTest_beta0.75$fdr_cutoff),unique ))
)
simuTest_beta1_mean <- data.frame(FDR = tapply(simuTest_beta1$fdr, paste0(simuTest_beta1$method,simuTest_beta1$fdr_cutoff), mean, na.rm=T),
Power = tapply(simuTest_beta1$power, paste0(simuTest_beta1$method,simuTest_beta1$fdr_cutoff), mean, na.rm=T),
method = tapply(simuTest_beta1$method, paste0(simuTest_beta1$method,simuTest_beta1$fdr_cutoff), function(x) unique(as.character(x)) ),
fdr_cutoff = as.factor(tapply(simuTest_beta1$fdr_cutoff, paste0(simuTest_beta1$method,simuTest_beta1$fdr_cutoff), unique))
)
p1 <- ggplot(simuTest_beta0.5_mean)+geom_point( aes(x = FDR, y = Power, colour = method, size = fdr_cutoff))+geom_line(aes(x = FDR, y = Power, colour = method))+theme_bw()+ggtitle("beta = 0.5")+theme(legend.position = "none", text = element_text(face = "bold", size = 18), axis.text = element_text(face = "bold", size = 15,colour = "black"), plot.title = element_text(hjust = 0.5) )
p2 <- ggplot(simuTest_beta0.75_mean )+geom_point( aes(x = FDR, y = Power, colour = method, size = fdr_cutoff))+geom_line(aes(x = FDR, y = Power, colour = method))+theme_bw()+ggtitle("beta = 0.75")+theme(legend.position = "none", text = element_text(face = "bold", size = 18), axis.text = element_text(face = "bold", size = 15,colour = "black"), plot.title = element_text(hjust = 0.5) )
p3 <- ggplot(simuTest_beta1_mean)+geom_point( aes(x = FDR, y = Power, colour = method, size = fdr_cutoff))+geom_line(aes(x = FDR, y = Power, colour = method))+theme_bw()+ggtitle("beta = 1")+theme(legend.position = "none", text = element_text(face = "bold", size = 18), axis.text = element_text(face = "bold", size = 15,colour = "black"), plot.title = element_text(hjust = 0.5) )
len <- ggplot(simuTest_beta1_mean, aes(x = FDR, y = Power, colour = method, size = fdr_cutoff))+geom_point()+theme_bw()+theme(plot.title = element_text(face = "bold", size = 10,hjust = 0.5), legend.title = element_text(face = "bold", size = 15),legend.text = element_text(face = "bold", size = 15) )
gridExtra::grid.arrange(p1,p2,p3,g_legend(len), ncol = 4)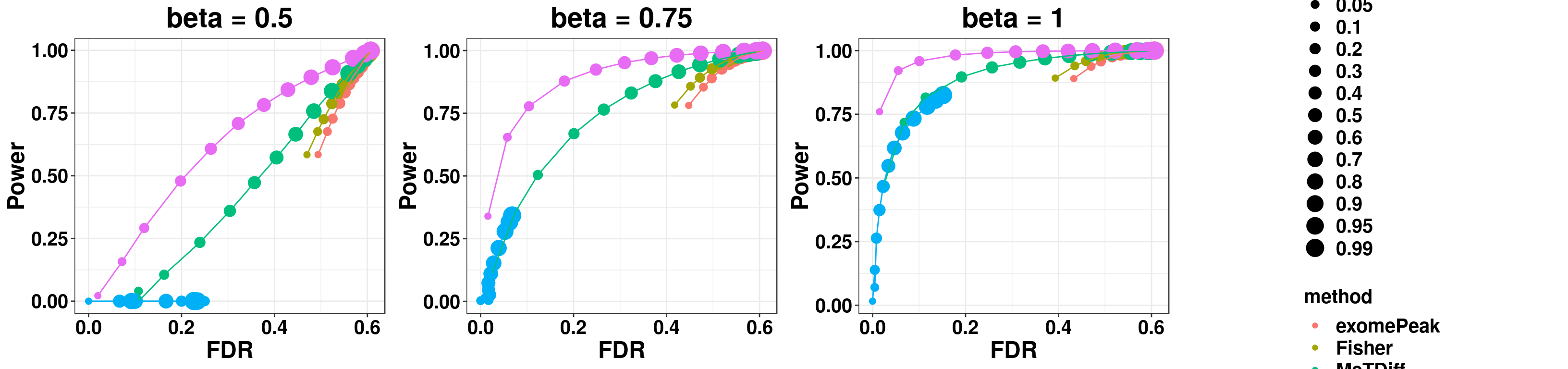
RADARmodel_combined <- dplyr::mutate( rbind(simuTest_beta0.5, simuTest_beta0.75,simuTest_beta1) , beta = c(rep("beta = 0.5",nrow(simuTest_beta1)),rep("beta = 0.75",nrow(simuTest_beta1)),rep("beta = 1",nrow(simuTest_beta1)) ) )
ggplot(RADARmodel_combined[RADARmodel_combined$fdr_cutoff == 0.1,] , aes(x = method, y = fdr, colour = method))+geom_boxplot()+facet_wrap(beta~.) + geom_abline(slope = 0,intercept = 0.1,lty=2)+ylab("FDR")+xlab("Methods")+ggtitle("Without Covariates") +theme_bw()+theme(legend.position = "none", text = element_text(face = "bold",colour = "black"), axis.text = element_text(size = 12,colour = "black"), axis.title = element_text(size = 16), strip.text = element_text(size = 16) , plot.title = element_text(hjust = 0.5) )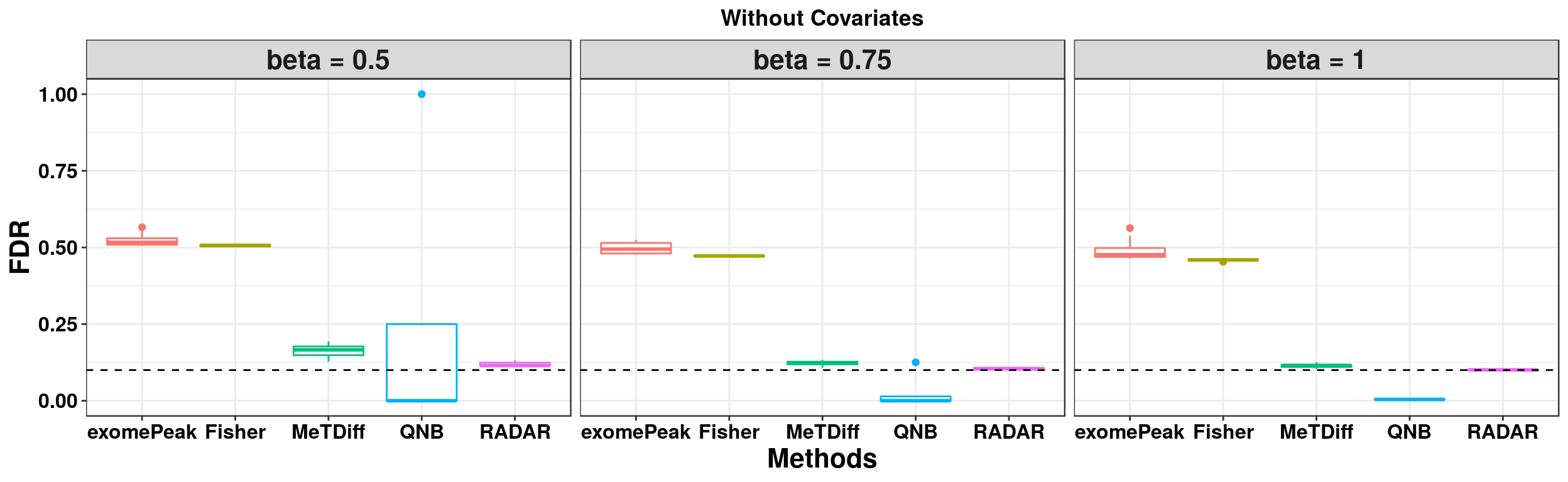
ggplot(RADARmodel_combined[RADARmodel_combined$fdr_cutoff == 0.1,] , aes(x = method, y = power, colour = method))+geom_boxplot()+facet_wrap(beta~.) + geom_abline(slope = 0,intercept = 1,lty=2)+ylab("Sensitivity")+xlab("Methods") +ggtitle("Without Covariates")+theme_bw()+theme(legend.position = "none", text = element_text(face = "bold",colour = "black"), axis.text = element_text(size = 12,colour = "black"), axis.title = element_text(size = 16), strip.text = element_text(size = 16) , plot.title = element_text(hjust = 0.5) )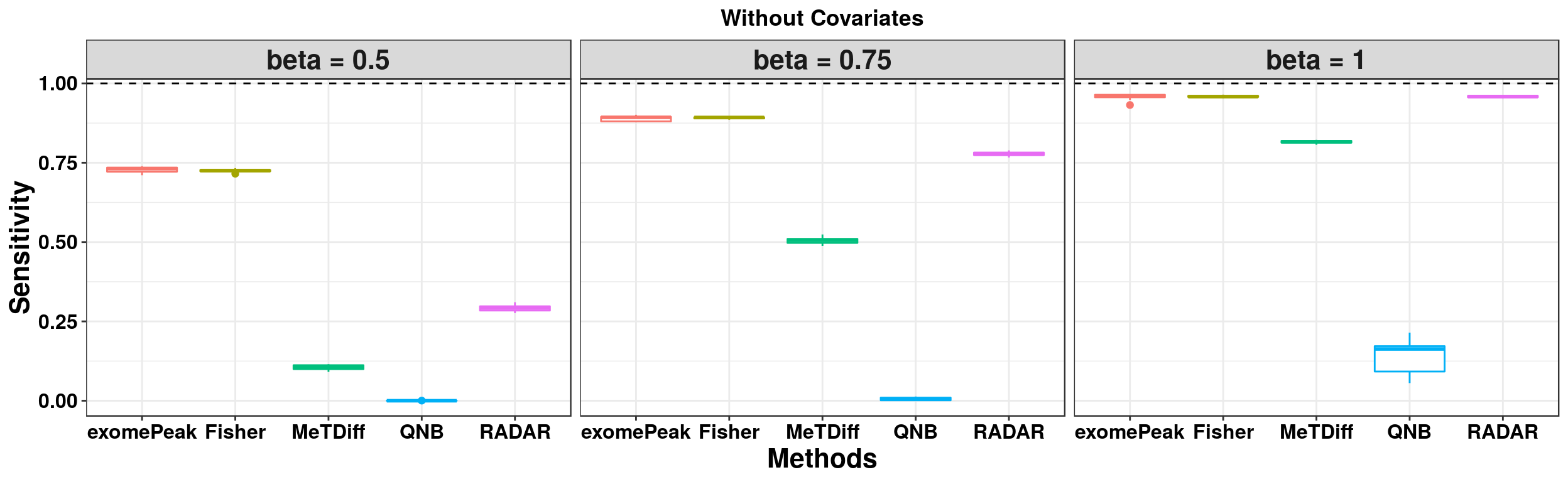
ggplot(RADARmodel_combined[RADARmodel_combined$fdr_cutoff == 0.1,] , aes(x = fdr, y = power, colour = method, shape = method))+geom_point(size=4)+facet_wrap(beta~.) + geom_vline(xintercept = 0.1,lty=2)+ylab("Sensitivity")+xlab("FDR")+ggtitle("RADAR Model Without Covariates") +theme_bw()+theme( text = element_text(face = "bold",colour = "black"), axis.text = element_text(size = 12,colour = "black"), axis.title = element_text(size = 16), strip.text = element_text(size = 16), plot.title = element_text(hjust = 0.5), legend.text = element_text(size = 15) )+scale_y_continuous(limits = c(0,1))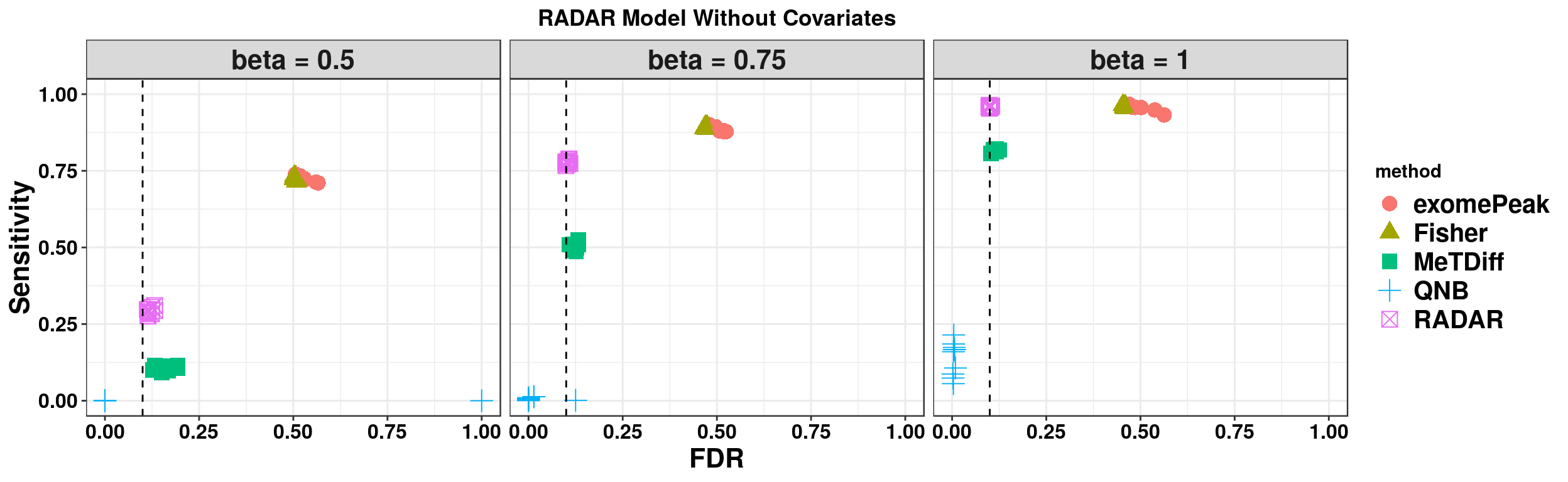
Complex case
We now test a more complex scenario where there are 8 cases and 8 controls and in each group, half of the samples are in different gender, which is a covariate that is often encountered in real study.
registerDoParallel(10)
simuTest_cov_beta0.5 <- foreach(i = 1:10, .combine = rbind)%dopar%{
iteration <- RADAR_simulateData2(n_Sites = 26324,beta = 0.5, replicate = 8, gene_specific_intercept_mean = 5, covariate_effect_size = 2 , seed = i*3)
simu.radar <- MeRIP.RADAR()
variable(simu.radar) <- iteration$variable
simu.radar@Peak_adjExpr <- cbind(iteration$IP_control, iteration$IP_treat)
rownames(simu.radar@Peak_adjExpr) <- paste0("peak",1:26324)
samplenames(simu.radar) <- paste0("s",1:16)
simu.radar@Peak_input <- cbind(CtlInput, TreatInput)
colnames(simu.radar@Peak_input ) <- paste0("s",1:16)
enrichRaio <- t( apply(simu.radar@Peak_adjExpr, 1, function(x){x/mean(x)}*max(1,rnorm(1,5.6,2) ) ) )
simu.radar@Peak_ip <- round( simu.radar@Peak_input * enrichRaio )
simu.radar <- diffIP( simu.radar )
simu.radar@test.est[,"fdr"] <- p.adjust(simu.radar@test.est[,"p_value"],method = "fdr")
simu.Metdiff <- MeTDiffTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.qnb <- QNB::qnbtest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16],plot.dispersion = FALSE)
simu.fisher <- fisherTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.exomePeak <- Bltest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
result <- NULL
for(q in c(0.01, 0.05, 0.1, 0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9,0.95,0.99)){
tmpPower <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" =simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), iteration$df_site_index ) )/length(iteration$df_site_index)})
tmpFDR <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length(which( x < q ) )})
tmpFalsePosRate <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length( setdiff( 1:26324, iteration$df_site_index ) ) })
result <- rbind(result, data.frame( power = tmpPower, fdr = tmpFDR, fpr = tmpFalsePosRate, method = c("RADAR","MeTDiff","exomePeak","Fisher","QNB") , fdr_cutoff = q ) )
}
result
}
simuTest_cov_beta0.75 <- foreach(i = 1:10, .combine = rbind)%dopar%{
iteration <- RADAR_simulateData2(n_Sites = 26324,beta = 0.75, replicate = 8,psi = 5, gene_specific_intercept_mean = 5, covariate_effect_size = 2 , seed = i*3)
simu.radar <- MeRIP.RADAR()
variable(simu.radar) <- iteration$variable
simu.radar@Peak_adjExpr <- cbind(iteration$IP_control, iteration$IP_treat)
rownames(simu.radar@Peak_adjExpr) <- paste0("peak",1:26324)
samplenames(simu.radar) <- paste0("s",1:16)
simu.radar@Peak_input <- cbind(CtlInput, TreatInput)
colnames(simu.radar@Peak_input ) <- paste0("s",1:16)
enrichRaio <- t( apply(simu.radar@Peak_adjExpr, 1, function(x){x/mean(x)}*max(1,rnorm(1,5.6,2) ) ) )
simu.radar@Peak_ip <- round( simu.radar@Peak_input * enrichRaio )
simu.radar <- diffIP( simu.radar )
simu.radar@test.est[,"fdr"] <- p.adjust(simu.radar@test.est[,"p_value"],method = "fdr")
simu.Metdiff <- MeTDiffTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.qnb <- QNB::qnbtest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16],plot.dispersion = FALSE)
simu.fisher <- fisherTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.exomePeak <- Bltest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
result <- NULL
for(q in c(0.01, 0.05, 0.1, 0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9,0.95,0.99)){
tmpPower <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" =simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), iteration$df_site_index ) )/length(iteration$df_site_index)})
tmpFDR <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length(which( x < q ) )})
tmpFalsePosRate <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length( setdiff( 1:26324, iteration$df_site_index ) ) })
result <- rbind(result, data.frame( power = tmpPower, fdr = tmpFDR, fpr = tmpFalsePosRate, method = c("RADAR","MeTDiff","exomePeak","Fisher","QNB") , fdr_cutoff = q ) )
}
result
}
simuTest_cov_beta1 <- foreach(i = 1:10, .combine = rbind)%dopar%{
iteration <- RADAR_simulateData2(n_Sites = 26324,beta = 1, replicate = 8,psi = 5, gene_specific_intercept_mean = 5, covariate_effect_size = 2 , seed = i*3)
simu.radar <- MeRIP.RADAR()
variable(simu.radar) <- iteration$variable
simu.radar@Peak_adjExpr <- cbind(iteration$IP_control, iteration$IP_treat)
rownames(simu.radar@Peak_adjExpr) <- paste0("peak",1:26324)
samplenames(simu.radar) <- paste0("s",1:16)
simu.radar@Peak_input <- cbind(CtlInput, TreatInput)
colnames(simu.radar@Peak_input ) <- paste0("s",1:16)
enrichRaio <- t( apply(simu.radar@Peak_adjExpr, 1, function(x){x/mean(x)}*max(1,rnorm(1,5.6,2) ) ) )
simu.radar@Peak_ip <- round( simu.radar@Peak_input * enrichRaio )
simu.radar <- diffIP( simu.radar )
simu.radar@test.est[,"fdr"] <- p.adjust(simu.radar@test.est[,"p_value"],method = "fdr")
simu.Metdiff <- MeTDiffTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.qnb <- QNB::qnbtest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16],plot.dispersion = FALSE)
