Ovarian_cancer
Zijie Zhang
2018-08-21
Count reads
library(RADAR)
samplenames <- paste0("OvCa",c("1850", "1917", "2005" ,"2013", "2053" ,"2064", "2072" ,"2186", "2221", "2261", "2270" ,"2343", "2380"))
RADAR <- countReads(samplenames = samplenames,
gtf = "~/Database/genome/hg38/hg38_UCSC.gtf",
bamFolder = "~/PoissonGamma_Method/Ovary_Cancer/bam_files",
outputDir = "~/PoissonGamma_Method/Ovary_Cancer",
modification = "m6A",
binSize = 50,
strandToKeep = "opposite",
paired = TRUE,
threads = 40
)
save(RADAR,file = "~/PoissonGamma_Method/Ovary_Cancer/radar.RData")Summary of read count
library(RADAR)
load( "~/Tools/RADARmannual/data/OvarianCancer_MeRIP.RADAR.RData" )
#load( "~/PoissonGamma_Method/Ovary_Cancer/RADAR_analysis.RData" ) # load work space with analysis result for quick plot update
summary(RADAR)## MeRIP.RADAR dataset of 13 samples.
## Read count quantified in 50-bp consecutive bins on the transcript.
## The total read count for Input and IP samples are (Million reads):
## OvCa1850 OvCa1917 OvCa2005 OvCa2013 OvCa2053 OvCa2064 OvCa2072
## Input 5.45 6.77 6.71 7.03 7.21 4.99 8.14
## IP 12.67 14.87 14.19 15.09 15.19 13.50 13.99
## OvCa2186 OvCa2221 OvCa2261 OvCa2270 OvCa2343 OvCa2380
## Input 4.81 3.37 7.73 6.10 6.18 4.68
## IP 11.73 10.18 14.59 11.04 13.54 12.18
## Input gene level read count available.
## There are 2 predictor variables/covariates. Can access by function variable(MeRIPdata).
## Differential methylation tested by PoissonGamma test (RADAR).
## Multiple test corrected by Benjamini & Hochberg.
## Neighboring significant bins merged. There are 2266 reported differential loci at FDR < 0.1 and logFoldChange > 0.5.RADAR <- normalizeLibrary(RADAR)
RADAR <- adjustExprLevel(RADAR)
RADAR <- filterBins(RADAR, minCountsCutOff = 15)
age <- c(45,54,45,59,35,36,51,73,68,64,70,68,62)