simu.fisher <- fisherTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.exomePeak <- Bltest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
result <- NULL
for(q in c(0.01, 0.05, 0.1, 0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9,0.95,0.99)){
tmpPower <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" =simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), iteration$df_site_index ) )/length(iteration$df_site_index)})
tmpFDR <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length(which( x < q ) )})
tmpFalsePosRate <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length( setdiff( 1:26324, iteration$df_site_index ) ) })
result <- rbind(result, data.frame( power = tmpPower, fdr = tmpFDR, fpr = tmpFalsePosRate, method = c("RADAR","MeTDiff","exomePeak","Fisher","QNB") , fdr_cutoff = q ) )
}
result
}
save.image("~/Tools/RADARmannual/data/simulationAnalysis.RData")g_legend <- function(a.gplot){
tmp <- ggplot_gtable(ggplot_build(a.gplot))
leg <- which(sapply(tmp$grobs, function(x) x$name) == "guide-box")
legend <- tmp$grobs[[leg]]
return(legend)}
p11 <- ggplot(simuTest_cov_beta0.5, aes(x = fdr, y = power, colour = method))+geom_point()+theme_bw()+ggtitle("beta = 0.5")+theme(legend.position = "none", text = element_text(face = "bold"))
p22 <- ggplot(simuTest_cov_beta0.75, aes(x = fdr, y = power, colour = method))+geom_point()+theme_bw()+ggtitle("beta = 0.75")+theme(legend.position = "none", text = element_text(face = "bold"))
p33 <- ggplot(simuTest_cov_beta1, aes(x = fdr, y = power, colour = method))+geom_point()+theme_bw()+ggtitle("beta = 1")+theme(legend.position = "none", text = element_text(face = "bold"))
len <- ggplot(simuTest_cov_beta1, aes(x = fdr, y = power, colour = method))+geom_point()+theme(plot.title = element_text(face = "bold", size = 10,hjust = 0.5), legend.title = element_text(face = "bold", size = 15),legend.text = element_text(face = "bold", size = 15) )
gridExtra::grid.arrange(p11,p22,p33,g_legend(len), ncol = 4)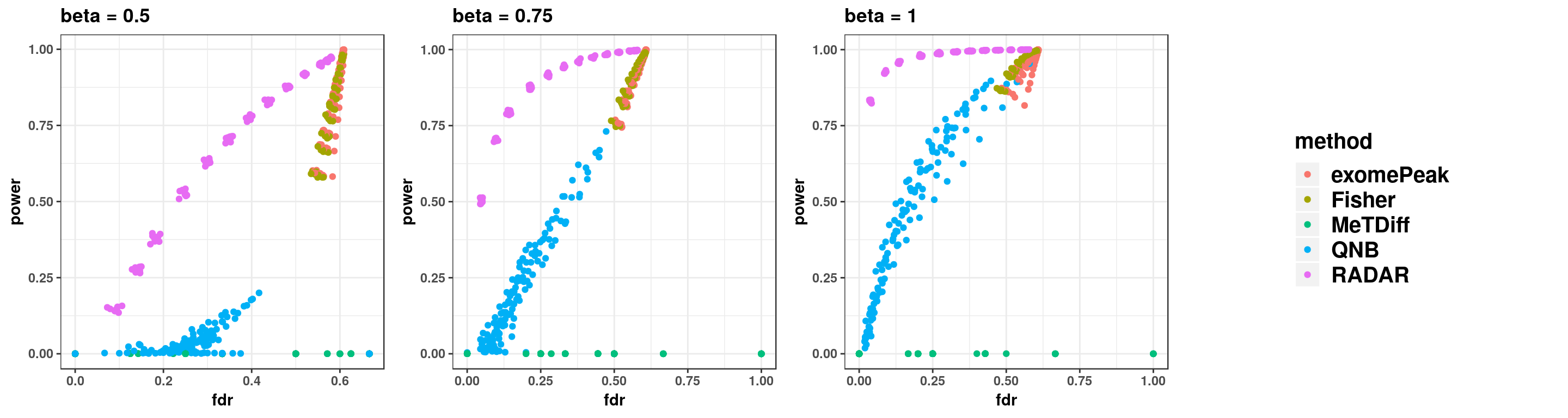
simuTest_cov_beta0.5_mean <- data.frame(FDR = tapply(simuTest_cov_beta0.5$fdr, paste0(simuTest_cov_beta0.5$method,simuTest_cov_beta0.5$fdr_cutoff), mean, na.rm=TRUE),
Power = tapply(simuTest_cov_beta0.5$power, paste0(simuTest_cov_beta0.5$method,simuTest_cov_beta0.5$fdr_cutoff), mean, na.rm=TRUE),
method = tapply(simuTest_cov_beta0.5$method, paste0(simuTest_cov_beta0.5$method,simuTest_cov_beta0.5$fdr_cutoff), function(x) unique(as.character(x)) ),
fdr_cutoff = as.factor(tapply(simuTest_cov_beta0.5$fdr_cutoff, paste0(simuTest_cov_beta0.5$method,simuTest_cov_beta0.5$fdr_cutoff),unique) )
)
simuTest_cov_beta0.75_mean <- data.frame(FDR = tapply(simuTest_cov_beta0.75$fdr, paste0(simuTest_cov_beta0.75$method,simuTest_cov_beta0.75$fdr_cutoff), mean, na.rm=TRUE),
Power = tapply(simuTest_cov_beta0.75$power, paste0(simuTest_cov_beta0.75$method,simuTest_cov_beta0.75$fdr_cutoff), mean, na.rm=TRUE),
method = tapply(simuTest_cov_beta0.75$method, paste0(simuTest_cov_beta0.75$method,simuTest_cov_beta0.75$fdr_cutoff), function(x) unique(as.character(x)) ),
fdr_cutoff = as.factor(tapply(simuTest_cov_beta0.75$fdr_cutoff, paste0(simuTest_cov_beta0.75$method,simuTest_cov_beta0.75$fdr_cutoff),unique ))
)
simuTest_cov_beta1_mean <- data.frame(FDR = tapply(simuTest_cov_beta1$fdr, paste0(simuTest_cov_beta1$method,simuTest_cov_beta1$fdr_cutoff), mean, na.rm=TRUE),
Power = tapply(simuTest_cov_beta1$power, paste0(simuTest_cov_beta1$method,simuTest_cov_beta1$fdr_cutoff), mean, na.rm=TRUE),
method = tapply(simuTest_cov_beta1$method, paste0(simuTest_cov_beta1$method,simuTest_cov_beta1$fdr_cutoff), function(x) unique(as.character(x)) ),
fdr_cutoff = as.factor(tapply(simuTest_cov_beta1$fdr_cutoff, paste0(simuTest_cov_beta1$method,simuTest_cov_beta1$fdr_cutoff), unique))
)
p1 <- ggplot(simuTest_cov_beta0.5_mean)+geom_point( aes(x = FDR, y = Power, colour = method, size = fdr_cutoff))+geom_line(aes(x = FDR, y = Power, colour = method))+theme_bw()+ggtitle("beta = 0.5")+theme(legend.position = "none", text = element_text(face = "bold", size = 18), axis.text = element_text(face = "bold", size = 15,colour = "black"), plot.title = element_text(hjust = 0.5) )
p2 <- ggplot(simuTest_cov_beta0.75_mean)+geom_point( aes(x = FDR, y = Power, colour = method, size = fdr_cutoff))+geom_line(aes(x = FDR, y = Power, colour = method))+theme_bw()+ggtitle("beta = 0.75")+theme(legend.position = "none", text = element_text(face = "bold", size = 18), axis.text = element_text(face = "bold", size = 15,colour = "black"), plot.title = element_text(hjust = 0.5) )
p3 <- ggplot(simuTest_cov_beta1_mean)+geom_point( aes(x = FDR, y = Power, colour = method, size = fdr_cutoff))+geom_line(aes(x = FDR, y = Power, colour = method))+theme_bw()+ggtitle("beta = 1")+theme(legend.position = "none", text = element_text(face = "bold", size = 18), axis.text = element_text(face = "bold", size = 15,colour = "black"), plot.title = element_text(hjust = 0.5) )
len <- ggplot(simuTest_cov_beta1_mean, aes(x = FDR, y = Power, colour = method, size = fdr_cutoff))+geom_point()+theme_bw()+theme(plot.title = element_text(face = "bold", size = 10,hjust = 0.5), legend.title = element_text(face = "bold", size = 15),legend.text = element_text(face = "bold", size = 15) )
gridExtra::grid.arrange(p1,p2,p3,g_legend(len), ncol = 4)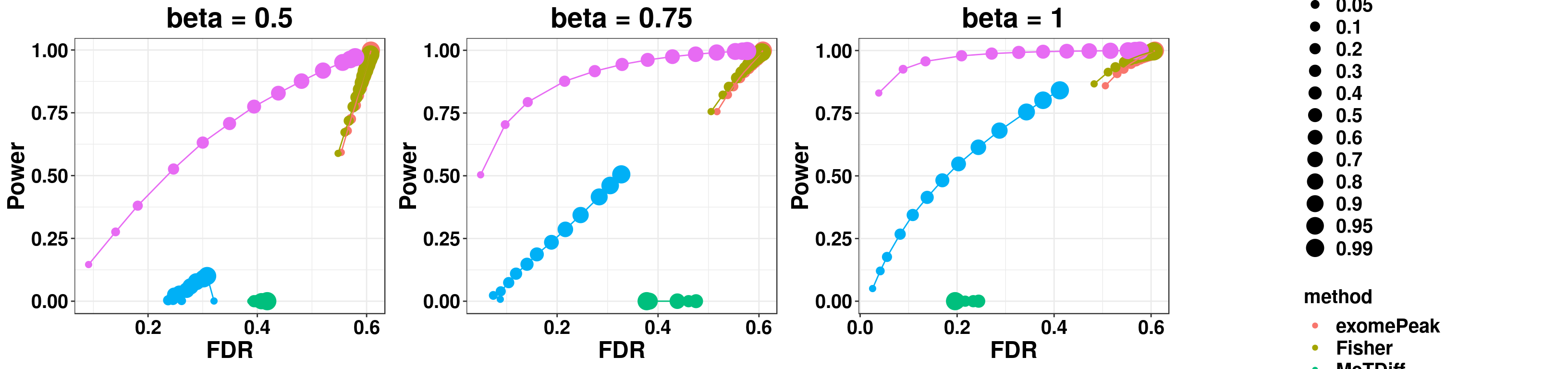
RADARmodel_cov_combined <- dplyr::mutate( rbind(simuTest_cov_beta0.5, simuTest_cov_beta0.75,simuTest_cov_beta1) , beta = c(rep("beta = 0.5",nrow(simuTest_cov_beta0.5)),rep("beta = 0.75",nrow(simuTest_cov_beta0.5)),rep("beta = 1",nrow(simuTest_cov_beta0.5)) ) )
ggplot(RADARmodel_cov_combined[RADARmodel_cov_combined$fdr_cutoff == 0.1,] , aes(x = method, y = fdr, colour = method))+geom_boxplot()+facet_wrap(beta~.) + geom_abline(slope = 0,intercept = 0.1,lty=2)+ylab("FDR")+xlab("Methods")+ ggtitle("With Covariates") +theme_bw()+theme(legend.position = "none", text = element_text(face = "bold",colour = "black"), axis.text = element_text(size = 12,colour = "black"), axis.title = element_text(size = 16), strip.text = element_text(size = 16), plot.title = element_text(hjust = 0.5) )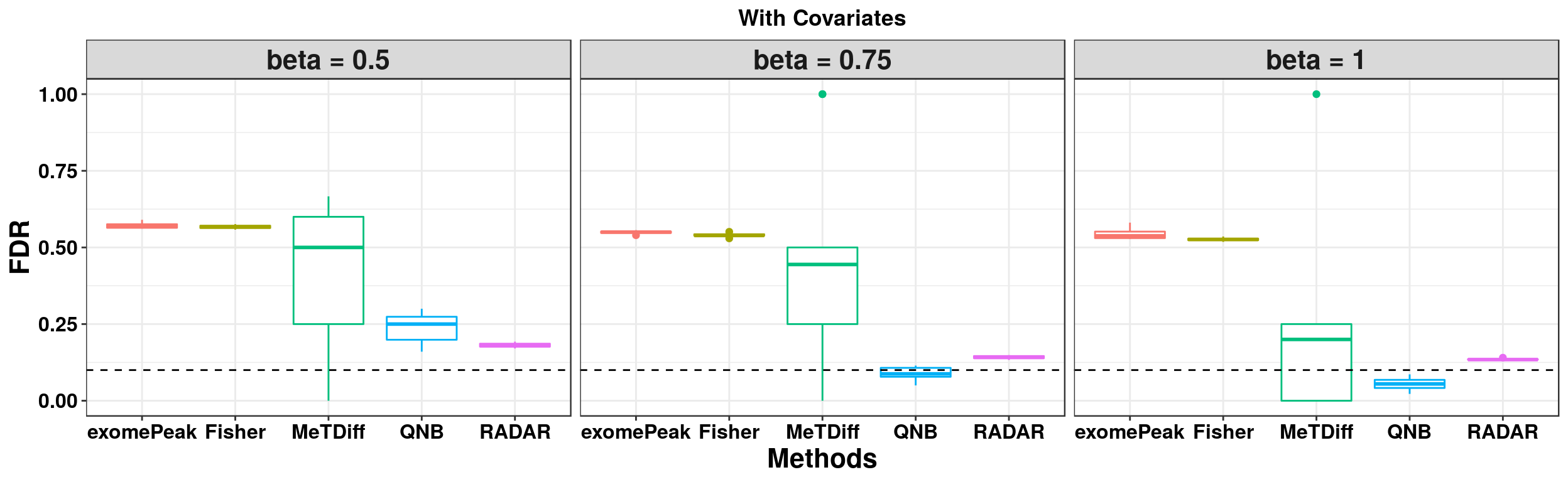
ggplot(RADARmodel_cov_combined[RADARmodel_cov_combined$fdr_cutoff == 0.1,] , aes(x = method, y = power, colour = method))+geom_boxplot()+facet_wrap(beta~.) + geom_abline(slope = 0,intercept = 1,lty=2)+ylab("Sensitivity")+xlab("Methods")+ggtitle("With Covariates") +theme_bw()+theme(legend.position = "none", text = element_text(face = "bold",colour = "black"), axis.text = element_text(size = 12,colour = "black"), axis.title = element_text(size = 16), strip.text = element_text(size = 16), plot.title = element_text(hjust = 0.5) )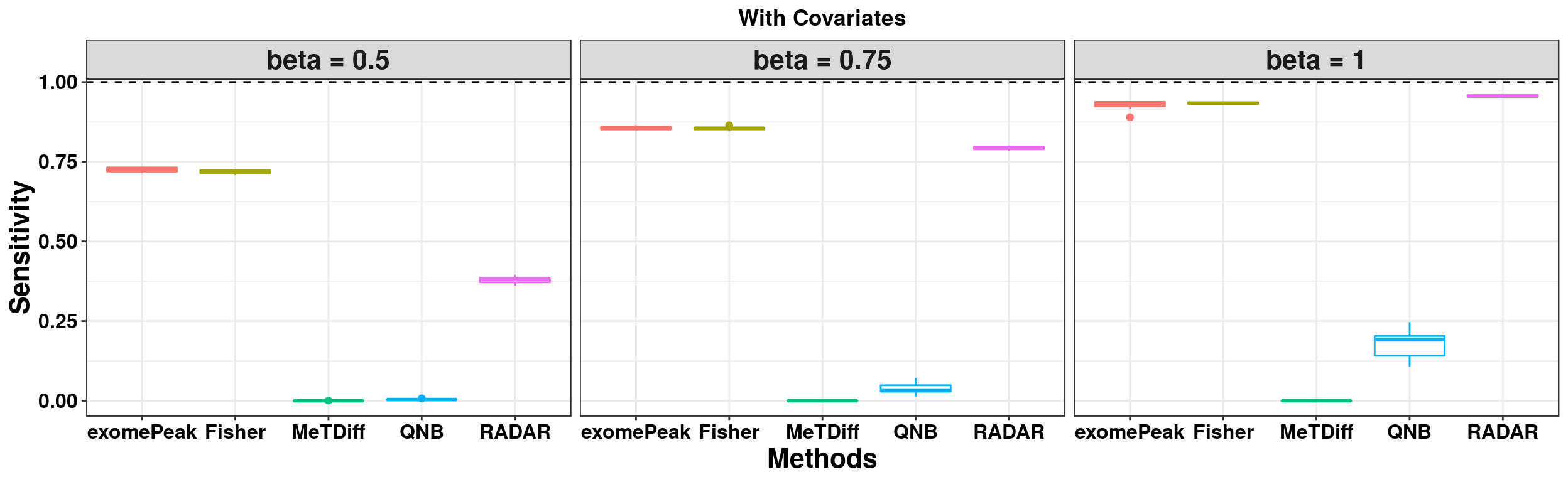
ggplot(RADARmodel_cov_combined[RADARmodel_cov_combined$fdr_cutoff == 0.1,] , aes(x = fdr, y = power, colour = method, shape = method))+geom_point( size = 4)+facet_wrap(beta~.) + geom_vline(xintercept = 0.1,lty=2)+ylab("Sensitivity")+xlab("FDR")+ggtitle("RADAR Model With Covariates") +theme_bw()+theme( text = element_text(face = "bold",colour = "black"), axis.text = element_text(size = 12,colour = "black"), axis.title = element_text(size = 16), strip.text = element_text(size = 16), plot.title = element_text(hjust = 0.5), legend.text = element_text(size = 15) )+scale_y_continuous(limits = c(0,1))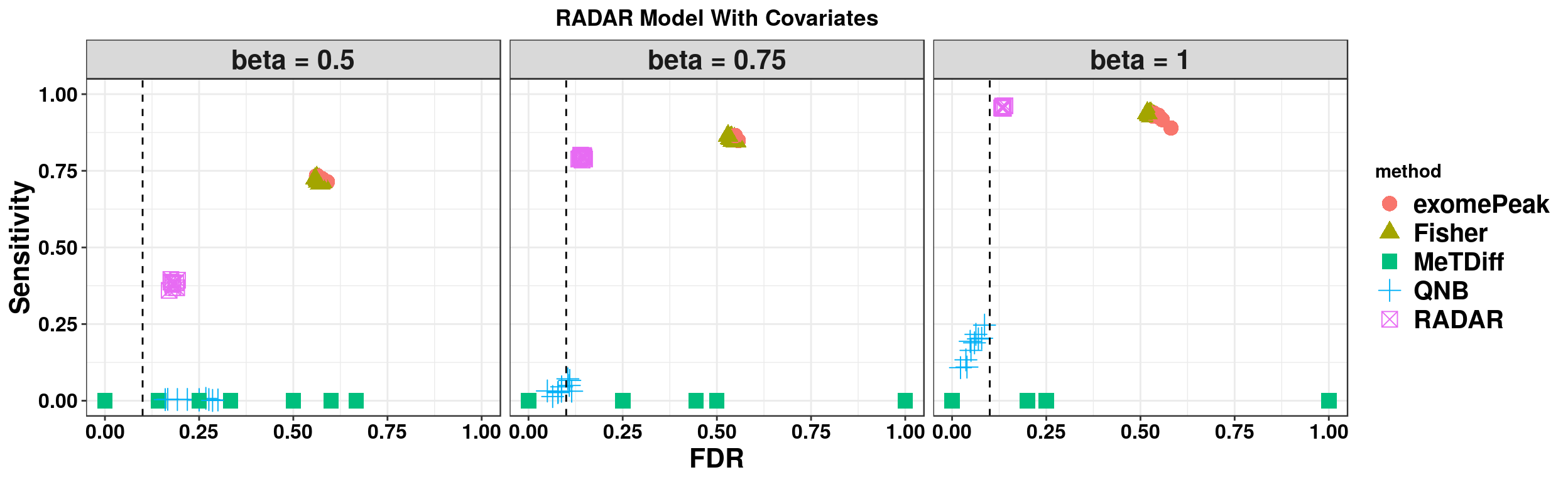
QNB model
Simple case
simulateData_with_more_replicates <- function(n_Sites=20000,replicate=8,min_expression=1,max_expression=3,
dif_express=1/4,per_me=1/3,dif_me=1/4,lib_s=1/10,var_input=1,var_ip = 2, beta=1 ){
#expression
q <- 10^runif(n_Sites,min_expression,max_expression)
p <- runif(n_Sites,0.2,0.9)
y <- rnorm(n_Sites,0,dif_express)
e1 <- exp(y)
y <- rnorm(n_Sites,0,dif_express)
e2 <- exp(y)
####e: gene/site-specific factor, e1 for condition 1, e2 for condition 2
T <- rnorm(4*replicate,0,lib_s)
S <- 2^T
####s quantifies the impact of sample-specific library size (or the sequencing depth)
s_t1 <- S[1:replicate]
s_t2 <- S[(replicate+1):(2*replicate)]
s_c1 <- S[(2*replicate+1):(3*replicate)]
s_c2 <- S[(3*replicate+1):(4*replicate)]
#mu
mu_t1 <- (e1*q*p)%*%t(as.numeric(s_t1))
mu_t2 <- (e2*q*p)%*%t(as.numeric(s_t2))
mu_c1 <- (e1*q*(1-p))%*%t(as.numeric(s_c1))
mu_c2 <- (e2*q*(1-p))%*%t(as.numeric(s_c2))
#generating differentially methylated sites
x <- rep(beta,round(n_Sites*per_me))
dif <- ifelse( 1:round(n_Sites*per_me) %in% sample(1:round(n_Sites*per_me),round(n_Sites*per_me)/2),exp(-x) ,exp(x) )
####adding DE from site round(n_Sites*(1-per_me))+1 to n_Sites
#mu_t1[((round(n_Sites*(1-per_me))+1):n_Sites),] <- mu_t1[((round(n_Sites*(1-per_me))+1):n_Sites),]/dif
mu_t2[((round(n_Sites*(1-per_me))+1):n_Sites),] <- mu_t2[((round(n_Sites*(1-per_me))+1):n_Sites),]*dif
####ATTENTION: the following codes aim to find out the differentially expressed sites
starting_site<-round(n_Sites*(1-per_me))+1
closing_site<-n_Sites
#all_sites<-1/dif^2 #### this would be the foldchange for any site in the two conditions because mu_t1 is divided by dif and mu_t2 multiplies dif.
df_sites<-c(starting_site:closing_site) ####manually set up the threshold for DE sites, above which we can say that the site is DE site.
#variance
# get all variance
var_t1 <- mu_t1+var_ip*p*q
var_t2 <- mu_t2+var_ip*p*q
var_c1 <- mu_c1+var_input*p*q
var_c2 <- mu_c2+var_input*p*q
size_t1=mu_t1^2/(var_t1-mu_t1)
size_t2=mu_t2^2/(var_t2-mu_t2)
size_c1=mu_c1^2/(var_c1-mu_c1)
size_c2=mu_c2^2/(var_c2-mu_c2)
meth1=matrix(0,n_Sites,replicate)
meth2=matrix(0,n_Sites,replicate)
unmeth1=matrix(0,n_Sites,replicate)
unmeth2=matrix(0,n_Sites,replicate)
for(i in 1:n_Sites){
meth1[i,]=rnbinom(replicate,mu=mu_t1[i,],size=size_t1[i,])
meth2[i,]=rnbinom(replicate,mu=mu_t2[i,],size=size_t2[i,])
unmeth1[i,]=rnbinom(replicate,mu=mu_c1[i,],size=size_c1[i,])
unmeth2[i,]=rnbinom(replicate,mu=mu_c2[i,],size=size_c2[i,])
}
res=list(5)
res[[paste0("IP",1)]]=meth1
res[[paste0("IP",2)]]=meth2
res[[paste0("Input",1)]]=unmeth1
res[[paste0("Input",2)]]=unmeth2
res[["df_site_index"]]=df_sites
return (res)
}registerDoParallel(10)
simuTest_simpleQNB_beta0.5 <- foreach(i = 1:10, .combine = rbind)%dopar%{
iteration <-simulateData_with_more_replicates(n_Sites=20000,replicate=8,min_expression=1,max_expression=3,
dif_express=1/2,per_me=1/4,dif_me=1/2,lib_s=1/10,var_input = 0.5, var_ip = 2, beta=0.5)
simu.radar <- MeRIP.RADAR()
variable(simu.radar) <- data.frame( condition = c(rep(0,8), rep(1,8)) )
simu.radar@Peak_input <- cbind(iteration$Input1, iteration$Input2)
simu.radar@Peak_ip <- cbind(iteration$IP1, iteration$IP2)
rownames(simu.radar@Peak_input) <- rownames(simu.radar@Peak_ip) <- paste0("peak",1:20000)
samplenames(simu.radar) <- paste0("s",1:16)
## perform library normalization for RADAR pipline
size.input <- DESeq2::estimateSizeFactorsForMatrix(simu.radar@Peak_input)
norm.Peak_input <-t( t(simu.radar@Peak_input) / size.input )
enrich <- simu.radar@Peak_ip/norm.Peak_input
enrich <- enrich[!apply(enrich,1, function(x){any(is.na(x)) | any(is.infinite(x))}),]
size.enrich.deseq2 <- DESeq2::estimateSizeFactorsForMatrix(enrich[,1:length(simu.radar@samplenames)])
norm.Peak_ip <-t( t(simu.radar@Peak_ip)/size.enrich.deseq2 )
simu.radar@sizeFactor <- data.frame(input=size.input,ip=size.enrich.deseq2)
simu.radar@norm.Peak_ip <- round(norm.Peak_ip)
simu.radar <- adjustExprLevel(simu.radar,adjustBy = "peak")
simu.radar <- diffIP( simu.radar )
simu.Metdiff <-MeTDiffTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.qnb <- QNB::qnbtest(control_ip = round( simu.radar@Peak_ip[,1:8]) ,
treated_ip = round(simu.radar@Peak_ip[,9:16] ),
control_input = round( simu.radar@Peak_input[,1:8] ),
treated_input = round( simu.radar@Peak_input[,9:16] ),plot.dispersion = FALSE)
simu.fisher <- fisherTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.exomePeak <- Bltest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
result <- NULL
for(q in c(0.01, 0.05, 0.1, 0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9,0.95,0.99)){
tmpPower <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" =simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), iteration$df_site_index ) )/length(iteration$df_site_index)})
tmpFDR <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:20000, iteration$df_site_index ) ) )/length(which( x < q ) )})
tmpFalsePosRate <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:20000, iteration$df_site_index ) ) )/length( setdiff( 1:20000, iteration$df_site_index ) ) })
result <- rbind(result, data.frame( power = tmpPower, fdr = tmpFDR, fpr = tmpFalsePosRate, method = c("RADAR","MeTDiff","exomePeak","Fisher","QNB") , fdr_cutoff = q ) )
}
result
}
simuTest_simpleQNB_beta0.75 <- foreach(i = 1:10, .combine = rbind)%dopar%{
iteration <-simulateData_with_more_replicates(n_Sites=20000,replicate=8,min_expression=1,max_expression=3,
dif_express=1/2,per_me=1/4,dif_me=1/2,lib_s=1/10,var_input = 0.5, var_ip = 2, beta=0.75)
simu.radar <- MeRIP.RADAR()
variable(simu.radar) <- data.frame( condition = c(rep(0,8), rep(1,8)) )
simu.radar@Peak_input <- cbind(iteration$Input1, iteration$Input2)
simu.radar@Peak_ip <- cbind(iteration$IP1, iteration$IP2)
rownames(simu.radar@Peak_input) <- rownames(simu.radar@Peak_ip) <- paste0("peak",1:20000)
samplenames(simu.radar) <- paste0("s",1:16)
## perform library normalization for RADAR pipline
size.input <- DESeq2::estimateSizeFactorsForMatrix(simu.radar@Peak_input)
norm.Peak_input <-t( t(simu.radar@Peak_input) / size.input )
enrich <- simu.radar@Peak_ip/norm.Peak_input
enrich <- enrich[!apply(enrich,1, function(x){any(is.na(x)) | any(is.infinite(x))}),]
size.enrich.deseq2 <- DESeq2::estimateSizeFactorsForMatrix(enrich[,1:length(simu.radar@samplenames)])
norm.Peak_ip <-t( t(simu.radar@Peak_ip)/size.enrich.deseq2 )
simu.radar@sizeFactor <- data.frame(input=size.input,ip=size.enrich.deseq2)
simu.radar@norm.Peak_ip <- round(norm.Peak_ip)
simu.radar <- adjustExprLevel(simu.radar,adjustBy = "peak")
simu.radar <- diffIP( simu.radar )
simu.Metdiff <-MeTDiffTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.qnb <- QNB::qnbtest(control_ip = round( simu.radar@Peak_ip[,1:8]) ,
treated_ip = round(simu.radar@Peak_ip[,9:16] ),
control_input = round( simu.radar@Peak_input[,1:8] ),
treated_input = round( simu.radar@Peak_input[,9:16] ),plot.dispersion = FALSE)
simu.fisher <- fisherTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.exomePeak <- Bltest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
result <- NULL
for(q in c(0.01, 0.05, 0.1, 0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9,0.95,0.99)){
tmpPower <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" =simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), iteration$df_site_index ) )/length(iteration$df_site_index)})
tmpFDR <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:20000, iteration$df_site_index ) ) )/length(which( x < q ) )})
tmpFalsePosRate <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:20000, iteration$df_site_index ) ) )/length( setdiff( 1:20000, iteration$df_site_index ) ) })
result <- rbind(result, data.frame( power = tmpPower, fdr = tmpFDR, fpr = tmpFalsePosRate, method = c("RADAR","MeTDiff","exomePeak","Fisher","QNB") , fdr_cutoff = q ) )
}
result
}
simuTest_simpleQNB_beta1 <- foreach(i = 1:10, .combine = rbind)%dopar%{
iteration <-simulateData_with_more_replicates(n_Sites=20000,replicate=8,min_expression=1,max_expression=3,
dif_express=1/2,per_me=1/4,dif_me=1/2,lib_s=1/10,var_input = 0.5, var_ip = 2, beta=1)
simu.radar <- MeRIP.RADAR()
variable(simu.radar) <- data.frame( condition = c(rep(0,8), rep(1,8)) )
simu.radar@Peak_input <- cbind(iteration$Input1, iteration$Input2)
simu.radar@Peak_ip <- cbind(iteration$IP1, iteration$IP2)
rownames(simu.radar@Peak_input) <- rownames(simu.radar@Peak_ip) <- paste0("peak",1:20000)
samplenames(simu.radar) <- paste0("s",1:16)
## perform library normalization for RADAR pipline
size.input <- DESeq2::estimateSizeFactorsForMatrix(simu.radar@Peak_input)
norm.Peak_input <-t( t(simu.radar@Peak_input) / size.input )
enrich <- simu.radar@Peak_ip/norm.Peak_input
enrich <- enrich[!apply(enrich,1, function(x){any(is.na(x)) | any(is.infinite(x))}),]
size.enrich.deseq2 <- DESeq2::estimateSizeFactorsForMatrix(enrich[,1:length(simu.radar@samplenames)])
norm.Peak_ip <-t( t(simu.radar@Peak_ip)/size.enrich.deseq2 )
simu.radar@sizeFactor <- data.frame(input=size.input,ip=size.enrich.deseq2)
simu.radar@norm.Peak_ip <- round(norm.Peak_ip)
simu.radar <- adjustExprLevel(simu.radar,adjustBy = "peak")
simu.radar <- diffIP( simu.radar )
simu.Metdiff <-MeTDiffTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.qnb <- QNB::qnbtest(control_ip = round( simu.radar@Peak_ip[,1:8]) ,
treated_ip = round(simu.radar@Peak_ip[,9:16] ),
control_input = round( simu.radar@Peak_input[,1:8] ),
treated_input = round( simu.radar@Peak_input[,9:16] ),plot.dispersion = FALSE)
simu.fisher <- fisherTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.exomePeak <- Bltest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
result <- NULL
for(q in c(0.01, 0.05, 0.1, 0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9,0.95,0.99)){
tmpPower <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" =simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), iteration$df_site_index ) )/length(iteration$df_site_index)})
tmpFDR <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:20000, iteration$df_site_index ) ) )/length(which( x < q ) )})
tmpFalsePosRate <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:20000, iteration$df_site_index ) ) )/length( setdiff( 1:20000, iteration$df_site_index ) ) })
result <- rbind(result, data.frame( power = tmpPower, fdr = tmpFDR, fpr = tmpFalsePosRate, method = c("RADAR","MeTDiff","exomePeak","Fisher","QNB") , fdr_cutoff = q ) )
}
result
}
save.image("~/Tools/RADARmannual/data/simulationAnalysis.RData")g_legend <- function(a.gplot){
tmp <- ggplot_gtable(ggplot_build(a.gplot))
leg <- which(sapply(tmp$grobs, function(x) x$name) == "guide-box")
legend <- tmp$grobs[[leg]]
return(legend)}
p11 <- ggplot(simuTest_simpleQNB_beta0.5, aes(x = fdr, y = power, colour = method))+geom_point()+theme_bw()+ggtitle("beta = 0.5")+theme(legend.position = "none", text = element_text(face = "bold"))
p22 <- ggplot(simuTest_simpleQNB_beta0.75, aes(x = fdr, y = power, colour = method))+geom_point()+theme_bw()+ggtitle("beta = 0.75")+theme(legend.position = "none", text = element_text(face = "bold"))
p33 <- ggplot(simuTest_simpleQNB_beta1, aes(x = fdr, y = power, colour = method))+geom_point()+theme_bw()+ggtitle("beta = 1")+theme(legend.position = "none", text = element_text(face = "bold"))
len <- ggplot(simuTest_simpleQNB_beta1, aes(x = fdr, y = power, colour = method))+geom_point()+theme(plot.title = element_text(face = "bold", size = 10,hjust = 0.5), legend.title = element_text(face = "bold", size = 15),legend.text = element_text(face = "bold", size = 15) )
gridExtra::grid.arrange(p11,p22,p33,g_legend(len), ncol = 4)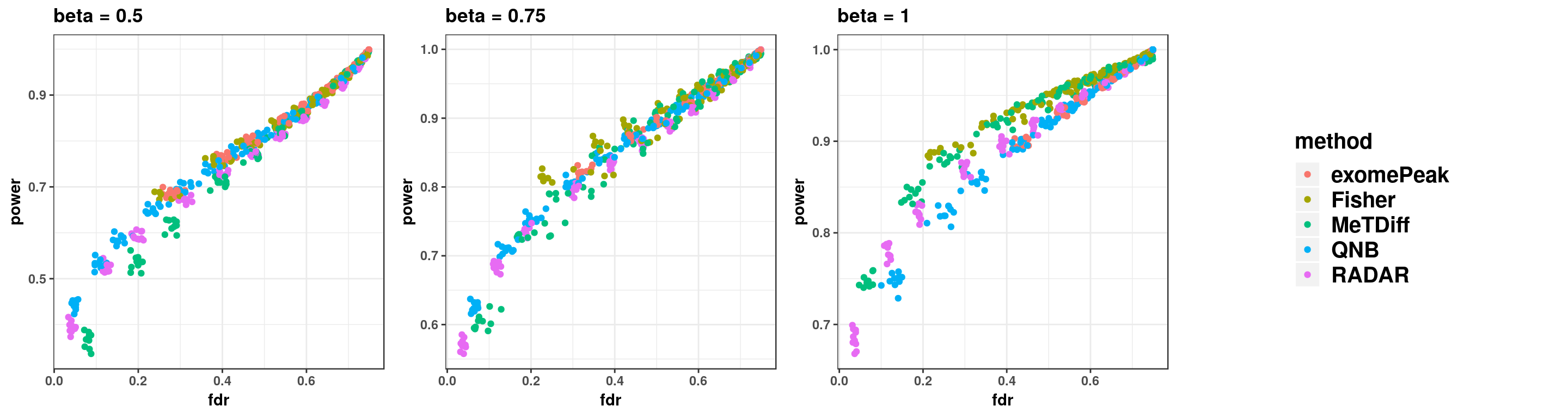
simuTest_simpleQNB_beta0.5_mean <- data.frame(FDR = tapply(simuTest_simpleQNB_beta0.5$fdr, paste0(simuTest_simpleQNB_beta0.5$method,simuTest_simpleQNB_beta0.5$fdr_cutoff), mean, na.rm=TRUE),
Power = tapply(simuTest_simpleQNB_beta0.5$power, paste0(simuTest_simpleQNB_beta0.5$method,simuTest_simpleQNB_beta0.5$fdr_cutoff), mean, na.rm=TRUE),
method = tapply(simuTest_simpleQNB_beta0.5$method, paste0(simuTest_simpleQNB_beta0.5$method,simuTest_simpleQNB_beta0.5$fdr_cutoff), function(x) unique(as.character(x)) ),