variable(RADAR)<-variable(RADAR_pos) <- data.frame( disease = c( rep("Ctl",7),rep("Tumor",6) ), age = age )
RADAR_pos <- RADAR::adjustExprLevel(RADAR, adjustBy = "pos")
RADAR_pos <- filterBins(RADAR_pos,minCountsCutOff = 15)Local Vs geneSum
Plot distribution of number of reads in each 50 bp bins
hist(log10(rowMeans(RADAR@reads[rowMeans(RADAR@reads[,grep("input",colnames(RADAR@reads))])>1,grep("input",colnames(RADAR@reads))]) ),xlab = "log10 read count",main = "Distribution of INPUT read count in bins",xlim = c(0,3), col =rgb(0.2,0.2,0.2,0.5),cex.main = 2,cex.axis =2,cex.lab=2)
axis(side = 1, lwd = 2,cex.axis =2)
axis(side = 2, lwd = 2,cex.axis =2)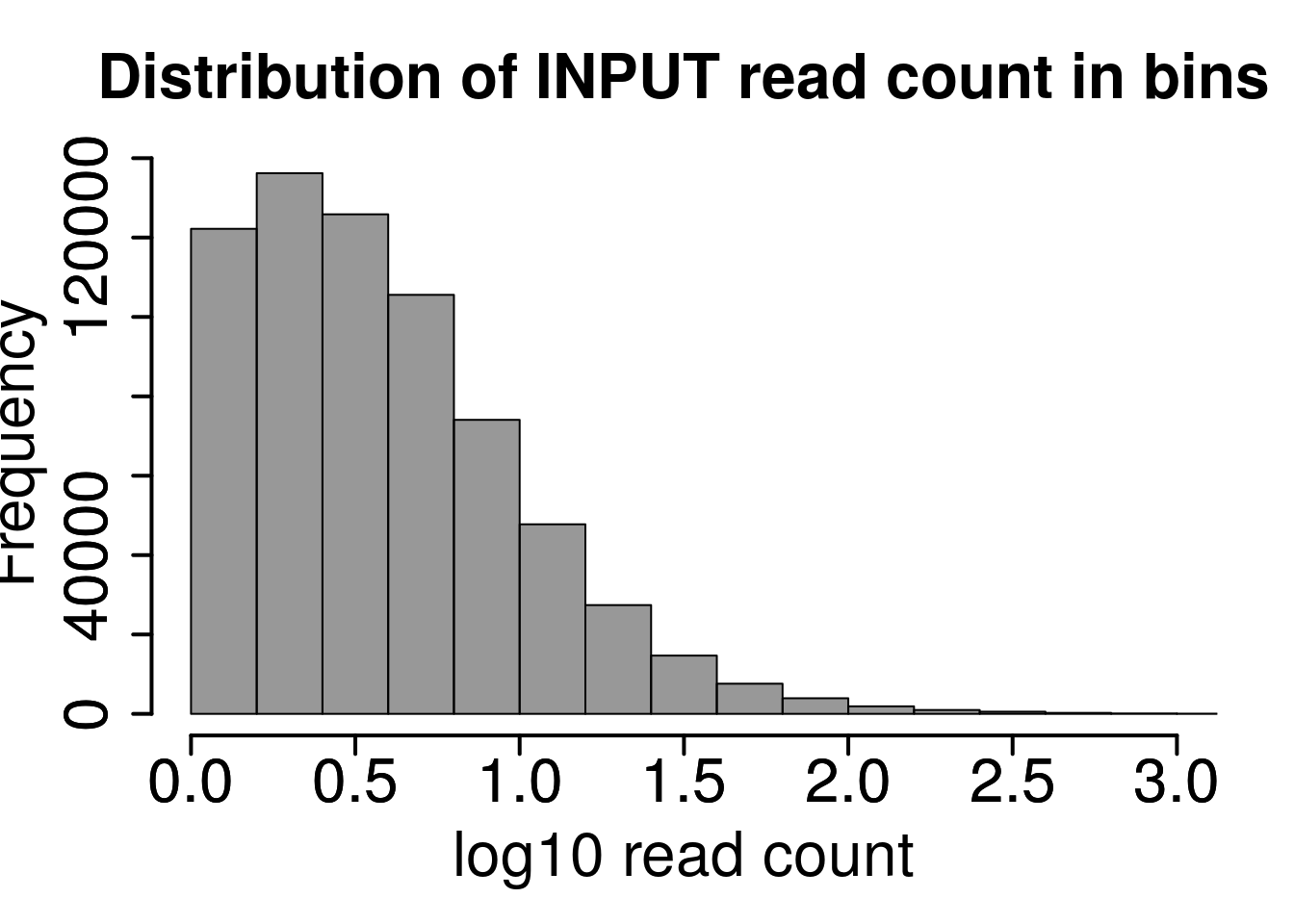
local read count V.S. geneSum
Compute the within group variability of INPUT geneSum VS INPUT local read count.
var.coef <- function(x){sd(as.numeric(x))/mean(as.numeric(x))}
## filter expressed genes
geneSum <- RADAR@geneSum[rowSums(RADAR@geneSum)>16,]
## within group variability
relative.var <- t( apply(geneSum,1,tapply,unlist(variable(RADAR)[,1]),var.coef) )
geneSum.var <- c(relative.var)
## For each gene used above, random sample a 50bp bin within this gene
set.seed(1)
r.bin50 <- tapply(rownames(RADAR@norm.input)[which(RADAR@geneBins$gene %in% rownames(geneSum))],as.character( RADAR@geneBins$gene[which(RADAR@geneBins$gene %in% rownames(geneSum))]) ,function(x){
n <- sample(1:length(x),1)
return(x[n])
})
relative.var <- apply(RADAR@norm.input[r.bin50,],1,tapply,unlist(variable(RADAR)[,1]),var.coef)
bin50.var <- c(relative.var)
bin50.var <- bin50.var[!is.na(bin50.var)]
## 100bp bins
r.bin100 <- tapply(rownames(RADAR@norm.input)[which(RADAR@geneBins$gene %in% rownames(geneSum))],as.character( RADAR@geneBins$gene[which(RADAR@geneBins$gene %in% rownames(geneSum))]) ,function(x){