fdr_cutoff = as.factor(tapply(simuTest_simpleQNB_beta0.5$fdr_cutoff, paste0(simuTest_simpleQNB_beta0.5$method,simuTest_simpleQNB_beta0.5$fdr_cutoff),unique) )
)
simuTest_simpleQNB_beta0.75_mean <- data.frame(FDR = tapply(simuTest_simpleQNB_beta0.75$fdr, paste0(simuTest_simpleQNB_beta0.75$method,simuTest_simpleQNB_beta0.75$fdr_cutoff), mean, na.rm=TRUE),
Power = tapply(simuTest_simpleQNB_beta0.75$power, paste0(simuTest_simpleQNB_beta0.75$method,simuTest_simpleQNB_beta0.75$fdr_cutoff), mean, na.rm=TRUE),
method = tapply(simuTest_simpleQNB_beta0.75$method, paste0(simuTest_simpleQNB_beta0.75$method,simuTest_simpleQNB_beta0.75$fdr_cutoff), function(x) unique(as.character(x)) ),
fdr_cutoff = as.factor(tapply(simuTest_simpleQNB_beta0.75$fdr_cutoff, paste0(simuTest_simpleQNB_beta0.75$method,simuTest_simpleQNB_beta0.75$fdr_cutoff),unique ))
)
simuTest_simpleQNB_beta1_mean <- data.frame(FDR = tapply(simuTest_simpleQNB_beta1$fdr, paste0(simuTest_simpleQNB_beta1$method,simuTest_simpleQNB_beta1$fdr_cutoff), mean, na.rm=TRUE),
Power = tapply(simuTest_simpleQNB_beta1$power, paste0(simuTest_simpleQNB_beta1$method,simuTest_simpleQNB_beta1$fdr_cutoff), mean, na.rm=TRUE),
method = tapply(simuTest_simpleQNB_beta1$method, paste0(simuTest_simpleQNB_beta1$method,simuTest_simpleQNB_beta1$fdr_cutoff), function(x) unique(as.character(x)) ),
fdr_cutoff = as.factor(tapply(simuTest_simpleQNB_beta1$fdr_cutoff, paste0(simuTest_simpleQNB_beta1$method,simuTest_simpleQNB_beta1$fdr_cutoff), unique))
)
p1 <- ggplot(simuTest_simpleQNB_beta0.5_mean )+geom_point( aes(x = FDR, y = Power, colour = method, size = fdr_cutoff))+geom_line(aes(x = FDR, y = Power, colour = method))+theme_bw()+ggtitle("beta = 0.5")+theme(legend.position = "none", text = element_text(face = "bold", size = 18), axis.text = element_text(face = "bold", size = 15,colour = "black"), plot.title = element_text(hjust = 0.5) )
p2 <- ggplot(simuTest_simpleQNB_beta0.75_mean )+geom_point( aes(x = FDR, y = Power, colour = method, size = fdr_cutoff))+geom_line(aes(x = FDR, y = Power, colour = method))+theme_bw()+ggtitle("beta = 0.75")+theme(legend.position = "none", text = element_text(face = "bold", size = 18), axis.text = element_text(face = "bold", size = 15,colour = "black"), plot.title = element_text(hjust = 0.5) )
p3 <- ggplot(simuTest_simpleQNB_beta1_mean )+geom_point( aes(x = FDR, y = Power, colour = method, size = fdr_cutoff))+geom_line(aes(x = FDR, y = Power, colour = method))+theme_bw()+ggtitle("beta = 1")+theme(legend.position = "none", text = element_text(face = "bold", size = 18), axis.text = element_text(face = "bold", size = 15,colour = "black"), plot.title = element_text(hjust = 0.5) )
len <- ggplot(simuTest_simpleQNB_beta1_mean, aes(x = FDR, y = Power, colour = method, size = fdr_cutoff))+geom_point()+theme_bw()+theme(plot.title = element_text(face = "bold", size = 10,hjust = 0.5), legend.title = element_text(face = "bold", size = 15),legend.text = element_text(face = "bold", size = 15) )
gridExtra::grid.arrange(p1,p2,p3,g_legend(len), ncol = 4)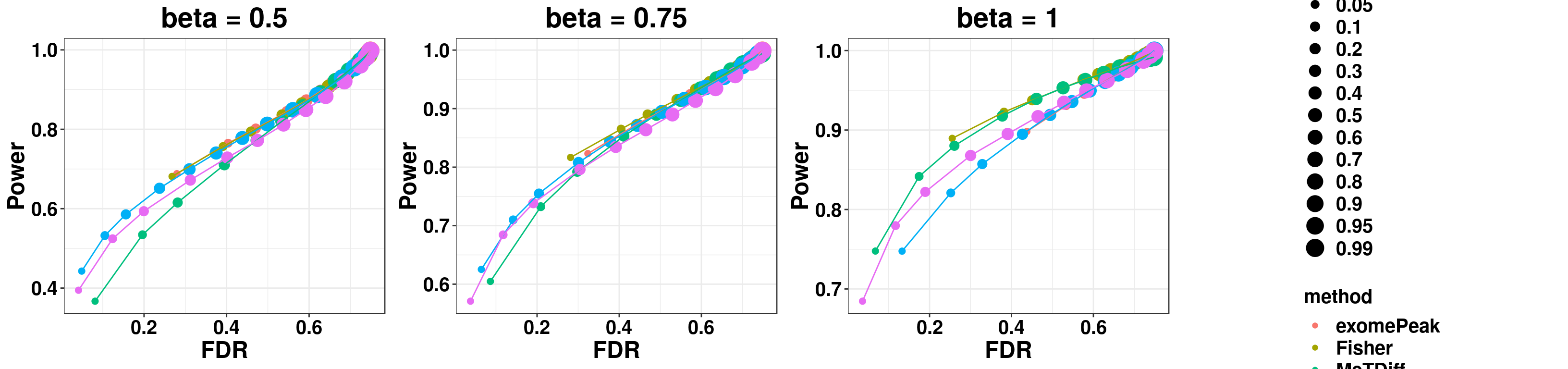
QNBmodel_combined <- dplyr::mutate( rbind(simuTest_simpleQNB_beta0.5, simuTest_simpleQNB_beta0.75,simuTest_simpleQNB_beta1) , beta = c(rep("beta = 0.5",nrow(simuTest_simpleQNB_beta0.5)),rep("beta = 0.75",nrow(simuTest_simpleQNB_beta0.75)),rep("beta = 1",nrow(simuTest_simpleQNB_beta1)) ) )
ggplot(QNBmodel_combined[QNBmodel_combined$fdr_cutoff == 0.1,] , aes(x = method, y = fdr, colour = method))+geom_boxplot()+facet_wrap(beta~.) + geom_abline(slope = 0,intercept = 0.1,lty=2)+ylab("FDR")+xlab("Methods")+ ggtitle("QNB Model Without Covariates") +theme_bw()+theme(legend.position = "none", text = element_text(face = "bold",colour = "black"), axis.text = element_text(size = 12,colour = "black"), axis.title = element_text(size = 16), strip.text = element_text(size = 16), plot.title = element_text(hjust = 0.5) )+scale_y_continuous(limits = c(0,1))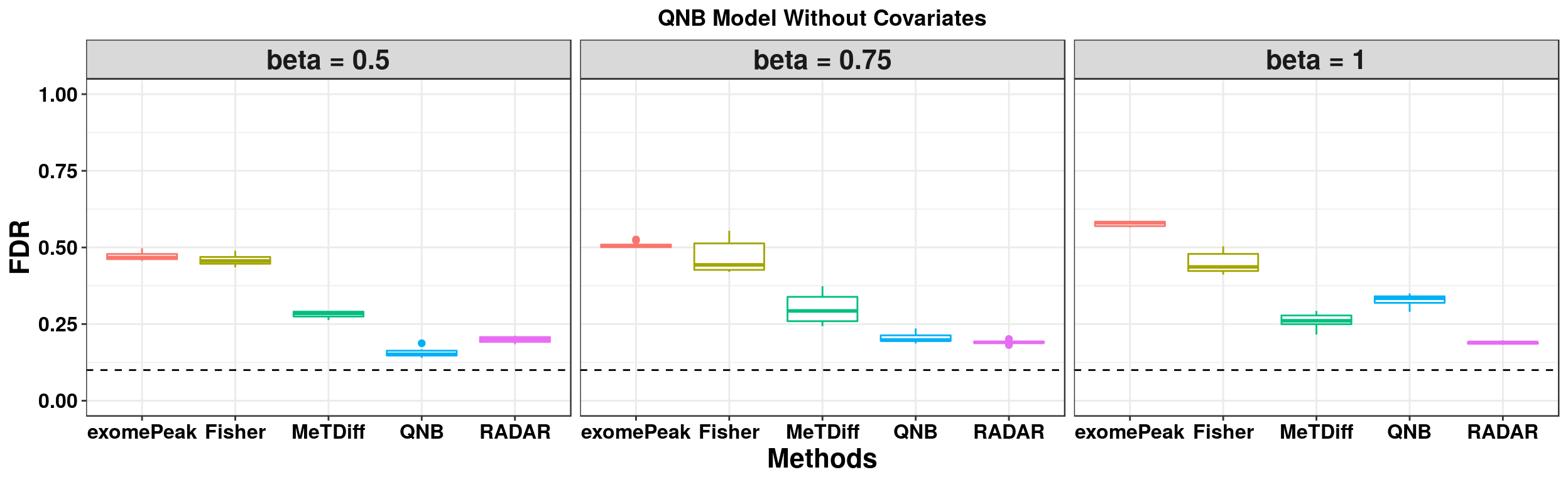
ggplot(QNBmodel_combined[QNBmodel_combined$fdr_cutoff == 0.1,] , aes(x = method, y = power, colour = method))+geom_boxplot()+facet_wrap(beta~.) + geom_abline(slope = 0,intercept = 1,lty=2)+ylab("Sensitivity")+xlab("Methods")+ggtitle("QNB Model Without Covariates") +theme_bw()+theme(legend.position = "none", text = element_text(face = "bold",colour = "black"), axis.text = element_text(size = 12,colour = "black"), axis.title = element_text(size = 16), strip.text = element_text(size = 16), plot.title = element_text(hjust = 0.5) )+scale_y_continuous(limits = c(0,1))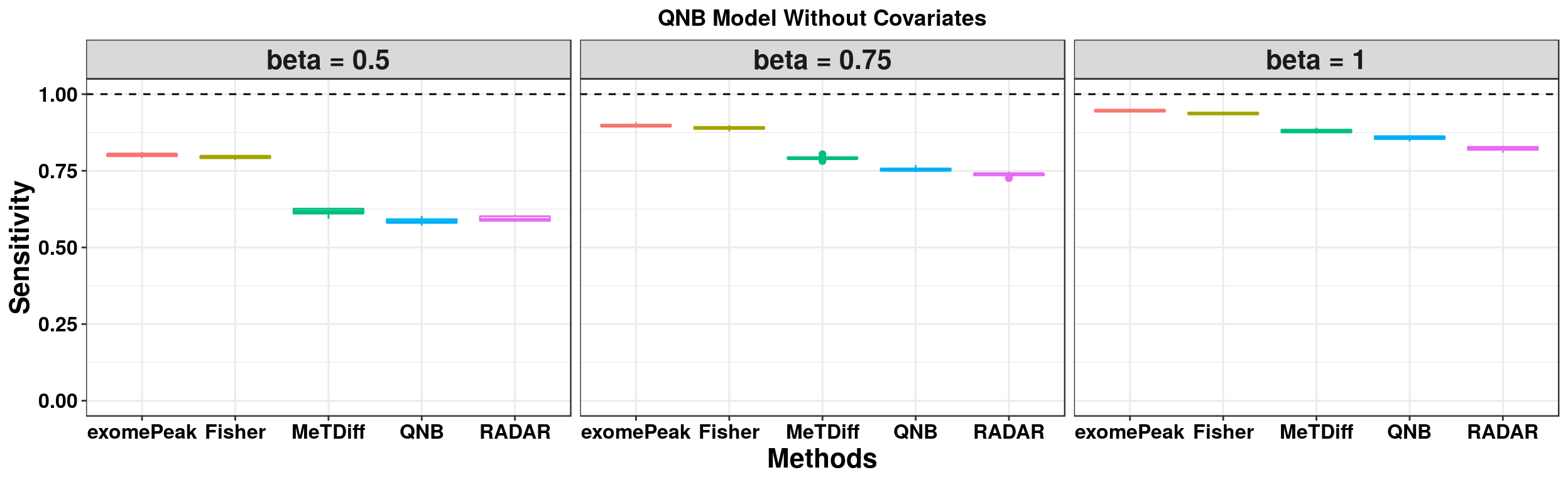
ggplot(QNBmodel_combined[QNBmodel_combined$fdr_cutoff == 0.1,] , aes(x = fdr, y = power, colour = method, shape = method))+geom_point(size = 4)+facet_wrap(beta~.) + geom_vline(xintercept = 0.1,lty=2)+ylab("Sensitivity")+xlab("FDR")+ggtitle("QNB Model Without Covariates") +theme_bw()+theme( text = element_text(face = "bold",colour = "black"), axis.text = element_text(size = 12,colour = "black"), axis.title = element_text(size = 16), strip.text = element_text(size = 16), plot.title = element_text(hjust = 0.5), legend.text = element_text(size = 15) )+scale_y_continuous(limits = c(0,1))+scale_x_continuous(limits = c(0,1))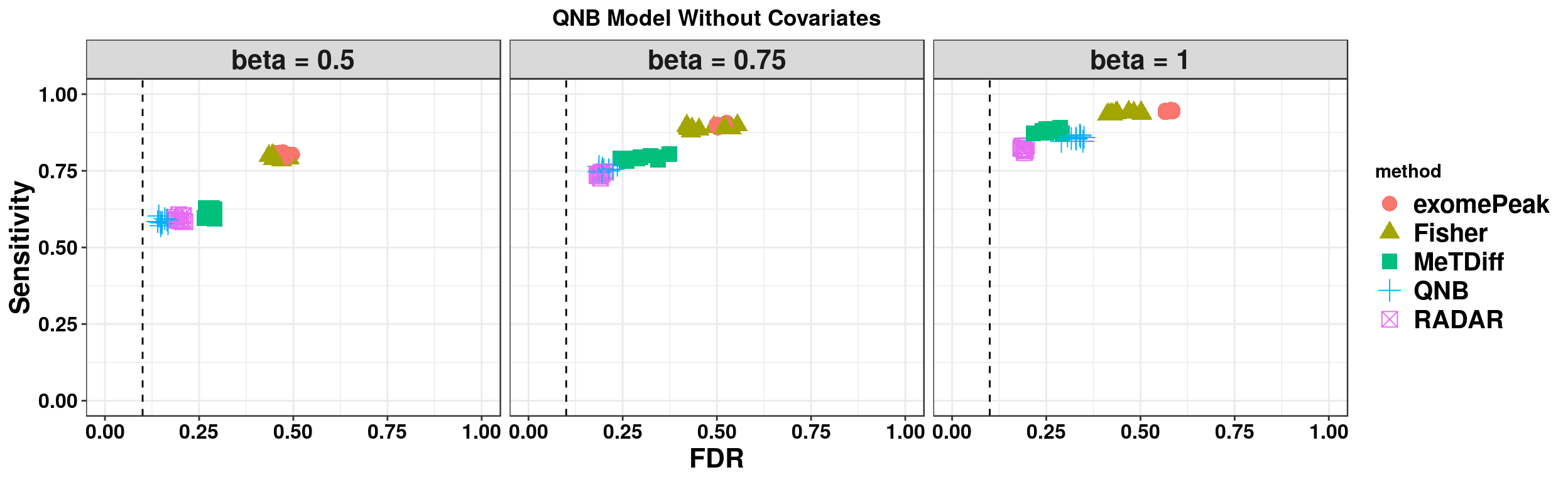
Complex case
simulateData_with_more_replicates_difficult <- function(n_Sites=20000,replicate=8,min_expression=1,max_expression=3,
dif_express=1/4,per_me=1/4,beta=1, lib_s=1/10,var_input = 1, var_ip = 2, batch1effectSize = 1, seed = 1212){
set.seed(seed)
#expression
q <- 10^runif(n_Sites,min_expression,max_expression)
p <- runif(n_Sites,0,1) # define the enrichment level at this site
y <- rnorm(n_Sites,0,dif_express)
e1 <- exp(y)
y <- rnorm(n_Sites,0,dif_express)
e2 <- exp(y)
####e: gene/site-specific factor, e1 for condition 1, e2 for condition 2
T <- rnorm(4*replicate,0,lib_s)
S <- 2^T
####s quantifies the impact of sample-specific library size (or the sequencing depth)
s_t1 <- S[1:replicate]
s_t2 <- S[(replicate+1):(2*replicate)]
s_c1 <- S[(2*replicate+1):(3*replicate)]
s_c2 <- S[(3*replicate+1):(4*replicate)]
#mu
mu_t1 <- (e1*q*p)%*%t(as.numeric(s_t1))
mu_t2 <- (e2*q*p)%*%t(as.numeric(s_t2))
mu_c1 <- (e1*q*(1-p))%*%t(as.numeric(s_c1))
mu_c2 <- (e2*q*(1-p))%*%t(as.numeric(s_c2))
####add batch effects/covariates (assume three batches)
n_batch<-2
batch1_replicates<-sort( c( sample(1:replicate,3),sample((replicate+1):(2*replicate),5) ) )
index_remainder1<-setdiff(c( 1:(2*replicate) ),batch1_replicates)
batchEffect <- ifelse( 1:n_Sites %in% sample(1:n_Sites,n_Sites/2) ,-batch1effectSize,batch1effectSize)
batchFold <- exp(batchEffect)
#### generate batch effects for batch1
mu_total_t<-cbind(mu_t1, mu_t2) ####eight IP replicates + eight input replicates for condition 1
mu_total_c<-cbind(mu_c1, mu_c2) ####eight IP replicates + eight input replicates for condition 2
for(j in batch1_replicates){
mu_total_t[,j] <- mu_total_t[,j]*batchFold
}#### add batch1 effect
mu_t1<-mu_total_t[,1:replicate]
mu_c1<-mu_total_c[,1:replicate]
mu_t2<-mu_total_t[,(replicate+1):(2*replicate)]
mu_c2<-mu_total_c[,(replicate+1):(2*replicate)]
#generating differentially methylated sites
x <- rep(beta, round(n_Sites*per_me) )
dif <- ifelse( 1:round(n_Sites*per_me) %in% sample(1:round(n_Sites*per_me),round(n_Sites*per_me)/2),exp(-x) ,exp(x) )
####adding DM from site round(n_Sites*(1-per_me))+1 to n_Sites
#mu_t1[((round(n_Sites*(1-per_me))+1):n_Sites),] <- mu_t1[((round(n_Sites*(1-per_me))+1):n_Sites),]/dif
mu_t2[((round(n_Sites*(1-per_me))+1):n_Sites),] <- mu_t2[((round(n_Sites*(1-per_me))+1):n_Sites),]*dif
####ATTENTION: the following codes aim to find out the differentially methylated sites
starting_site<-round(n_Sites*(1-per_me))+1
closing_site<-n_Sites
#all_sites<-1/dif^2 #### this would be the foldchange for any site in the two conditions because mu_t1 is divided by dif and mu_t2 multiplies dif.
df_sites<-c(starting_site:closing_site) ####manually set up the threshold for DE sites, above which we can say that the site is DE site.
#variance
# get all variance
var_t1 <- mu_t1+var_ip*p*q
var_t2 <- mu_t2+var_ip*p*q
var_c1 <- mu_c1+var_input*p*q
var_c2 <- mu_c2+var_input*p*q
size_t1=mu_t1^2/(var_t1-mu_t1)
size_t2=mu_t2^2/(var_t2-mu_t2)
size_c1=mu_c1^2/(var_c1-mu_c1)
size_c2=mu_c2^2/(var_c2-mu_c2)
meth1=matrix(0,n_Sites,replicate)
meth2=matrix(0,n_Sites,replicate)
unmeth1=matrix(0,n_Sites,replicate)
unmeth2=matrix(0,n_Sites,replicate)
for(i in 1:n_Sites){
meth1[i,]=pmax(rnbinom(replicate,mu=mu_t1[i,],size=size_t1[i,]),1)
meth2[i,]=pmax(rnbinom(replicate,mu=mu_t2[i,],size=size_t2[i,]),1)
unmeth1[i,]=pmax(rnbinom(replicate,mu=mu_c1[i,],size=size_c1[i,]),1)
unmeth2[i,]=pmax(rnbinom(replicate,mu=mu_c2[i,],size=size_c2[i,]),1)
}
res=list(6)
meth1[is.na(meth1)]<-0
res[[paste0("IP",1)]]=meth1
meth2[is.na(meth2)]<-0
res[[paste0("IP",2)]]=meth2
unmeth1[is.na(unmeth1)]<-0
res[[paste0("Input",1)]]=unmeth1
unmeth2[is.na(unmeth2)]<-0
res[[paste0("Input",2)]]=unmeth2
res[["df_site_index"]]=df_sites
res[["covariates"]] = data.frame(batch2 = ifelse(1:(2*replicate) %in% batch1_replicates ,1,0) )
return (res)
}Run the test on complex QNB model
registerDoParallel(10)
simuTest_difficultQNB_beta0.5 <- foreach(i = 1:10, .combine = rbind)%dopar%{
iteration <-simulateData_with_more_replicates_difficult(n_Sites=20000,replicate=8,min_expression=1,max_expression=3,
dif_express=1/2,per_me=1/4, beta = 0.5,lib_s=0.1,var_input = 0.1, var_ip = 2, seed = i*3, batch1effectSize = 2 )
simu.radar <- MeRIP.RADAR()
variable(simu.radar) <- cbind( data.frame( condition = c(rep(0,8), rep(1,8)) ), iteration$covariates )
simu.radar@Peak_input <- cbind(iteration$Input1, iteration$Input2)
simu.radar@Peak_ip <- cbind(iteration$IP1, iteration$IP2)
rownames(simu.radar@Peak_input) <- rownames(simu.radar@Peak_ip) <- paste0("peak",1:20000)
samplenames(simu.radar) <- paste0("s",1:16)
## perform library normalization for RADAR pipline
size.input <- DESeq2::estimateSizeFactorsForMatrix(simu.radar@Peak_input)
norm.Peak_input <-t( t(simu.radar@Peak_input) / size.input )
enrich <- simu.radar@Peak_ip/norm.Peak_input
enrich <- enrich[!apply(enrich,1, function(x){any(is.na(x)) | any(is.infinite(x))}),]
size.enrich.deseq2 <- DESeq2::estimateSizeFactorsForMatrix(enrich[,1:length(simu.radar@samplenames)])
norm.Peak_ip <-t( t(simu.radar@Peak_ip)/size.enrich.deseq2 )