n <- sample(1:(length(x)-2),1)
return(x[n:(n+1)])
})
relative.var <- lapply(r.bin100, function(x){ tapply( colSums(RADAR@norm.input[x,]),unlist(variable(RADAR)[,1]),var.coef ) })
bin100.var <- unlist(relative.var)
bin100.var <- bin100.var[!is.na(bin100.var)]
## 200bp bins
r.bin200 <- tapply(rownames(RADAR@norm.input)[which(RADAR@geneBins$gene %in% rownames(geneSum))],as.character( RADAR@geneBins$gene[which(RADAR@geneBins$gene %in% rownames(geneSum))]) ,function(x){
if(length(x)>4){
n <- sample(1:(length(x)-3),1)
return(x[n:(n+3)])
}else{
return(NULL)
}
})
r.bin200 <- r.bin200[which(!unlist(lapply(r.bin200,is.null)) ) ]
relative.var <- lapply(r.bin200, function(x){ tapply( colSums(RADAR@norm.input[x,]),unlist(variable(RADAR)[,1]),var.coef ) })
bin200.var <- unlist(relative.var)
bin200.var <- bin200.var[!is.na(bin200.var)]
relative.var <- data.frame(group=c(rep("geneSum",length(geneSum.var)),rep("bin-50bp",length(bin50.var)),rep("bin-100bp",length(bin100.var)),rep("bin-200bp",length(bin200.var))), variance=c(geneSum.var,bin50.var,bin100.var,bin200.var))ggplot(relative.var,aes(variance,colour=group))+geom_density()+xlab("Relative variation")+
theme_bw() + theme(panel.border = element_blank(), panel.grid.major = element_blank(),
panel.grid.minor = element_blank(), axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=20, face="bold", hjust=0.5,family = "arial"),
axis.title.y=element_text(size=20, face="bold", vjust=0.4, angle=90,family = "arial"),
legend.position = c(0.88,0.88),legend.title=element_blank(),legend.text = element_text(size = 14, face = "bold",family = "arial"),
axis.text = element_text(size = 15,face = "bold",family = "arial",colour = "black") )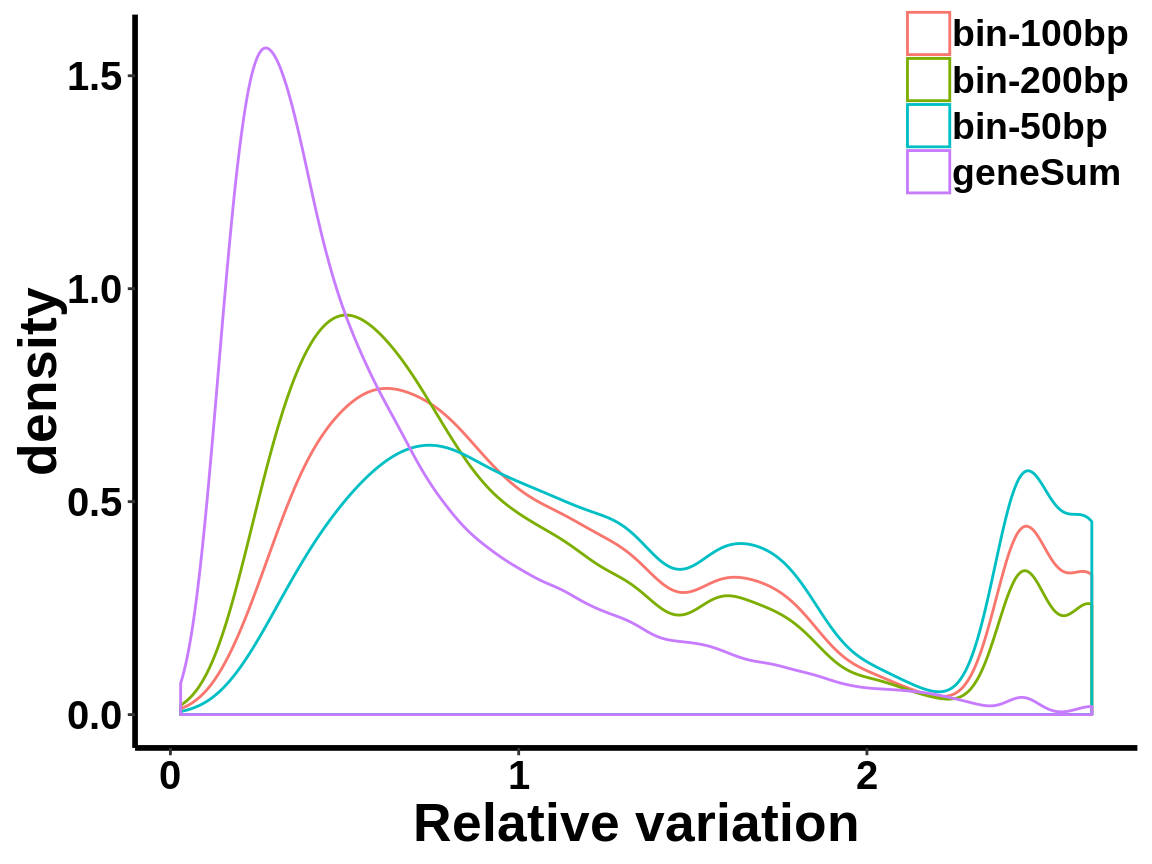
Check mean variance relationship of the data.
ip_var <- t(apply(RADAR@ip_adjExpr_filtered,1,tapply,unlist(variable(RADAR)[,1]),var))
#ip_var <- ip_coef_var[!apply(ip_var,1,function(x){return(any(is.na(x)))}),]
ip_mean <- t(apply(RADAR@ip_adjExpr_filtered,1,tapply,unlist(variable(RADAR)[,1]),mean))
###
gene_var <- t(apply(RADAR@geneSum,1,tapply,unlist(variable(RADAR)[,1]),var))
#gene_var <- gene_var[!apply(gene_var,1,function(x){return(any(is.na(x)))}),]
gene_mean <- t(apply(RADAR@geneSum,1,tapply,unlist(variable(RADAR)[,1]),mean))
all_var <- list('RNA-seq'=c(gene_var),'m6A-IP'=c(ip_var))
nn<-sapply(all_var, length)
rs<-cumsum(nn)
re<-rs-nn+1
group <- factor(rep(names(all_var), nn), levels=names(all_var))
all_var.df <- data.frame(variance = c(c(gene_var),c(ip_var)),mean= c(c(gene_mean),c(ip_mean)),label = group)ggplot(data = all_var.df,aes(x=mean,y=variance,colour = group,shape = group))+geom_point(size = I(0.2))+stat_smooth(se = T,show.legend = F)+stat_smooth(se = F)+theme_bw() +xlab("Mean")+ ylab("Variance")+theme(panel.border = element_blank(), panel.grid.major = element_blank(),