simu.radar@sizeFactor <- data.frame(input=size.input,ip=size.enrich.deseq2)
simu.radar@norm.Peak_ip <- round(norm.Peak_ip)
simu.radar <- adjustExprLevel(simu.radar,adjustBy = "peak")
#plotPCAfromMatrix(simu.radar@Peak_adjExpr, group = factor( unlist(variable(simu.radar)) ), loglink = T)
simu.radar <- diffIP( simu.radar)
simu.Metdiff <-MeTDiffTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.qnb <- QNB::qnbtest(control_ip = round( simu.radar@Peak_ip[,1:8]) ,
treated_ip = round(simu.radar@Peak_ip[,9:16] ),
control_input = round( simu.radar@Peak_input[,1:8] ),
treated_input = round( simu.radar@Peak_input[,9:16] ),plot.dispersion = FALSE)
simu.fisher <- fisherTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.exomePeak <- Bltest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
result <- NULL
for(q in c(0.01, 0.05, 0.1, 0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9,0.95,0.99)){
tmpPower <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" =simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), iteration$df_site_index ) )/length(iteration$df_site_index)})
tmpFDR <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:20000, iteration$df_site_index ) ) )/length(which( x < q ) )})
tmpFalsePosRate <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:20000, iteration$df_site_index ) ) )/length( setdiff( 1:20000, iteration$df_site_index ) ) })
result <- rbind(result, data.frame( power = tmpPower, fdr = tmpFDR, fpr = tmpFalsePosRate, method = c("RADAR","MeTDiff","exomePeak","Fisher","QNB") , fdr_cutoff = q ) )
}
result
}
simuTest_difficultQNB_beta0.75 <- foreach(i = 1:10, .combine = rbind)%dopar%{
iteration <-simulateData_with_more_replicates_difficult(n_Sites=20000,replicate=8,min_expression=1,max_expression=3,
dif_express=1/2,per_me=1/4, beta = 0.75,lib_s=0.1,var_input = 0.1, var_ip = 2, seed = i*3, batch1effectSize = 2 )
simu.radar <- MeRIP.RADAR()
variable(simu.radar) <- cbind( data.frame( condition = c(rep(0,8), rep(1,8)) ), iteration$covariates )
simu.radar@Peak_input <- cbind(iteration$Input1, iteration$Input2)
simu.radar@Peak_ip <- cbind(iteration$IP1, iteration$IP2)
rownames(simu.radar@Peak_input) <- rownames(simu.radar@Peak_ip) <- paste0("peak",1:20000)
samplenames(simu.radar) <- paste0("s",1:16)
## perform library normalization for RADAR pipline
size.input <- DESeq2::estimateSizeFactorsForMatrix(simu.radar@Peak_input)
norm.Peak_input <-t( t(simu.radar@Peak_input) / size.input )
enrich <- simu.radar@Peak_ip/norm.Peak_input
enrich <- enrich[!apply(enrich,1, function(x){any(is.na(x)) | any(is.infinite(x))}),]
size.enrich.deseq2 <- DESeq2::estimateSizeFactorsForMatrix(enrich[,1:length(simu.radar@samplenames)])
norm.Peak_ip <-t( t(simu.radar@Peak_ip)/size.enrich.deseq2 )
simu.radar@sizeFactor <- data.frame(input=size.input,ip=size.enrich.deseq2)
simu.radar@norm.Peak_ip <- round(norm.Peak_ip)
simu.radar <- adjustExprLevel(simu.radar,adjustBy = "peak")
#plotPCAfromMatrix(simu.radar@Peak_adjExpr, group = factor( unlist(variable(simu.radar)) ), loglink = T)
simu.radar <- diffIP( simu.radar)
simu.Metdiff <-MeTDiffTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.qnb <- QNB::qnbtest(control_ip = round( simu.radar@Peak_ip[,1:8]) ,
treated_ip = round(simu.radar@Peak_ip[,9:16] ),
control_input = round( simu.radar@Peak_input[,1:8] ),
treated_input = round( simu.radar@Peak_input[,9:16] ),plot.dispersion = FALSE)
simu.fisher <- fisherTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.exomePeak <- Bltest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
result <- NULL
for(q in c(0.01, 0.05, 0.1, 0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9,0.95,0.99)){
tmpPower <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" =simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), iteration$df_site_index ) )/length(iteration$df_site_index)})
tmpFDR <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:20000, iteration$df_site_index ) ) )/length(which( x < q ) )})
tmpFalsePosRate <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:20000, iteration$df_site_index ) ) )/length( setdiff( 1:20000, iteration$df_site_index ) ) })
result <- rbind(result, data.frame( power = tmpPower, fdr = tmpFDR, fpr = tmpFalsePosRate, method = c("RADAR","MeTDiff","exomePeak","Fisher","QNB") , fdr_cutoff = q ) )
}
result
}
simuTest_difficultQNB_beta1 <- foreach(i = 1:10, .combine = rbind)%dopar%{
iteration <-simulateData_with_more_replicates_difficult(n_Sites=20000,replicate=8,min_expression=1,max_expression=3,
dif_express=1/2,per_me=1/4, beta = 1,lib_s=0.1,var_input = 0.1, var_ip = 2, seed = i*3, batch1effectSize = 2 )
simu.radar <- MeRIP.RADAR()
variable(simu.radar) <- cbind( data.frame( condition = c(rep(0,8), rep(1,8)) ), iteration$covariates )
simu.radar@Peak_input <- cbind(iteration$Input1, iteration$Input2)
simu.radar@Peak_ip <- cbind(iteration$IP1, iteration$IP2)
rownames(simu.radar@Peak_input) <- rownames(simu.radar@Peak_ip) <- paste0("peak",1:20000)
samplenames(simu.radar) <- paste0("s",1:16)
## perform library normalization for RADAR pipline
size.input <- DESeq2::estimateSizeFactorsForMatrix(simu.radar@Peak_input)
norm.Peak_input <-t( t(simu.radar@Peak_input) / size.input )
enrich <- simu.radar@Peak_ip/norm.Peak_input
enrich <- enrich[!apply(enrich,1, function(x){any(is.na(x)) | any(is.infinite(x))}),]
size.enrich.deseq2 <- DESeq2::estimateSizeFactorsForMatrix(enrich[,1:length(simu.radar@samplenames)])
norm.Peak_ip <-t( t(simu.radar@Peak_ip)/size.enrich.deseq2 )
simu.radar@sizeFactor <- data.frame(input=size.input,ip=size.enrich.deseq2)
simu.radar@norm.Peak_ip <- round(norm.Peak_ip)
simu.radar <- adjustExprLevel(simu.radar,adjustBy = "peak")
#plotPCAfromMatrix(simu.radar@Peak_adjExpr, group = factor( unlist(variable(simu.radar)) ), loglink = T)
simu.radar <- diffIP( simu.radar)
simu.Metdiff <-MeTDiffTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.qnb <- QNB::qnbtest(control_ip = round( simu.radar@Peak_ip[,1:8]) ,
treated_ip = round(simu.radar@Peak_ip[,9:16] ),
control_input = round( simu.radar@Peak_input[,1:8] ),
treated_input = round( simu.radar@Peak_input[,9:16] ),plot.dispersion = FALSE)
simu.fisher <- fisherTest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
simu.exomePeak <- Bltest(control_ip = simu.radar@Peak_ip[,1:8],
treated_ip = simu.radar@Peak_ip[,9:16],
control_input = simu.radar@Peak_input[,1:8],
treated_input = simu.radar@Peak_input[,9:16] )
result <- NULL
for(q in c(0.01, 0.05, 0.1, 0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9,0.95,0.99)){
tmpPower <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" =simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), iteration$df_site_index ) )/length(iteration$df_site_index)})
tmpFDR <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:20000, iteration$df_site_index ) ) )/length(which( x < q ) )})
tmpFalsePosRate <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:20000, iteration$df_site_index ) ) )/length( setdiff( 1:20000, iteration$df_site_index ) ) })
result <- rbind(result, data.frame( power = tmpPower, fdr = tmpFDR, fpr = tmpFalsePosRate, method = c("RADAR","MeTDiff","exomePeak","Fisher","QNB") , fdr_cutoff = q ) )
}
result
}
save.image("~/Tools/RADARmannual/data/simulationAnalysis.RData")g_legend <- function(a.gplot){
tmp <- ggplot_gtable(ggplot_build(a.gplot))
leg <- which(sapply(tmp$grobs, function(x) x$name) == "guide-box")
legend <- tmp$grobs[[leg]]
return(legend)}
p11 <- ggplot(simuTest_difficultQNB_beta0.5, aes(x = fdr, y = power, colour = method))+geom_point()+theme_bw()+ggtitle("beta = 0.5")+theme(legend.position = "none", text = element_text(face = "bold"))
p22 <- ggplot(simuTest_difficultQNB_beta0.75, aes(x = fdr, y = power, colour = method))+geom_point()+theme_bw()+ggtitle("beta = 0.75")+theme(legend.position = "none", text = element_text(face = "bold"))
p33 <- ggplot(simuTest_difficultQNB_beta1, aes(x = fdr, y = power, colour = method))+geom_point()+theme_bw()+ggtitle("beta = 1")+theme(legend.position = "none", text = element_text(face = "bold"))
len <- ggplot(simuTest_difficultQNB_beta0.75, aes(x = fdr, y = power, colour = method))+geom_point()+theme(plot.title = element_text(face = "bold", size = 10,hjust = 0.5), legend.title = element_text(face = "bold", size = 15),legend.text = element_text(face = "bold", size = 15) )
gridExtra::grid.arrange(p11,p22,p33,g_legend(len), ncol = 4)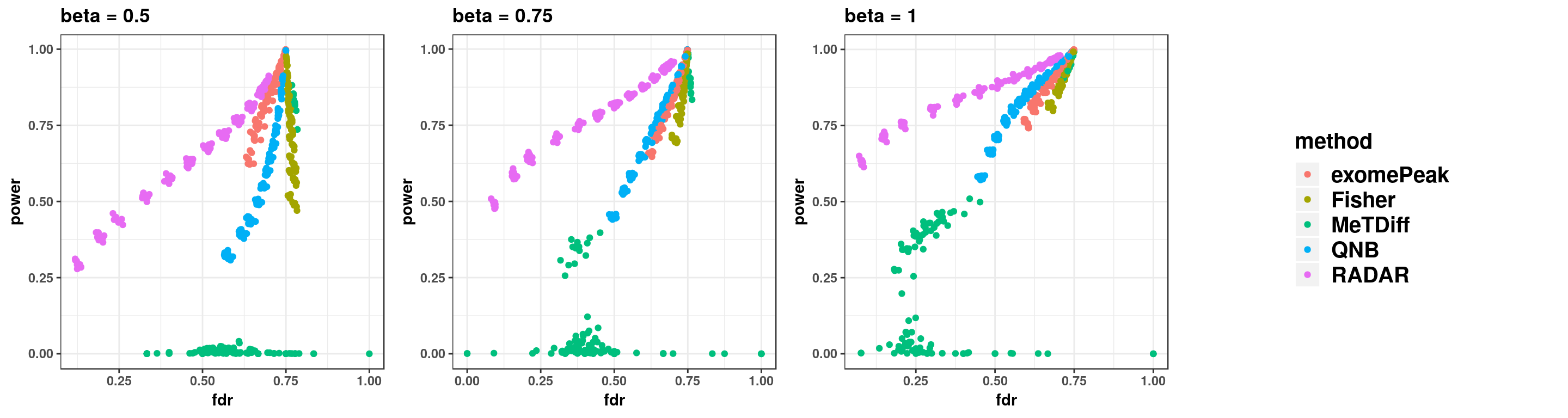
simuTest_difficultQNB_beta0.5_mean <- data.frame(FDR = tapply(simuTest_difficultQNB_beta0.5$fdr, paste0(simuTest_difficultQNB_beta0.5$method,simuTest_difficultQNB_beta0.5$fdr_cutoff), mean, na.rm=TRUE),
Power = tapply(simuTest_difficultQNB_beta0.5$power, paste0(simuTest_difficultQNB_beta0.5$method,simuTest_difficultQNB_beta0.5$fdr_cutoff), mean, na.rm=TRUE),
method = tapply(simuTest_difficultQNB_beta0.5$method, paste0(simuTest_difficultQNB_beta0.5$method,simuTest_difficultQNB_beta0.5$fdr_cutoff), function(x) unique(as.character(x)) ),
fdr_cutoff = as.factor(tapply(simuTest_difficultQNB_beta0.5$fdr_cutoff, paste0(simuTest_difficultQNB_beta0.5$method,simuTest_difficultQNB_beta0.5$fdr_cutoff),unique) )
)
simuTest_difficultQNB_beta0.75_mean <- data.frame(FDR = tapply(simuTest_difficultQNB_beta0.75$fdr, paste0(simuTest_difficultQNB_beta0.75$method,simuTest_difficultQNB_beta0.75$fdr_cutoff), mean, na.rm=TRUE),
Power = tapply(simuTest_difficultQNB_beta0.75$power, paste0(simuTest_difficultQNB_beta0.75$method,simuTest_difficultQNB_beta0.75$fdr_cutoff), mean, na.rm=TRUE),
method = tapply(simuTest_difficultQNB_beta0.75$method, paste0(simuTest_difficultQNB_beta0.75$method,simuTest_difficultQNB_beta0.75$fdr_cutoff), function(x) unique(as.character(x)) ),
fdr_cutoff = as.factor(tapply(simuTest_difficultQNB_beta0.75$fdr_cutoff, paste0(simuTest_difficultQNB_beta0.75$method,simuTest_difficultQNB_beta0.75$fdr_cutoff),unique ))
)
simuTest_difficultQNB_beta1_mean <- data.frame(FDR = tapply(simuTest_difficultQNB_beta1$fdr, paste0(simuTest_difficultQNB_beta1$method,simuTest_difficultQNB_beta1$fdr_cutoff), mean, na.rm=TRUE),
Power = tapply(simuTest_difficultQNB_beta1$power, paste0(simuTest_difficultQNB_beta1$method,simuTest_difficultQNB_beta1$fdr_cutoff), mean, na.rm=TRUE),
method = tapply(simuTest_difficultQNB_beta1$method, paste0(simuTest_difficultQNB_beta1$method,simuTest_difficultQNB_beta1$fdr_cutoff), function(x) unique(as.character(x)) ),
fdr_cutoff = as.factor(tapply(simuTest_difficultQNB_beta1$fdr_cutoff, paste0(simuTest_difficultQNB_beta1$method,simuTest_difficultQNB_beta1$fdr_cutoff), unique))
)
p1 <- ggplot(simuTest_difficultQNB_beta0.5_mean )+geom_point( aes(x = FDR, y = Power, colour = method, size = fdr_cutoff))+geom_line(aes(x = FDR, y = Power, colour = method))+theme_bw()+ggtitle("beta = 0.5")+theme(legend.position = "none", text = element_text(face = "bold", size = 18), axis.text = element_text(face = "bold", size = 15,colour = "black"), plot.title = element_text(hjust = 0.5) )
p2 <- ggplot(simuTest_difficultQNB_beta0.75_mean )+geom_point( aes(x = FDR, y = Power, colour = method, size = fdr_cutoff))+geom_line(aes(x = FDR, y = Power, colour = method))+theme_bw()+ggtitle("beta = 0.75")+theme(legend.position = "none", text = element_text(face = "bold", size = 18), axis.text = element_text(face = "bold", size = 15,colour = "black"), plot.title = element_text(hjust = 0.5) )
p3 <- ggplot(simuTest_difficultQNB_beta1_mean )+geom_point( aes(x = FDR, y = Power, colour = method, size = fdr_cutoff))+geom_line(aes(x = FDR, y = Power, colour = method))+theme_bw()+ggtitle("beta = 1")+theme(legend.position = "none", text = element_text(face = "bold", size = 18), axis.text = element_text(face = "bold", size = 15,colour = "black"), plot.title = element_text(hjust = 0.5) )
len <- ggplot(simuTest_difficultQNB_beta1_mean, aes(x = FDR, y = Power, colour = method, size = fdr_cutoff))+geom_point()+theme_bw()+theme(plot.title = element_text(face = "bold", size = 10,hjust = 0.5), legend.title = element_text(face = "bold", size = 15),legend.text = element_text(face = "bold", size = 15) )
gridExtra::grid.arrange(p1,p2,p3,g_legend(len), ncol = 4)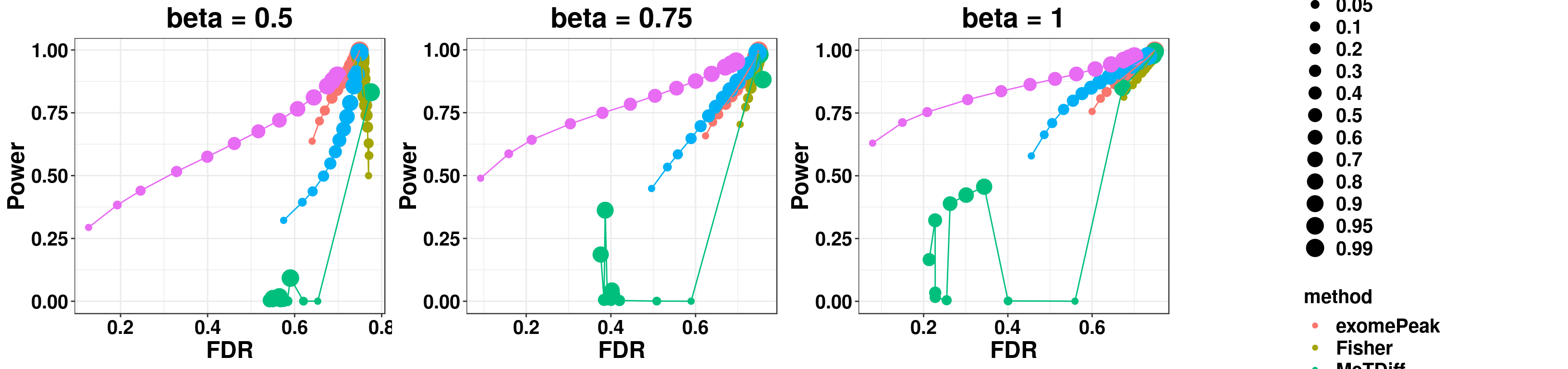
QNBmodel_cov_combined <- dplyr::mutate( rbind(simuTest_difficultQNB_beta0.5, simuTest_difficultQNB_beta0.75,simuTest_difficultQNB_beta1) , beta = c(rep("beta = 0.5",nrow(simuTest_difficultQNB_beta0.5)),rep("beta = 0.75",nrow(simuTest_difficultQNB_beta0.75)),rep("beta = 1",nrow(simuTest_difficultQNB_beta1)) ) )
ggplot(QNBmodel_cov_combined[QNBmodel_cov_combined$fdr_cutoff == 0.1,] , aes(x = method, y = fdr, colour = method))+geom_boxplot()+facet_wrap(beta~.) + geom_abline(slope = 0,intercept = 0.1,lty=2)+ylab("FDR")+xlab("Methods")+ ggtitle("QNB Model With Covariates") +theme_bw()+theme(legend.position = "none", text = element_text(face = "bold",colour = "black"), axis.text = element_text(size = 12,colour = "black"), axis.title = element_text(size = 16), strip.text = element_text(size = 16), plot.title = element_text(hjust = 0.5) )+scale_y_continuous(limits = c(0,1))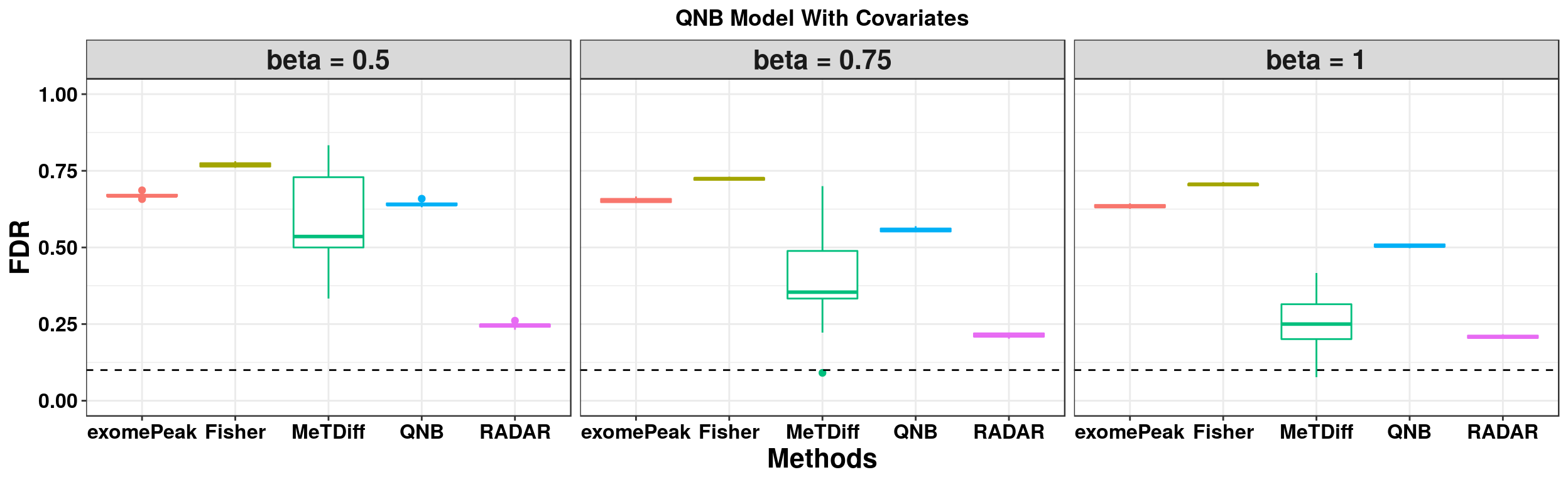
ggplot(QNBmodel_cov_combined[QNBmodel_cov_combined$fdr_cutoff == 0.1,] , aes(x = method, y = power, colour = method))+geom_boxplot()+facet_wrap(beta~.) + geom_abline(slope = 0,intercept = 1,lty=2)+ylab("Sensitivity")+xlab("Methods")+ggtitle("QNB Model With Covariates") +theme_bw()+theme(legend.position = "none", text = element_text(face = "bold",colour = "black"), axis.text = element_text(size = 12,colour = "black"), axis.title = element_text(size = 16), strip.text = element_text(size = 16), plot.title = element_text(hjust = 0.5) )+scale_y_continuous(limits = c(0,1))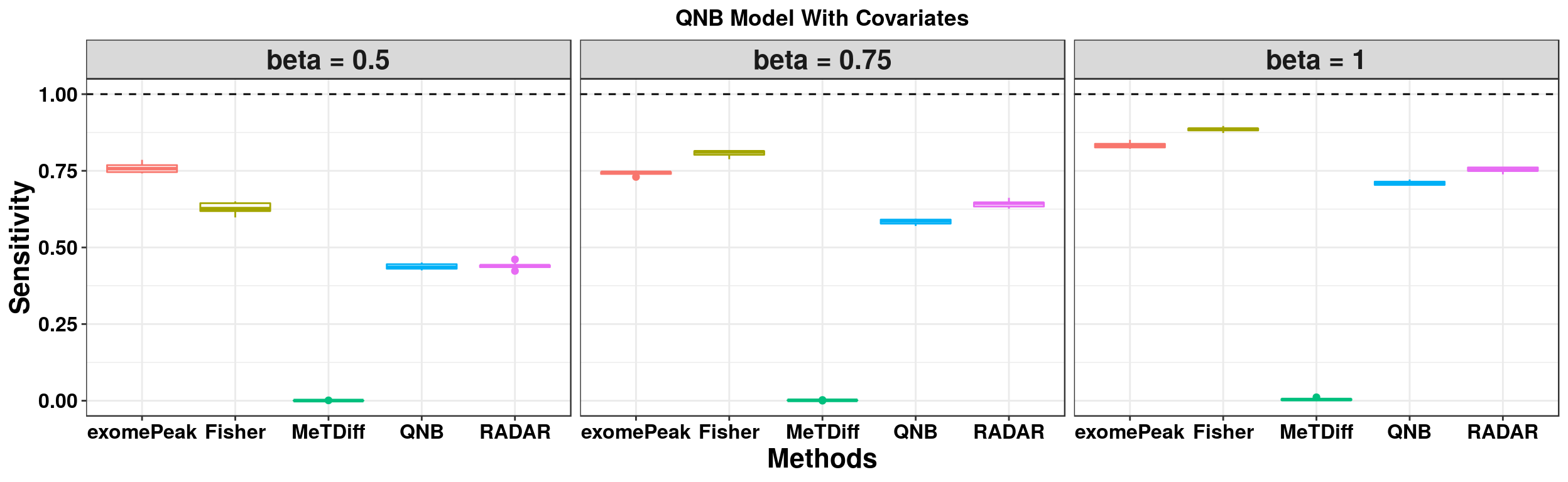
ggplot(QNBmodel_cov_combined[QNBmodel_cov_combined$fdr_cutoff == 0.1,] , aes(x = fdr, y = power, colour = method, shape = method))+geom_point(size = 4)+facet_wrap(beta~.) + geom_vline(xintercept = 0.1,lty=2)+ylab("Sensitivity")+xlab("FDR")+ggtitle("QNB Model With Covariates") +theme_bw()+theme( text = element_text(face = "bold",colour = "black"), axis.text = element_text(size = 12,colour = "black"), axis.title = element_text(size = 16), strip.text = element_text(size = 16), plot.title = element_text(hjust = 0.5), legend.text = element_text(size = 15) )+scale_y_continuous(limits = c(0,1))+scale_x_continuous(limits = c(0,1))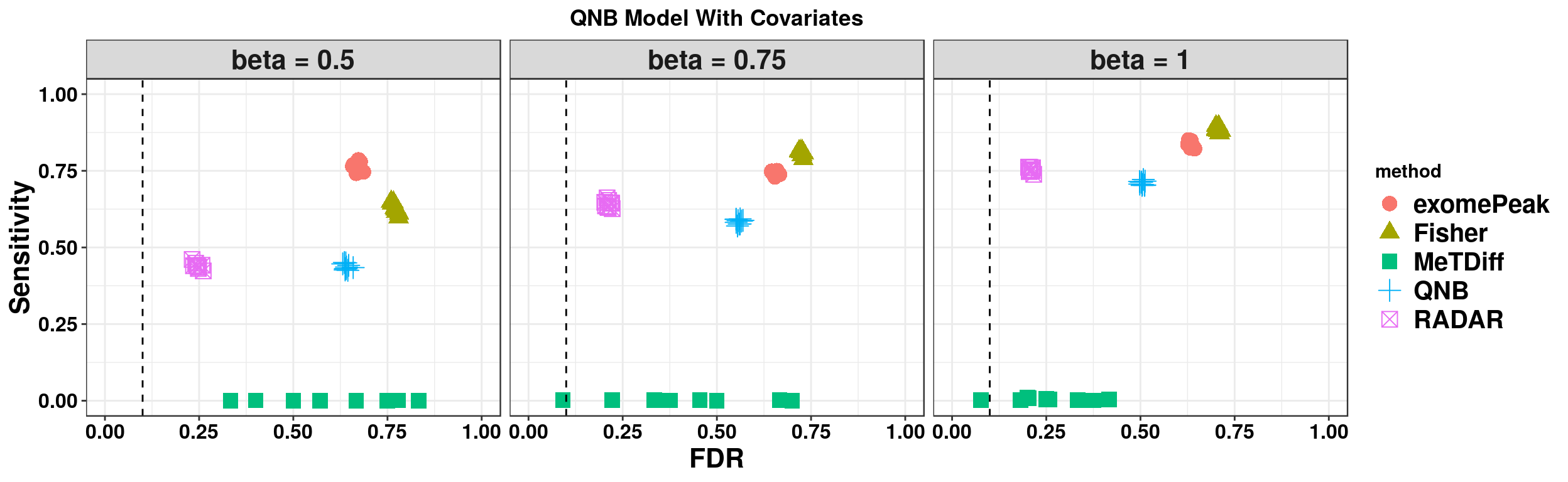
Check estimation
iteration <- RADAR_simulateData2(n_Sites = 26324,beta = 0.75, replicate = 8, psi = 5 , gene_specific_intercept_mean = 5, covariate_effect_size = 0 , seed = i*3)
simu.radar <- MeRIP.RADAR()
variable(simu.radar) <- data.frame( iteration$variable$predictor )
simu.radar@ip_adjExpr_filtered <- cbind(iteration$IP_control, iteration$IP_treat)
rownames(simu.radar@ip_adjExpr_filtered) <- paste0("peak",1:26324)
samplenames(simu.radar) <- paste0("s",1:16)
simu.radar@norm.input <- cbind(CtlInput[,1:8], TreatInput[,1:8])
colnames(simu.radar@norm.input ) <- paste0("s",1:16)
enrichRaio <- t( apply(simu.radar@ip_adjExpr_filtered, 1, function(x){x/mean(x)}*max(1,rnorm(1,5.6,2) ) ) )
simu.radar@norm.ip <- round( simu.radar@norm.input * enrichRaio )
simu.radar <- diffIP_parallel( simu.radar , thread = 20)
simu.Metdiff <-MeTDiffTest(control_ip = simu.radar@norm.ip[,1:8],
treated_ip = simu.radar@norm.ip[,9:16],
control_input = simu.radar@norm.input[,1:8],
treated_input = simu.radar@norm.input[,9:16] )
simu.qnb <- QNB::qnbtest(control_ip = simu.radar@norm.ip[,1:8],
treated_ip = simu.radar@norm.ip[,9:16],
control_input = simu.radar@norm.input[,1:8],
treated_input = simu.radar@norm.input[,9:16] ,plot.dispersion = FALSE)
simu.fisher <- fisherTest(control_ip = simu.radar@norm.ip[,1:8],
treated_ip = simu.radar@norm.ip[,9:16],
control_input = simu.radar@norm.input[,1:8],
treated_input = simu.radar@norm.input[,9:16] )
simu.exomePeak <- Bltest(control_ip = simu.radar@norm.ip[,1:8],
treated_ip = simu.radar@norm.ip[,9:16],
control_input = simu.radar@norm.input[,1:8],
treated_input = simu.radar@norm.input[,9:16])
simu.result <- data.frame(pvalue = c(simu.radar@test.est[,"p_value"],simu.Metdiff$pvalue,simu.qnb$pvalue,simu.fisher$pvalue,simu.exomePeak$pvalue),
beta = c(simu.radar@test.est[,"beta"],simu.Metdiff$logFC,log(2^(simu.qnb$log2.OR)),simu.fisher$logFC,simu.exomePeak$logFC),
truth = rep( 1:26324%in%iteration$df_site_index , 5),
method = factor( c(rep("RADAR",26324),rep("MeTDiff",26324),rep("QNB",26324),rep("Fisher",26324),rep("exomePeak",26324)),levels = c( c("RADAR", "MeTDiff", "QNB", "Fisher", "exomePeak"))),
model = "simple random effect model"
)
iteration <- RADAR_simulateData2(n_Sites = 26324,beta = 0.75, replicate = 8, psi = 5 , gene_specific_intercept_mean = 5, covariate_effect_size = 2 , seed = i*3)
simu.radar <- MeRIP.RADAR()
variable(simu.radar) <- iteration$variable
simu.radar@ip_adjExpr_filtered <- cbind(iteration$IP_control, iteration$IP_treat)
rownames(simu.radar@ip_adjExpr_filtered) <- paste0("peak",1:26324)
samplenames(simu.radar) <- paste0("s",1:16)
simu.radar@norm.input <- cbind(CtlInput[,1:8], TreatInput[,1:8])
colnames(simu.radar@norm.input ) <- paste0("s",1:16)
enrichRaio <- t( apply(simu.radar@ip_adjExpr_filtered, 1, function(x){x/mean(x)}*max(1,rnorm(1,5.6,2) ) ) )
simu.radar@norm.ip <- round( simu.radar@norm.input * enrichRaio )
simu.radar <- diffIP_parallel( simu.radar , thread = 20)
simu.Metdiff <-MeTDiffTest(control_ip = simu.radar@norm.ip[,1:8],
treated_ip = simu.radar@norm.ip[,9:16],
control_input = simu.radar@norm.input[,1:8],
treated_input = simu.radar@norm.input[,9:16] )
simu.qnb <- QNB::qnbtest(control_ip = simu.radar@norm.ip[,1:8],
treated_ip = simu.radar@norm.ip[,9:16],
control_input = simu.radar@norm.input[,1:8],
treated_input = simu.radar@norm.input[,9:16] ,plot.dispersion = FALSE)
simu.fisher <- fisherTest(control_ip = simu.radar@norm.ip[,1:8],
treated_ip = simu.radar@norm.ip[,9:16],
control_input = simu.radar@norm.input[,1:8],
treated_input = simu.radar@norm.input[,9:16] )
simu.exomePeak <- Bltest(control_ip = simu.radar@norm.ip[,1:8],
treated_ip = simu.radar@norm.ip[,9:16],
control_input = simu.radar@norm.input[,1:8],
treated_input = simu.radar@norm.input[,9:16])
simu.result <- rbind( simu.result,
data.frame(pvalue = c(simu.radar@test.est[,"p_value"],simu.Metdiff$pvalue,simu.qnb$pvalue,simu.fisher$pvalue,simu.exomePeak$pvalue),
beta = c(simu.radar@test.est[,"beta"],simu.Metdiff$logFC,log(2^(simu.qnb$log2.OR)),simu.fisher$logFC,simu.exomePeak$logFC),
truth = rep( 1:26324%in%iteration$df_site_index , 5),
method = factor( c(rep("RADAR",26324),rep("MeTDiff",26324),rep("QNB",26324),rep("Fisher",26324),rep("exomePeak",26324)),levels = c( c("RADAR", "MeTDiff", "QNB", "Fisher", "exomePeak"))),
model = "difficult random effect model")
)
save.image("~/Tools/RADARmannual/data/simulationAnalysis.RData")ggplot(simu.result[!simu.result$truth,],aes(x = pvalue))+geom_histogram(breaks = seq(0,1,0.05),col=I("black"))+facet_grid(model~method)+ggtitle("Null sites")+theme_bw()+theme(axis.title = element_text(size = 20, face = "bold"),strip.text = element_text(size = 12, face = "bold"), axis.text =element_text(color = "black",size = 12, face = "bold") )+scale_x_continuous(breaks = seq(0,1,0.2))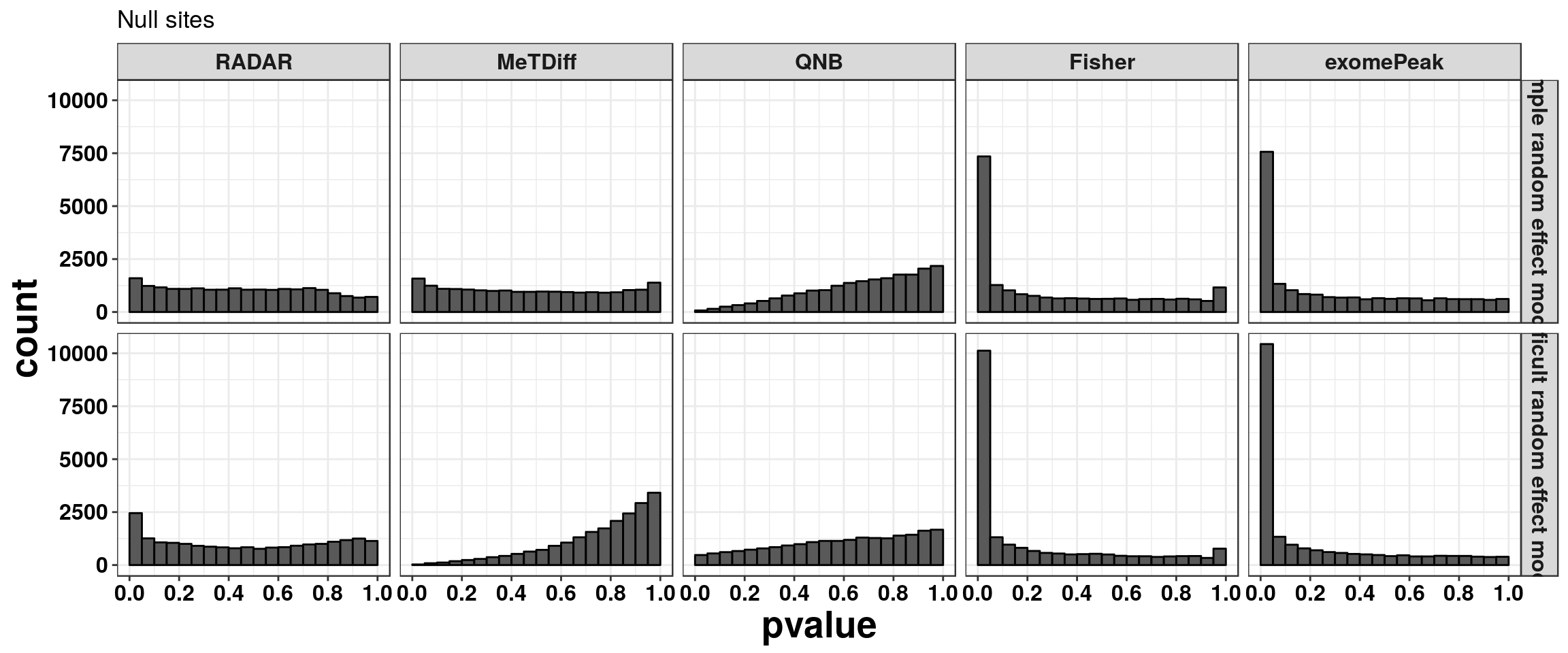
ggplot(simu.result[!simu.result$truth,],aes(x = beta))+geom_histogram(breaks = seq(-2,2,0.1),col=I("black"))+facet_grid(model~method)+ggtitle("Null sites")+theme_bw()+theme(axis.title = element_text(size = 20, face = "bold"),strip.text = element_text(size = 12, face = "bold"), axis.text =element_text(color = "black",size = 12, face = "bold") )+geom_vline(xintercept = 0, lty = "dashed")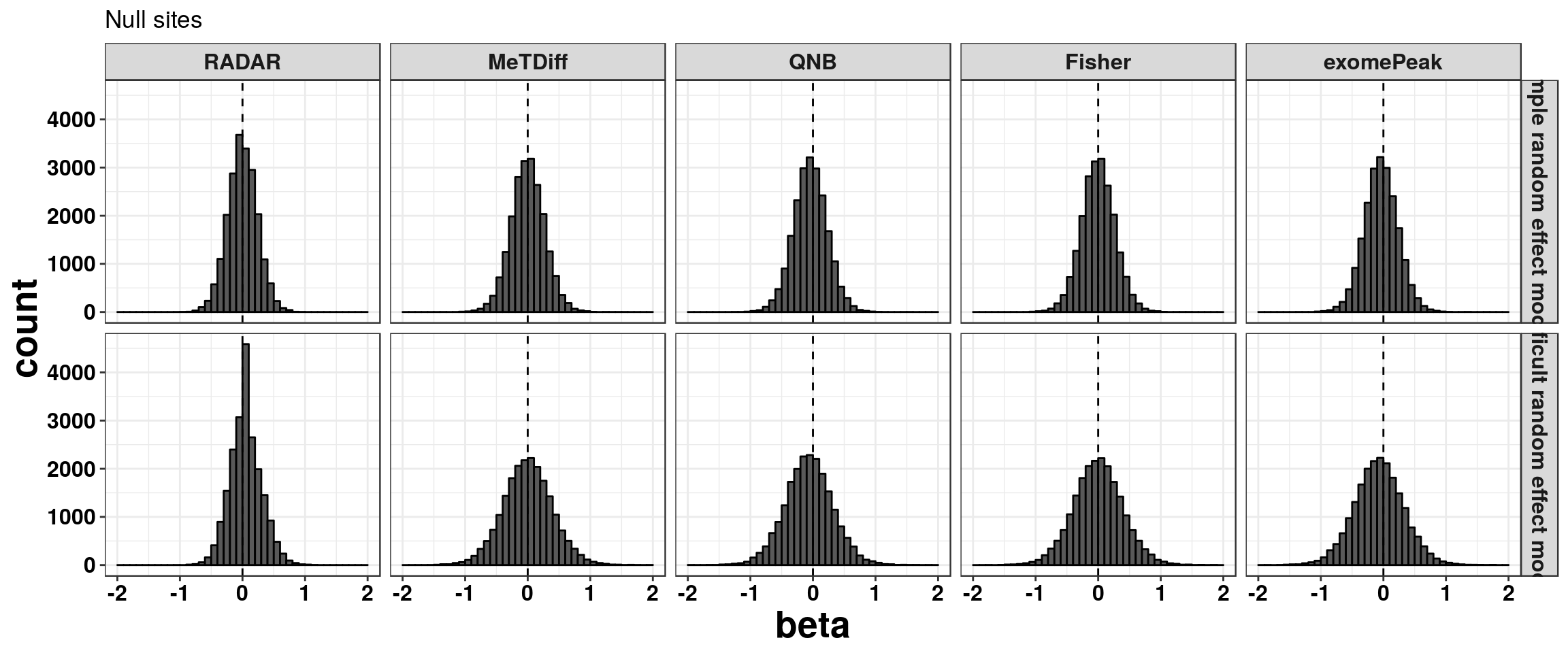
ggplot(simu.result[simu.result$truth,],aes(x = pvalue))+geom_histogram(breaks = seq(0,1,0.05),col=I("black"))+facet_grid(model~method)+ggtitle("True sites")+theme_bw()+theme(axis.title = element_text(size = 20, face = "bold"),strip.text = element_text(size = 12, face = "bold"), axis.text =element_text(color = "black",size = 12, face = "bold") )+scale_x_continuous(breaks = seq(0,1,0.2))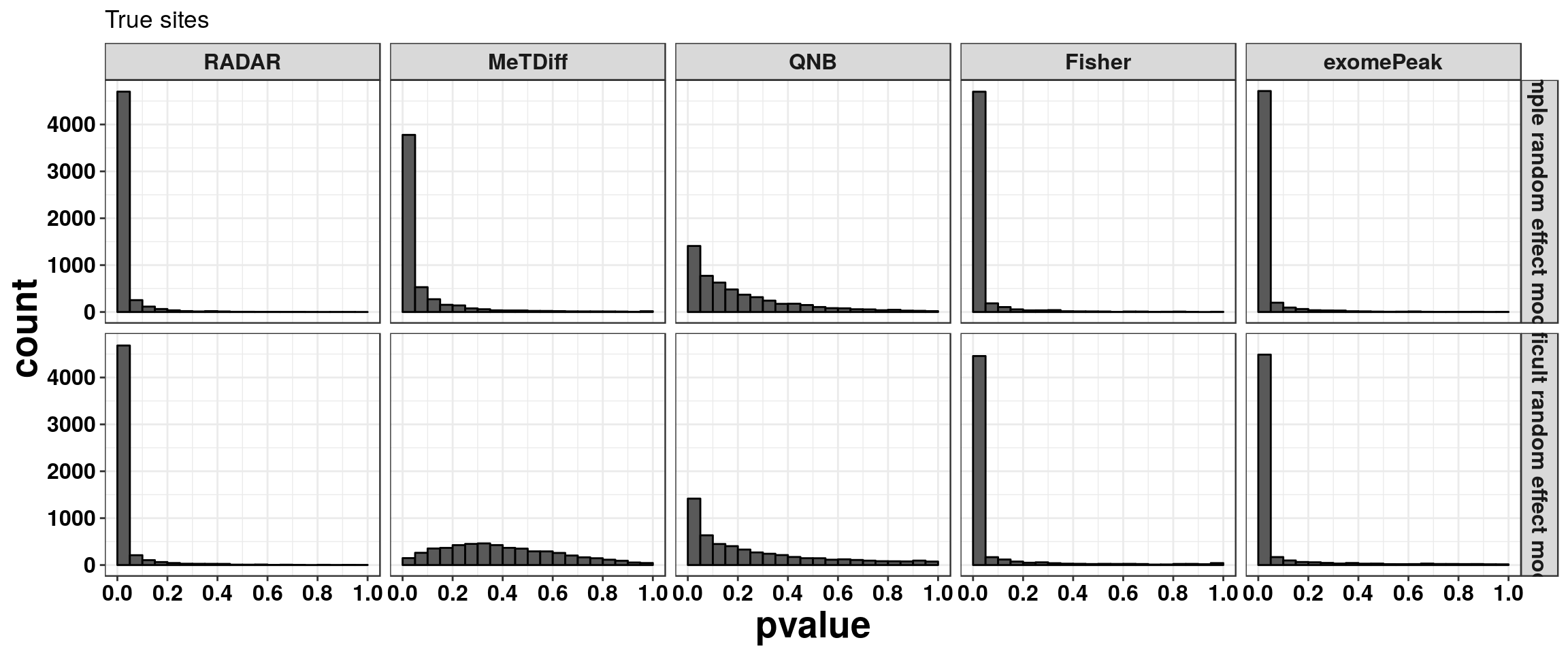
ggplot(simu.result[simu.result$truth,],aes(x = beta))+geom_histogram(breaks = seq(-2,2,0.1),col=I("black"))+facet_grid(model~method)+ggtitle("True sites")+theme_bw()+theme(axis.title = element_text(size = 20, face = "bold"),strip.text = element_text(size = 12, face = "bold"), axis.text =element_text(color = "black",size = 12, face = "bold") )+geom_vline(xintercept = -0.75, lty = "dashed")+geom_vline(xintercept = 0.75, lty = "dashed")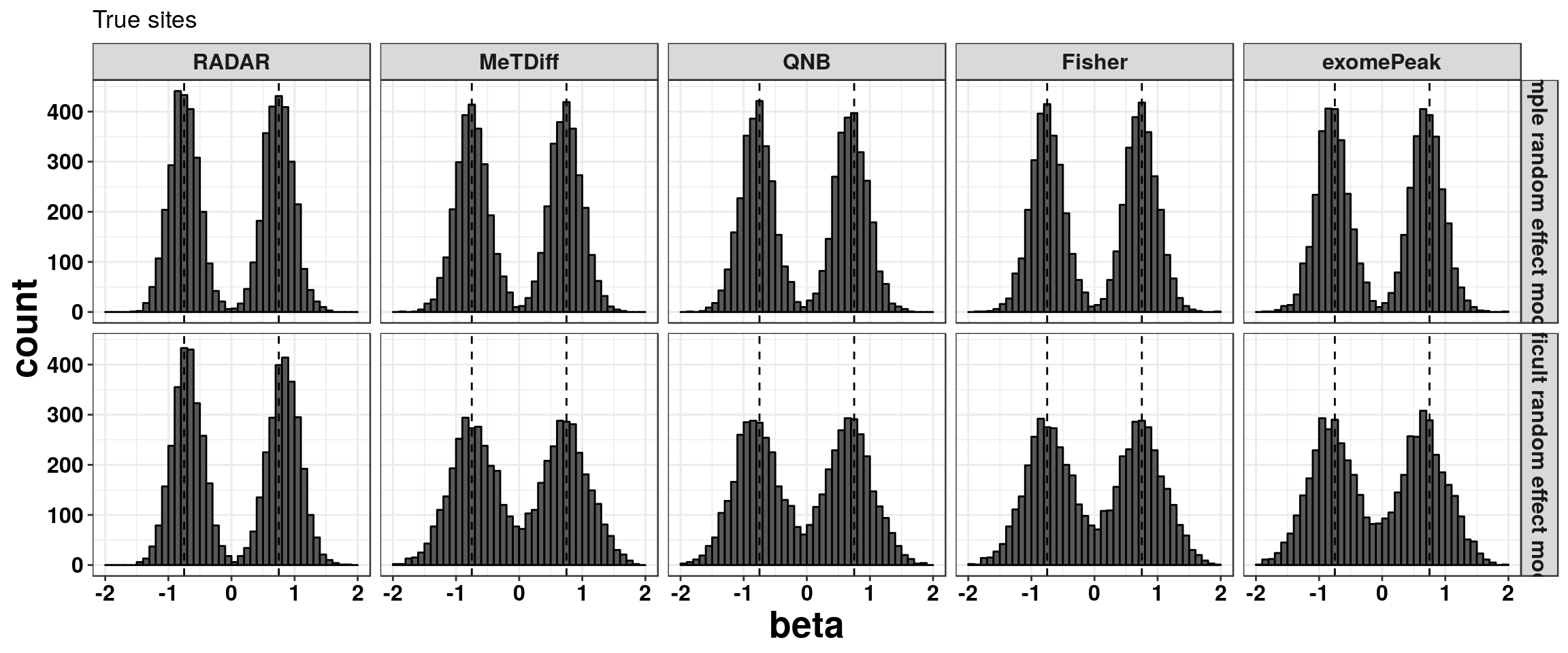
Analyze the impact of the number of replicates
Using the simple RADAR model, we simulate the impact of the N number on the power and false discovery rate.