panel.grid.minor = element_blank(), axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=22, face="bold", hjust=0.5,family = "arial"),
axis.title.y=element_text(size=22, face="bold", vjust=0.4, angle=90,family = "arial"),
legend.position = c(0.85,0.95),legend.title=element_blank(),legend.text = element_text(size = 20, face = "bold",family = "arial"),
axis.text = element_text(size = 15,face = "bold",family = "arial",colour = "black") ) +
scale_x_continuous(limits = c(0,7000))+scale_y_continuous(limits = c(0,5e6))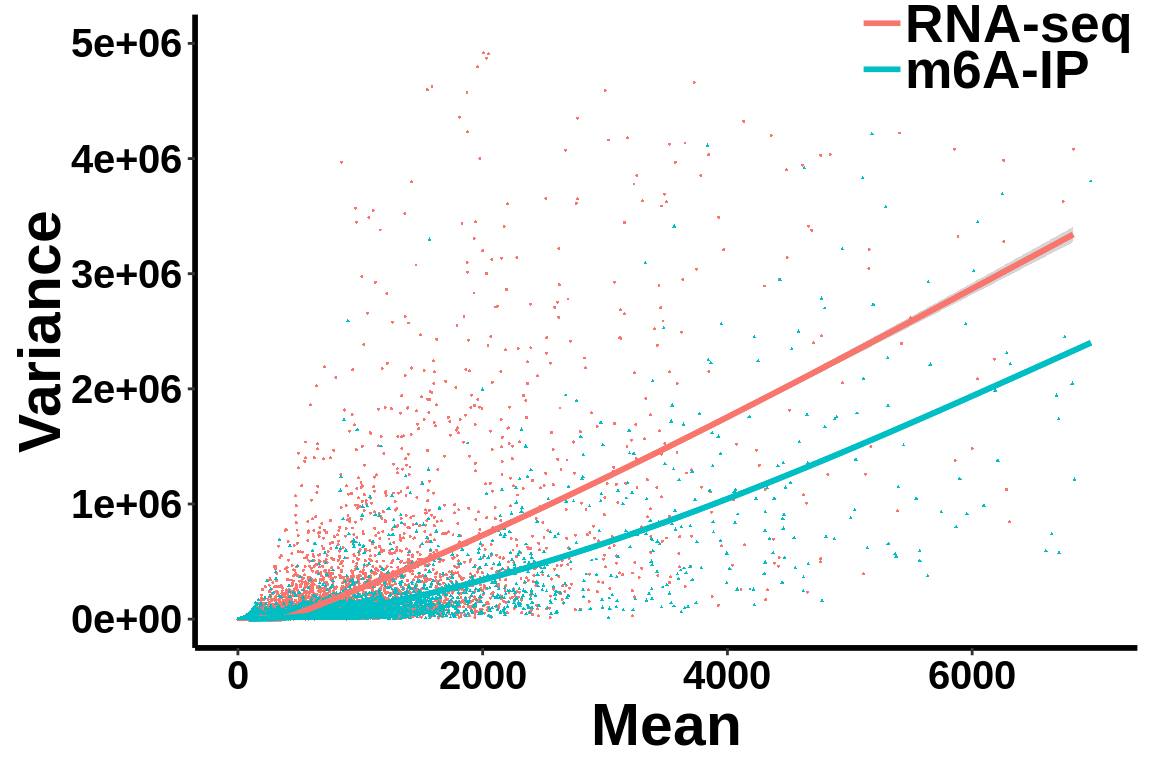
Compare MeRIP-seq IP data with regular RNA-seq data
var.coef <- function(x){sd(as.numeric(x))/mean(as.numeric(x))}
ip_coef_var <- t(apply(RADAR@ip_adjExpr_filtered,1,tapply,unlist(variable(RADAR)[,1]),var.coef))
ip_coef_var <- ip_coef_var[!apply(ip_coef_var,1,function(x){return(any(is.na(x)))}),]