registerDoParallel(10)
simuTest_beta0.75_N2 <- foreach(i = 1:10, .combine = rbind)%dopar%{
iteration <- RADAR_simulateData2(n_Sites = 26324,beta = 0.75, replicate = 2, psi = 5 , gene_specific_intercept_mean = 5, covariate_effect_size = 0 , seed = i*3)
simu.radar <- MeRIP.RADAR()
variable(simu.radar) <- data.frame( iteration$variable$predictor )
simu.radar@Peak_adjExpr <- cbind(iteration$IP_control, iteration$IP_treat)
rownames(simu.radar@Peak_adjExpr) <- paste0("peak",1:26324)
samplenames(simu.radar) <- paste0("s",1:4)
simu.radar@Peak_input <- cbind(CtlInput[,1:2], TreatInput[,1:2])
colnames(simu.radar@Peak_input ) <- paste0("s",1:4)
enrichRaio <- t( apply(simu.radar@Peak_adjExpr, 1, function(x){x/mean(x)}*max(1,rnorm(1,5.6,2) ) ) )
simu.radar@Peak_ip <- round( simu.radar@Peak_input * enrichRaio )
simu.radar <- diffIP( simu.radar )
simu.Metdiff <- MeTDiffTest(control_ip = simu.radar@Peak_ip[,1:2],
treated_ip = simu.radar@Peak_ip[,3:4],
control_input = simu.radar@Peak_input[,1:2],
treated_input = simu.radar@Peak_input[,3:4] )
simu.qnb <- QNB::qnbtest(control_ip = simu.radar@Peak_ip[,1:2],
treated_ip = simu.radar@Peak_ip[,3:4],
control_input = simu.radar@Peak_input[,1:2],
treated_input = simu.radar@Peak_input[,3:4],plot.dispersion = FALSE)
simu.fisher <- fisherTest(control_ip = simu.radar@Peak_ip[,1:2],
treated_ip = simu.radar@Peak_ip[,3:4],
control_input = simu.radar@Peak_input[,1:2],
treated_input = simu.radar@Peak_input[,3:4] )
simu.exomePeak <- Bltest(control_ip = simu.radar@Peak_ip[,1:2],
treated_ip = simu.radar@Peak_ip[,3:4],
control_input = simu.radar@Peak_input[,1:2],
treated_input = simu.radar@Peak_input[,3:4] )
result <- NULL
for(q in c(0.01, 0.05, 0.1, 0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9,0.95,0.99)){
tmpPower <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" =simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), iteration$df_site_index ) )/length(iteration$df_site_index)})
tmpFDR <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length(which( x < q ) )})
tmpFalsePosRate <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length( setdiff( 1:26324, iteration$df_site_index ) ) })
result <- rbind(result, data.frame( power = tmpPower, fdr = tmpFDR, fpr = tmpFalsePosRate, method = c("RADAR","MeTDiff","exomePeak","Fisher","QNB") , fdr_cutoff = q ) )
}
result
}
simuTest_beta0.75_N3 <- foreach(i = 1:10, .combine = rbind)%dopar%{
iteration <- RADAR_simulateData2(n_Sites = 26324,beta = 0.75, replicate = 3, psi = 5 , gene_specific_intercept_mean = 5, covariate_effect_size = 0 , seed = i*3)
simu.radar <- MeRIP.RADAR()
variable(simu.radar) <- data.frame( iteration$variable$predictor )
simu.radar@Peak_adjExpr <- cbind(iteration$IP_control, iteration$IP_treat)
rownames(simu.radar@Peak_adjExpr) <- paste0("peak",1:26324)
samplenames(simu.radar) <- paste0("s",1:6)
simu.radar@Peak_input <- cbind(CtlInput[,1:3], TreatInput[,1:3])
colnames(simu.radar@Peak_input ) <- paste0("s",1:6)
enrichRaio <- t( apply(simu.radar@Peak_adjExpr, 1, function(x){x/mean(x)}*max(1,rnorm(1,5.6,2) ) ) )
simu.radar@Peak_ip <- round( simu.radar@Peak_input * enrichRaio )
simu.radar <- diffIP( simu.radar )
simu.Metdiff <- MeTDiffTest(control_ip = simu.radar@Peak_ip[,1:3],
treated_ip = simu.radar@Peak_ip[,4:6],
control_input = simu.radar@Peak_input[,1:3],
treated_input = simu.radar@Peak_input[,4:6] )
simu.qnb <- QNB::qnbtest(control_ip = simu.radar@Peak_ip[,1:3],
treated_ip = simu.radar@Peak_ip[,4:6],
control_input = simu.radar@Peak_input[,1:3],
treated_input = simu.radar@Peak_input[,4:6],plot.dispersion = FALSE)
simu.fisher <- fisherTest(control_ip = simu.radar@Peak_ip[,1:3],
treated_ip = simu.radar@Peak_ip[,4:6],
control_input = simu.radar@Peak_input[,1:3],
treated_input = simu.radar@Peak_input[,4:6] )
simu.exomePeak <- Bltest(control_ip = simu.radar@Peak_ip[,1:3],
treated_ip = simu.radar@Peak_ip[,4:6],
control_input = simu.radar@Peak_input[,1:3],
treated_input = simu.radar@Peak_input[,4:6] )
result <- NULL
for(q in c(0.01, 0.05, 0.1, 0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9,0.95,0.99)){
tmpPower <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" =simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), iteration$df_site_index ) )/length(iteration$df_site_index)})
tmpFDR <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length(which( x < q ) )})
tmpFalsePosRate <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length( setdiff( 1:26324, iteration$df_site_index ) ) })
result <- rbind(result, data.frame( power = tmpPower, fdr = tmpFDR, fpr = tmpFalsePosRate, method = c("RADAR","MeTDiff","exomePeak","Fisher","QNB") , fdr_cutoff = q ) )
}
result
}
simuTest_beta0.75_N4 <- foreach(i = 1:10, .combine = rbind)%dopar%{
iteration <- RADAR_simulateData2(n_Sites = 26324,beta = 0.75, replicate = 4, psi = 5 , gene_specific_intercept_mean = 5, covariate_effect_size = 0 , seed = i*3)
simu.radar <- MeRIP.RADAR()
variable(simu.radar) <- data.frame( iteration$variable$predictor )
simu.radar@Peak_adjExpr <- cbind(iteration$IP_control, iteration$IP_treat)
rownames(simu.radar@Peak_adjExpr) <- paste0("peak",1:26324)
samplenames(simu.radar) <- paste0("s",1:8)
simu.radar@Peak_input <- cbind(CtlInput[,1:4], TreatInput[,1:4])
colnames(simu.radar@Peak_input ) <- paste0("s",1:8)
enrichRaio <- t( apply(simu.radar@Peak_adjExpr, 1, function(x){x/mean(x)}*max(1,rnorm(1,5.6,2) ) ) )
simu.radar@Peak_ip <- round( simu.radar@Peak_input * enrichRaio )
simu.radar <- diffIP( simu.radar )
simu.Metdiff <- MeTDiffTest(control_ip = simu.radar@Peak_ip[,1:4],
treated_ip = simu.radar@Peak_ip[,5:8],
control_input = simu.radar@Peak_input[,1:4],
treated_input = simu.radar@Peak_input[,5:8] )
simu.qnb <- QNB::qnbtest(control_ip = simu.radar@Peak_ip[,1:4],
treated_ip = simu.radar@Peak_ip[,5:8],
control_input = simu.radar@Peak_input[,1:4],
treated_input = simu.radar@Peak_input[,5:8],plot.dispersion = FALSE)
simu.fisher <- fisherTest(control_ip = simu.radar@Peak_ip[,1:4],
treated_ip = simu.radar@Peak_ip[,5:8],
control_input = simu.radar@Peak_input[,1:4],
treated_input = simu.radar@Peak_input[,5:8] )
simu.exomePeak <- Bltest(control_ip = simu.radar@Peak_ip[,1:4],
treated_ip = simu.radar@Peak_ip[,5:8],
control_input = simu.radar@Peak_input[,1:4],
treated_input = simu.radar@Peak_input[,5:8] )
result <- NULL
for(q in c(0.01, 0.05, 0.1, 0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9,0.95,0.99)){
tmpPower <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" =simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), iteration$df_site_index ) )/length(iteration$df_site_index)})
tmpFDR <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length(which( x < q ) )})
tmpFalsePosRate <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length( setdiff( 1:26324, iteration$df_site_index ) ) })
result <- rbind(result, data.frame( power = tmpPower, fdr = tmpFDR, fpr = tmpFalsePosRate, method = c("RADAR","MeTDiff","exomePeak","Fisher","QNB") , fdr_cutoff = q ) )
}
result
}
simuTest_beta0.75_N5 <- foreach(i = 1:10, .combine = rbind)%dopar%{
iteration <- RADAR_simulateData2(n_Sites = 26324,beta = 0.75, replicate = 5, psi = 5 , gene_specific_intercept_mean = 5, covariate_effect_size = 0 , seed = i*3)
simu.radar <- MeRIP.RADAR()
variable(simu.radar) <- data.frame( iteration$variable$predictor )
simu.radar@Peak_adjExpr <- cbind(iteration$IP_control, iteration$IP_treat)
rownames(simu.radar@Peak_adjExpr) <- paste0("peak",1:26324)
samplenames(simu.radar) <- paste0("s",1:10)
simu.radar@Peak_input <- cbind(CtlInput[,1:5], TreatInput[,1:5])
colnames(simu.radar@Peak_input ) <- paste0("s",1:10)
enrichRaio <- t( apply(simu.radar@Peak_adjExpr, 1, function(x){x/mean(x)}*max(1,rnorm(1,5.6,2) ) ) )
simu.radar@Peak_ip <- round( simu.radar@Peak_input * enrichRaio )
simu.radar <- diffIP( simu.radar )
simu.Metdiff <- MeTDiffTest(control_ip = simu.radar@Peak_ip[,1:5],
treated_ip = simu.radar@Peak_ip[,6:10],
control_input = simu.radar@Peak_input[,1:5],
treated_input = simu.radar@Peak_input[,6:10] )
simu.qnb <- QNB::qnbtest(control_ip = simu.radar@Peak_ip[,1:5],
treated_ip = simu.radar@Peak_ip[,6:10],
control_input = simu.radar@Peak_input[,1:5],
treated_input = simu.radar@Peak_input[,6:10],plot.dispersion = FALSE)
simu.fisher <- fisherTest(control_ip = simu.radar@Peak_ip[,1:5],
treated_ip = simu.radar@Peak_ip[,6:10],
control_input = simu.radar@Peak_input[,1:5],
treated_input = simu.radar@Peak_input[,6:10] )
simu.exomePeak <- Bltest(control_ip = simu.radar@Peak_ip[,1:5],
treated_ip = simu.radar@Peak_ip[,6:10],
control_input = simu.radar@Peak_input[,1:5],
treated_input = simu.radar@Peak_input[,6:10] )
result <- NULL
for(q in c(0.01, 0.05, 0.1, 0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9,0.95,0.99)){
tmpPower <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" =simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), iteration$df_site_index ) )/length(iteration$df_site_index)})
tmpFDR <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length(which( x < q ) )})
tmpFalsePosRate <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length( setdiff( 1:26324, iteration$df_site_index ) ) })
result <- rbind(result, data.frame( power = tmpPower, fdr = tmpFDR, fpr = tmpFalsePosRate, method = c("RADAR","MeTDiff","exomePeak","Fisher","QNB") , fdr_cutoff = q ) )
}
result
}
simuTest_beta0.75_N6 <- foreach(i = 1:10, .combine = rbind)%dopar%{
iteration <- RADAR_simulateData2(n_Sites = 26324,beta = 0.75, replicate = 6, psi = 5 , gene_specific_intercept_mean = 5, covariate_effect_size = 0 , seed = i*3)
simu.radar <- MeRIP.RADAR()
variable(simu.radar) <- data.frame( iteration$variable$predictor )
simu.radar@Peak_adjExpr <- cbind(iteration$IP_control, iteration$IP_treat)
rownames(simu.radar@Peak_adjExpr) <- paste0("peak",1:26324)
samplenames(simu.radar) <- paste0("s",1:12)
simu.radar@Peak_input <- cbind(CtlInput[,1:6], TreatInput[,1:6])
colnames(simu.radar@Peak_input ) <- paste0("s",1:12)
enrichRaio <- t( apply(simu.radar@Peak_adjExpr, 1, function(x){x/mean(x)}*max(1,rnorm(1,5.6,2) ) ) )
simu.radar@Peak_ip <- round( simu.radar@Peak_input * enrichRaio )
simu.radar <- diffIP( simu.radar )
simu.Metdiff <- MeTDiffTest(control_ip = simu.radar@Peak_ip[,1:6],
treated_ip = simu.radar@Peak_ip[,7:12],
control_input = simu.radar@Peak_input[,1:6],
treated_input = simu.radar@Peak_input[,7:12] )
simu.qnb <- QNB::qnbtest(control_ip = simu.radar@Peak_ip[,1:6],
treated_ip = simu.radar@Peak_ip[,7:12],
control_input = simu.radar@Peak_input[,1:6],