#hist(c(ip_coef_var),main = "M14KO mouse liver\n m6A-IP",xlab = "within group coefficient of variation",breaks = 50)
###
gene_coef_var <- t(apply(RADAR@geneSum,1,tapply,unlist(variable(RADAR)[,1]),var.coef))
gene_coef_var <- gene_coef_var[!apply(gene_coef_var,1,function(x){return(any(is.na(x)))}),]
#hist(c(gene_coef_var),main = "M14KO mouse liver\n RNA-seq",xlab = "within group coefficient of variation",breaks = 50)
coef_var <- list('RNA-seq'=c(gene_coef_var),'m6A-IP'=c(ip_coef_var))
nn<-sapply(coef_var, length)
rs<-cumsum(nn)
re<-rs-nn+1
grp <- factor(rep(names(coef_var), nn), levels=names(coef_var))
coef_var.df <- data.frame(coefficient_var = c(c(gene_coef_var),c(ip_coef_var)),label = grp)ggplot(data = coef_var.df,aes(coefficient_var,colour = grp))+geom_density()+theme_bw() +theme(panel.border = element_blank(), panel.grid.major = element_blank(),
panel.grid.minor = element_blank(), axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=20, face="bold", hjust=0.5,family = "arial"),
axis.title.y=element_text(size=20, face="bold", vjust=0.4, angle=90,family = "arial"),
legend.position = c(0.9,0.9),legend.title=element_blank(),legend.text = element_text(size = 18, face = "bold",family = "arial"),
axis.text = element_text(size = 15,face = "bold",family = "arial",colour = "black") )+
xlab("Coefficient of variation")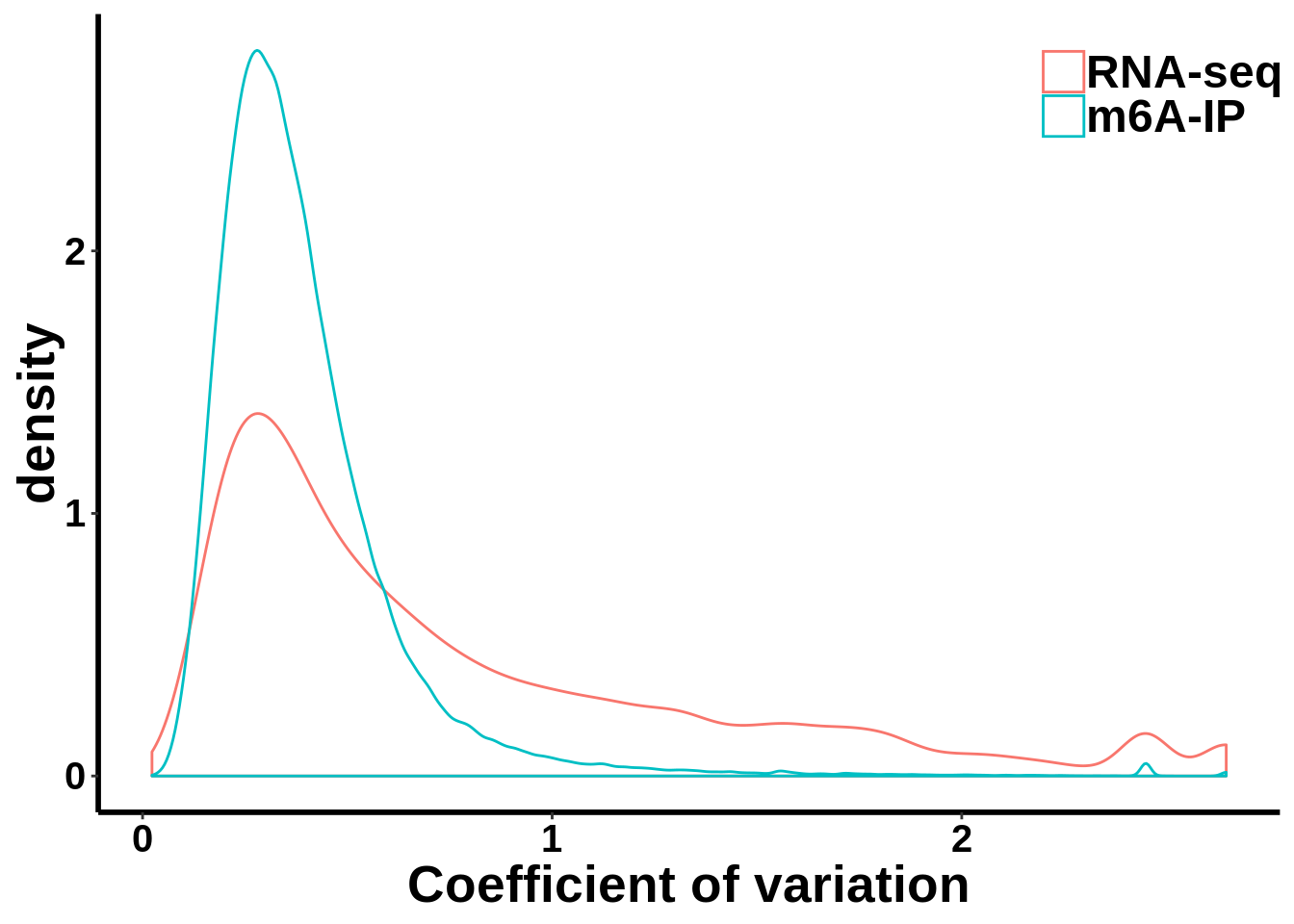
PCA analysis
We plot PCA colored by Ctl vs Tumor
plotPCAfromMatrix(RADAR@ip_adjExpr_filtered,group = unlist(variable(RADAR)[,1]) )+scale_color_discrete(name = "Group")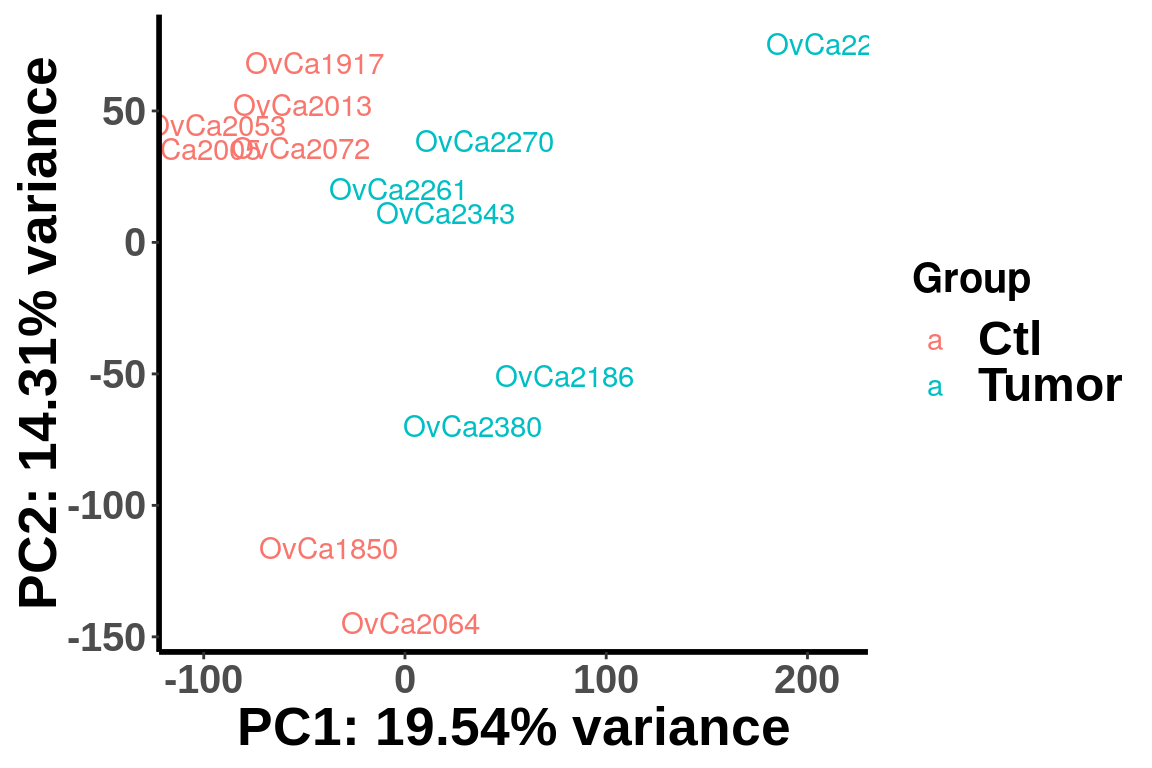
(Adjust expression level by local read count)
plotPCAfromMatrix(RADAR_pos@ip_adjExpr_filtered,group = unlist(variable(RADAR)[,1]) )+scale_color_discrete(name = "Group")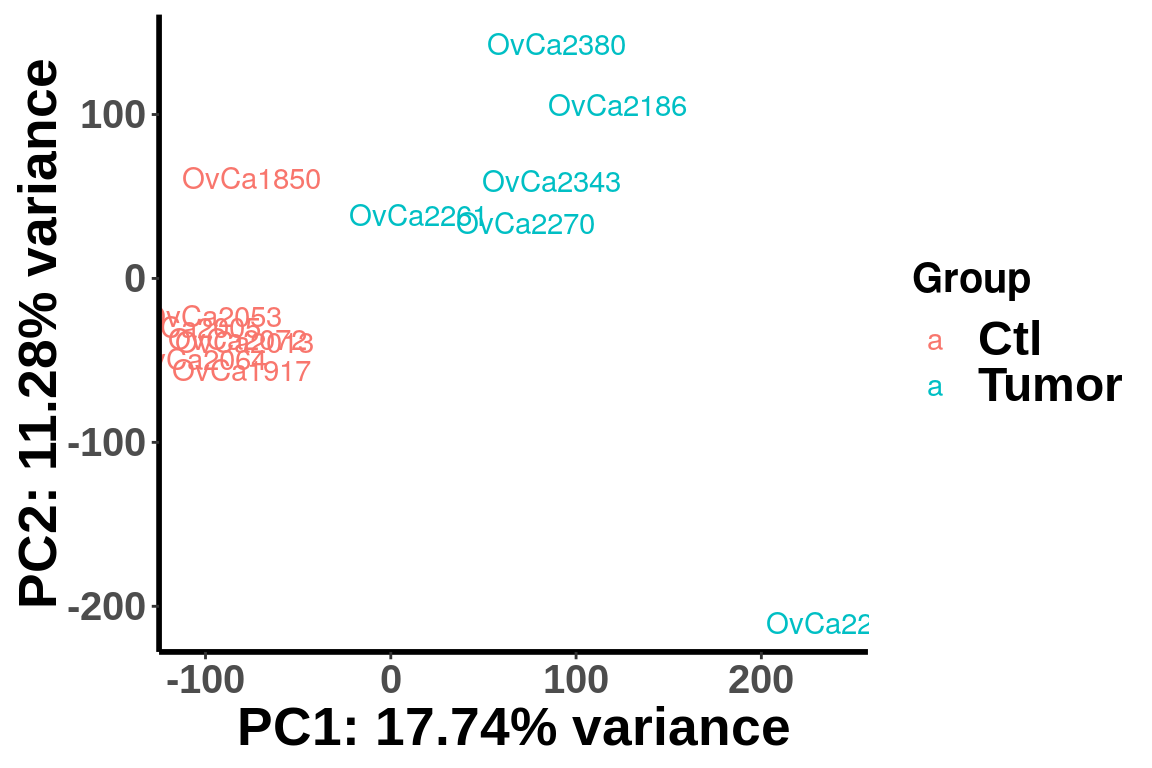
age <- c(45,54,45,59,35,36,51,73,68,64,70,68,62)
IP_counts <- RADAR@ip_adjExpr_filtered