treated_input = simu.radar@Peak_input[,7:12],plot.dispersion = FALSE)
simu.fisher <- fisherTest(control_ip = simu.radar@Peak_ip[,1:6],
treated_ip = simu.radar@Peak_ip[,7:12],
control_input = simu.radar@Peak_input[,1:6],
treated_input = simu.radar@Peak_input[,7:12] )
simu.exomePeak <- Bltest(control_ip = simu.radar@Peak_ip[,1:6],
treated_ip = simu.radar@Peak_ip[,7:12],
control_input = simu.radar@Peak_input[,1:6],
treated_input = simu.radar@Peak_input[,7:12] )
result <- NULL
for(q in c(0.01, 0.05, 0.1, 0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9,0.95,0.99)){
tmpPower <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" =simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), iteration$df_site_index ) )/length(iteration$df_site_index)})
tmpFDR <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length(which( x < q ) )})
tmpFalsePosRate <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length( setdiff( 1:26324, iteration$df_site_index ) ) })
result <- rbind(result, data.frame( power = tmpPower, fdr = tmpFDR, fpr = tmpFalsePosRate, method = c("RADAR","MeTDiff","exomePeak","Fisher","QNB") , fdr_cutoff = q ) )
}
result
}
simuTest_beta0.75_N7 <- foreach(i = 1:10, .combine = rbind)%dopar%{
iteration <- RADAR_simulateData2(n_Sites = 26324,beta = 0.75, replicate = 7, psi = 5 , gene_specific_intercept_mean = 5, covariate_effect_size = 0 , seed = i*3)
simu.radar <- MeRIP.RADAR()
variable(simu.radar) <- data.frame( iteration$variable$predictor )
simu.radar@Peak_adjExpr <- cbind(iteration$IP_control, iteration$IP_treat)
rownames(simu.radar@Peak_adjExpr) <- paste0("peak",1:26324)
samplenames(simu.radar) <- paste0("s",1:14)
simu.radar@Peak_input <- cbind(CtlInput[,1:7], TreatInput[,1:7])
colnames(simu.radar@Peak_input ) <- paste0("s",1:14)
enrichRaio <- t( apply(simu.radar@Peak_adjExpr, 1, function(x){x/mean(x)}*max(1,rnorm(1,5.6,2) ) ) )
simu.radar@Peak_ip <- round( simu.radar@Peak_input * enrichRaio )
simu.radar <- diffIP( simu.radar )
simu.Metdiff <- MeTDiffTest(control_ip = simu.radar@Peak_ip[,1:7],
treated_ip = simu.radar@Peak_ip[,8:14],
control_input = simu.radar@Peak_input[,1:7],
treated_input = simu.radar@Peak_input[,8:14] )
simu.qnb <- QNB::qnbtest(control_ip = simu.radar@Peak_ip[,1:7],
treated_ip = simu.radar@Peak_ip[,8:14],
control_input = simu.radar@Peak_input[,1:7],
treated_input = simu.radar@Peak_input[,8:14],plot.dispersion = FALSE)
simu.fisher <- fisherTest(control_ip = simu.radar@Peak_ip[,1:7],
treated_ip = simu.radar@Peak_ip[,8:14],
control_input = simu.radar@Peak_input[,1:7],
treated_input = simu.radar@Peak_input[,8:14] )
simu.exomePeak <- Bltest(control_ip = simu.radar@Peak_ip[,1:7],
treated_ip = simu.radar@Peak_ip[,8:14],
control_input = simu.radar@Peak_input[,1:7],
treated_input = simu.radar@Peak_input[,8:14] )
result <- NULL
for(q in c(0.01, 0.05, 0.1, 0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9,0.95,0.99)){
tmpPower <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" =simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), iteration$df_site_index ) )/length(iteration$df_site_index)})
tmpFDR <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length(which( x < q ) )})
tmpFalsePosRate <- apply(cbind("RADAR"=simu.radar@test.est[,"fdr"],"MeTDiff"=simu.Metdiff$fdr,"exomePeak" = simu.exomePeak$fdr,"Fisher"=simu.fisher$fdr,"QNB"=simu.qnb$padj),2, function(x){
length( intersect( which( x < q ), setdiff( 1:26324, iteration$df_site_index ) ) )/length( setdiff( 1:26324, iteration$df_site_index ) ) })
result <- rbind(result, data.frame( power = tmpPower, fdr = tmpFDR, fpr = tmpFalsePosRate, method = c("RADAR","MeTDiff","exomePeak","Fisher","QNB") , fdr_cutoff = q ) )
}
result
}
simuTest_beta0.75_N8 <- simuTest_beta0.75
save.image("~/Tools/RADARmannual/data/simulationAnalysis.RData")sampleSizeSimu <- dplyr::mutate( rbind(simuTest_beta0.75_N2,simuTest_beta0.75_N3,simuTest_beta0.75_N4,simuTest_beta0.75_N5,simuTest_beta0.75_N6,simuTest_beta0.75_N7,simuTest_beta0.75_N8),
sampleSize = c( rep("2",nrow(simuTest_beta0.75_N2)), rep("3",nrow(simuTest_beta0.75_N3) ), rep("4",nrow(simuTest_beta0.75_N4) ), rep("5",nrow(simuTest_beta0.75_N5) ), rep("6",nrow(simuTest_beta0.75_N6) ), rep("7",nrow(simuTest_beta0.75_N7) ) , rep("8",nrow(simuTest_beta0.75_N8) ) )
)
cbbPalette <- c("#000000", "#E69F00", "#56B4E9", "#009E73", "#F0E442", "#0072B2", "#D55E00", "#CC79A7")
sampleSizeSimu$method <- factor(sampleSizeSimu$method, levels = c("RADAR", "MeTDiff", "QNB", "Fisher", "exomePeak"))
ggplot(sampleSizeSimu[sampleSizeSimu$fdr_cutoff == 0.1,] , aes(x = fdr, y = power, colour = sampleSize))+geom_point(size = 3)+ facet_wrap(.~ method ) + geom_vline(xintercept = 0.1,lty=2)+ylab("Sensitivity")+xlab("FDR")+ggtitle("Simple RADAR Model with Varying Sample Size") +theme_bw()+theme(panel.grid.minor = element_blank(), text = element_text(face = "bold",colour = "black"), axis.text = element_text(size = 12,colour = "black"), axis.title = element_text(size = 16), strip.text = element_text(size = 16), plot.title = element_text(hjust = 0.5), legend.text = element_text(size = 15) )+scale_y_continuous(limits = c(0,1), breaks = seq(0,1,0.2))+scale_color_manual(values = cbbPalette)+scale_x_continuous(limits = c(0,1), breaks = seq(0,1,0.2))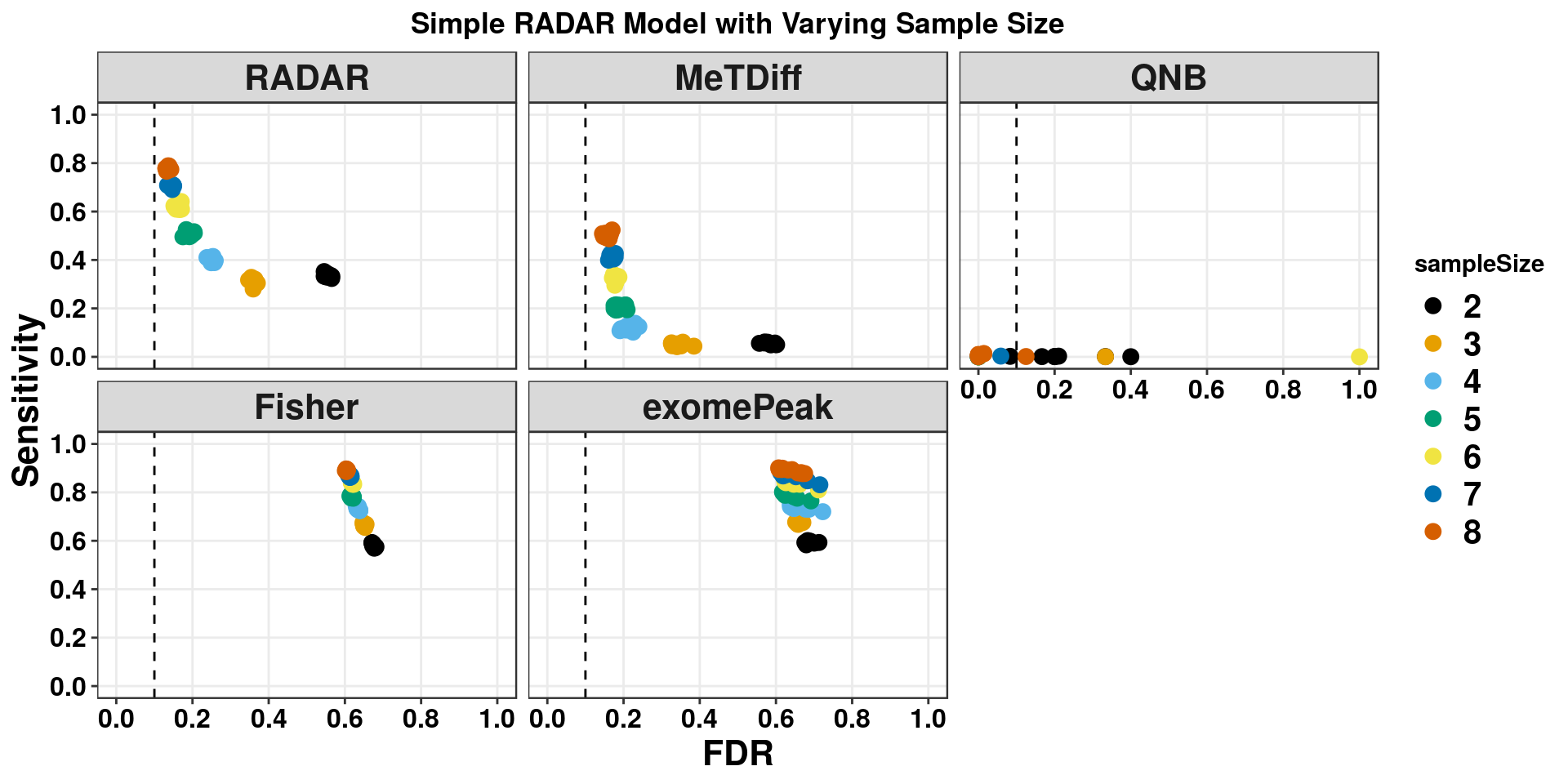
ggplot(sampleSizeSimu[sampleSizeSimu$method == "RADAR",] , aes(x = fpr, y = power, colour = sampleSize))+geom_smooth(se = FALSE)+ ylab("Sensitivity")+xlab("False Positive Rate")+ggtitle("Simple RADAR Model with Varying Sample Size") +theme_bw()+theme( text = element_text(face = "bold",colour = "black"), axis.text = element_text(size = 12,colour = "black"), axis.title = element_text(size = 16), strip.text = element_text(size = 16), plot.title = element_text(hjust = 0.5), legend.text = element_text(size = 15) )+scale_y_continuous(limits = c(0,1))+scale_color_manual(values = cbbPalette)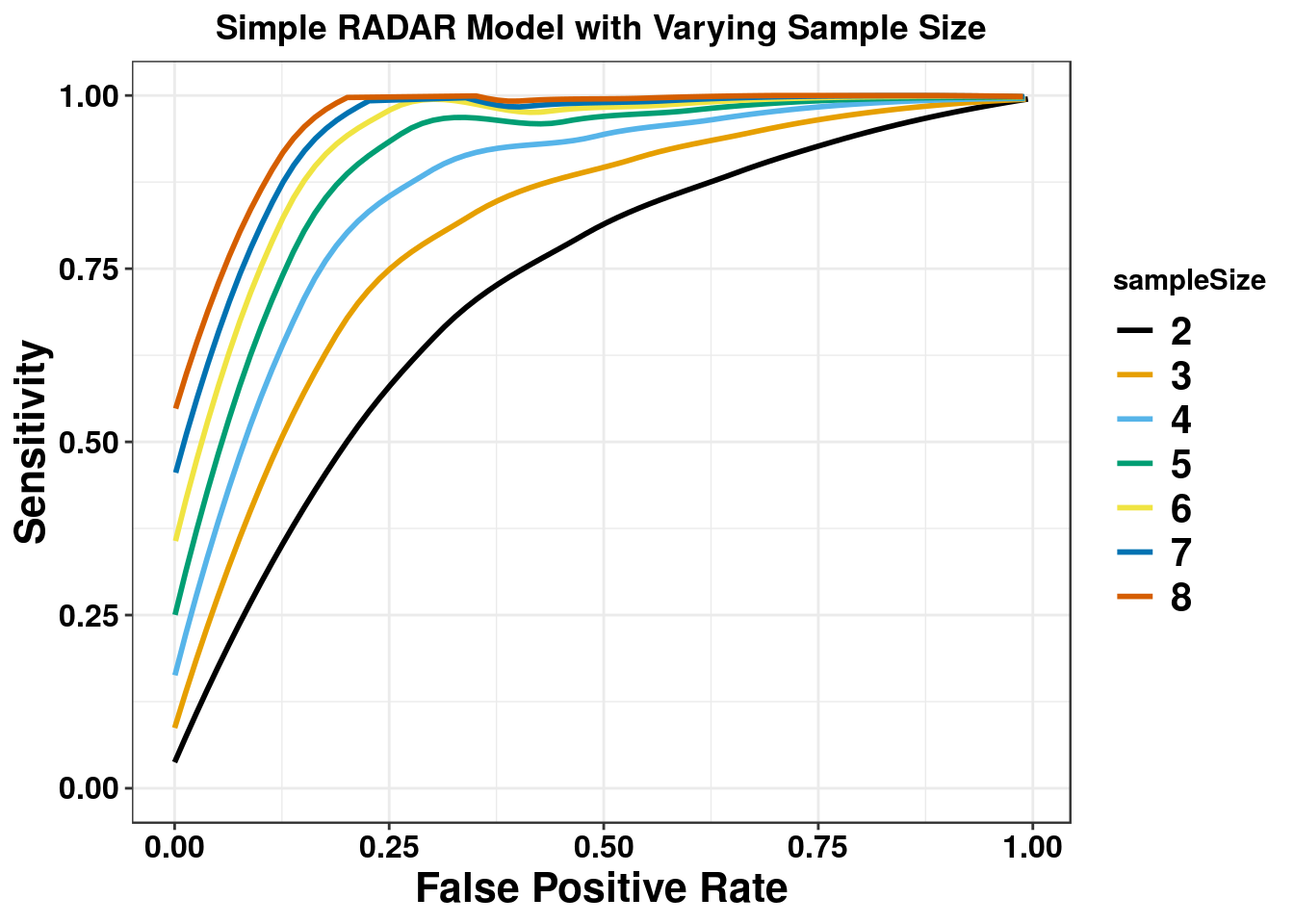
sampleSizeSimu_RADAR_mean <- data.frame( power = c( sapply( 2:8 , function(i){ tapply(sampleSizeSimu[sampleSizeSimu$method == "RADAR" & sampleSizeSimu$sampleSize == i,"power"],sampleSizeSimu[sampleSizeSimu$method == "RADAR" & sampleSizeSimu$sampleSize == i,"fdr_cutoff"],mean) } ) ),
fdr = c( sapply( 2:8 , function(i){ tapply(sampleSizeSimu[sampleSizeSimu$method == "RADAR" & sampleSizeSimu$sampleSize == i,"fdr"],sampleSizeSimu[sampleSizeSimu$method == "RADAR" & sampleSizeSimu$sampleSize == i,"fdr_cutoff"],mean) } ) ) ,
fpr =c( sapply( 2:8 , function(i){ tapply(sampleSizeSimu[sampleSizeSimu$method == "RADAR" & sampleSizeSimu$sampleSize == i,"fpr"],sampleSizeSimu[sampleSizeSimu$method == "RADAR" & sampleSizeSimu$sampleSize == i,"fdr_cutoff"],mean) } ) ) ,
sampleSize = as.character(c( sapply(2:8,rep, 13 ) ) )
)
ggplot(sampleSizeSimu_RADAR_mean , aes(x = fpr, y = power, colour = sampleSize))+geom_line()+ ylab("Sensitivity")+xlab("False Positive Rate")+ggtitle("Simple RADAR Model with Varying Sample Size") +theme_bw()+theme( text = element_text(face = "bold",colour = "black"), axis.text = element_text(size = 12,colour = "black"), axis.title = element_text(size = 16), strip.text = element_text(size = 16), plot.title = element_text(hjust = 0.5), legend.text = element_text(size = 15) )+scale_y_continuous(limits = c(0,1))+scale_color_manual(values = cbbPalette)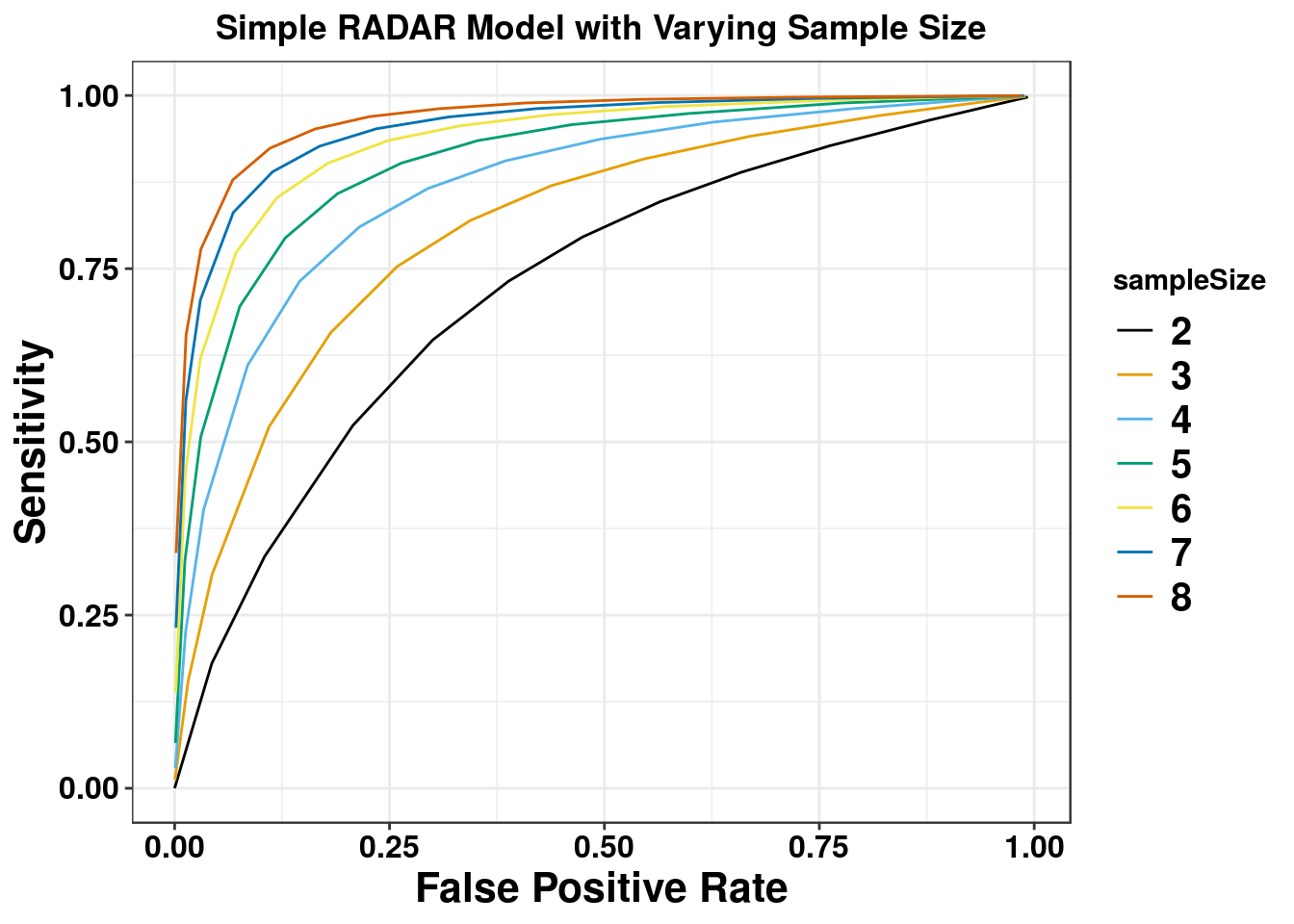
simple_auc <- function(TPR, FPR){
# inputs already sorted, best scores first
dFPR <- c(diff(FPR), 0)
dTPR <- c(diff(TPR), 0)
sum(TPR * dFPR) + sum(dTPR * dFPR)/2
}
cat(paste("auROC for N =",2:8,"is", foreach(s = 2:8, .combine = c )%do%{
simple_auc(TPR = sampleSizeSimu_RADAR_mean[sampleSizeSimu_RADAR_mean$sampleSize == s,"power"],
FPR = sampleSizeSimu_RADAR_mean[sampleSizeSimu_RADAR_mean$sampleSize == s,"fpr"]
)
} , "\n"))## auROC for N = 2 is 0.716349733239408
## auROC for N = 3 is 0.80005477650703
## auROC for N = 4 is 0.857025002337078
## auROC for N = 5 is 0.894359820306779
## auROC for N = 6 is 0.921213893295853
## auROC for N = 7 is 0.938942904182691
## auROC for N = 8 is 0.951227225034607sessionInfo()## R version 3.5.3 (2019-03-11)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 17.10
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/libopenblasp-r0.2.20.so
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats4 parallel stats graphics grDevices utils datasets
## [8] methods base
##
## other attached packages:
## [1] RADAR_0.3.0 qvalue_2.14.1
## [3] RcppArmadillo_0.9.400.2.0 Rcpp_1.0.1
## [5] RColorBrewer_1.1-2 gplots_3.0.1.1
## [7] doParallel_1.0.14 iterators_1.0.10
## [9] foreach_1.4.4 ggplot2_3.1.1
## [11] Rsamtools_1.34.1 Biostrings_2.50.2
## [13] XVector_0.22.0 GenomicFeatures_1.34.8
## [15] AnnotationDbi_1.44.0 Biobase_2.42.0
## [17] GenomicRanges_1.34.0 GenomeInfoDb_1.18.2
## [19] IRanges_2.16.0 S4Vectors_0.20.1
## [21] BiocGenerics_0.28.0
##
## loaded via a namespace (and not attached):
## [1] bitops_1.0-6 matrixStats_0.54.0
## [3] fs_1.3.0 bit64_0.9-7
## [5] progress_1.2.0 httr_1.4.0
## [7] backports_1.1.4 tools_3.5.3
## [9] R6_2.4.0 rpart_4.1-13
## [11] KernSmooth_2.23-15 Hmisc_4.2-0
## [13] DBI_1.0.0 lazyeval_0.2.2
## [15] colorspace_1.4-1 nnet_7.3-12
## [17] withr_2.1.2 gridExtra_2.3
## [19] tidyselect_0.2.5 prettyunits_1.0.2
## [21] DESeq2_1.22.2 bit_1.1-14
## [23] compiler_3.5.3 htmlTable_1.13.1
## [25] DelayedArray_0.8.0 labeling_0.3
## [27] rtracklayer_1.42.2 checkmate_1.9.1
## [29] caTools_1.17.1.2 scales_1.0.0
## [31] genefilter_1.64.0 stringr_1.4.0
## [33] digest_0.6.18 foreign_0.8-71
## [35] rmarkdown_1.12 base64enc_0.1-3
## [37] pkgconfig_2.0.2 htmltools_0.3.6
## [39] htmlwidgets_1.3 rlang_0.4.0
## [41] rstudioapi_0.10 RSQLite_2.1.1
## [43] BiocParallel_1.16.6 gtools_3.8.1
## [45] acepack_1.4.1 dplyr_0.8.0.1
## [47] RCurl_1.95-4.12 magrittr_1.5
## [49] GenomeInfoDbData_1.2.0 Formula_1.2-3
## [51] Matrix_1.2-17 munsell_0.5.0
## [53] stringi_1.4.3 yaml_2.2.0
## [55] SummarizedExperiment_1.12.0 zlibbioc_1.28.0
## [57] plyr_1.8.4 grid_3.5.3
## [59] blob_1.1.1 gdata_2.18.0
## [61] crayon_1.3.4 lattice_0.20-38
## [63] splines_3.5.3 annotate_1.60.1
## [65] hms_0.4.2 locfit_1.5-9.1
## [67] knitr_1.22 pillar_1.3.1
## [69] geneplotter_1.60.0 reshape2_1.4.3
## [71] codetools_0.2-16 biomaRt_2.38.0
## [73] XML_3.98-1.19 glue_1.3.1
## [75] evaluate_0.13 latticeExtra_0.6-28
## [77] data.table_1.12.2 gtable_0.3.0
## [79] purrr_0.3.2 assertthat_0.2.1
## [81] xfun_0.6 xtable_1.8-4
## [83] survival_2.44-1.1 tibble_2.1.1
## [85] GenomicAlignments_1.18.1 memoise_1.1.0
## [87] cluster_2.0.7-1This R Markdown site was created with workflowr