colnames(IP_counts)<- c(paste0("Ctl",c("1850", "1917", "2005" ,"2013", "2053" ,"2064", "2072")), paste0("Tumor",c("2186", "2221", "2261", "2270" ,"2343", "2380")) )
plotPCAfromMatrix(IP_counts[order(rowSums(RADAR@ip_adjExpr_filtered),decreasing = T)[1:15000],], group = age )+scale_colour_continuous(name = "Age",low = "blue",high = "red")+scale_color_continuous(name = "Age")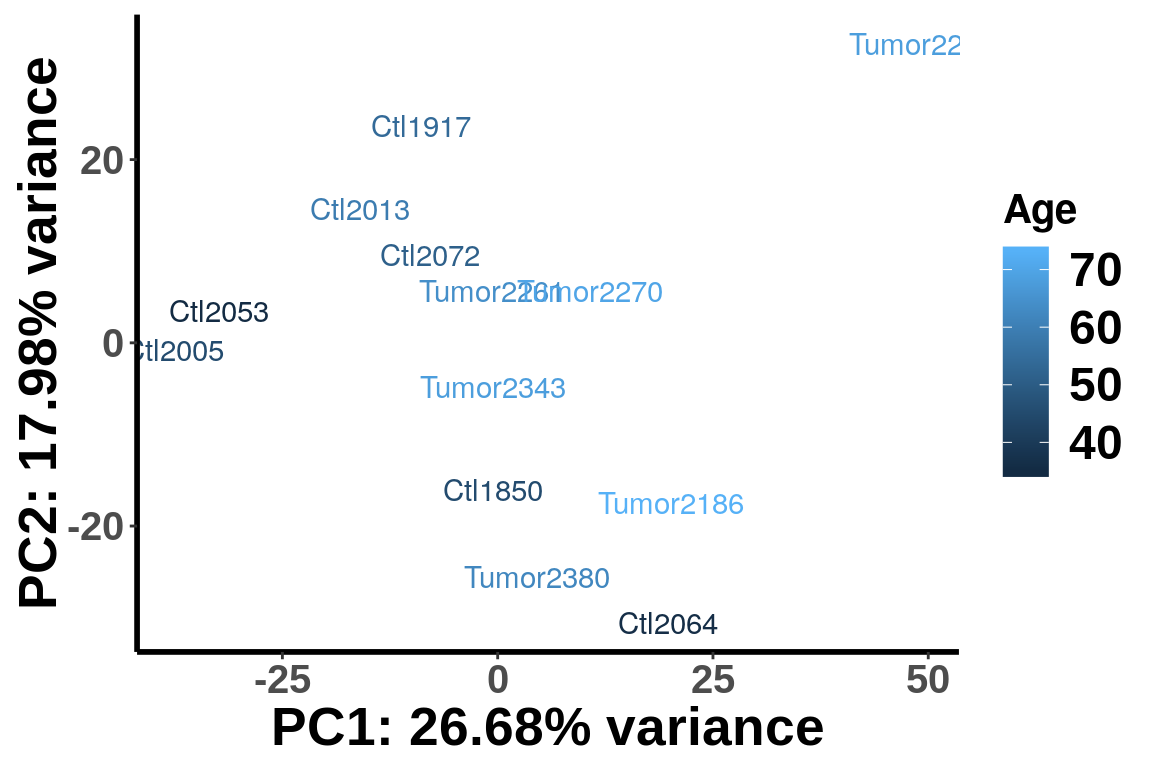
Regress out age and re-plot PCA
registerDoParallel(cores = 10)
m6A.cov.out <- foreach(i = 1:nrow(RADAR@ip_adjExpr_filtered), .combine = rbind) %dopar% {
Y = RADAR@ip_adjExpr_filtered[i,]
resi <- residuals( lm(log(Y+1) ~ age ) )
resi
}
rm(list=ls(name=foreach:::.foreachGlobals), pos=foreach:::.foreachGlobals)
rownames(m6A.cov.out) <- rownames(RADAR@ip_adjExpr_filtered)
plotPCAfromMatrix(m6A.cov.out[order(rowSums(RADAR@ip_adjExpr_filtered),decreasing = T)[1:15000],],unlist(variable(RADAR)[,1]),loglink = FALSE)+scale_color_discrete(name = "Group")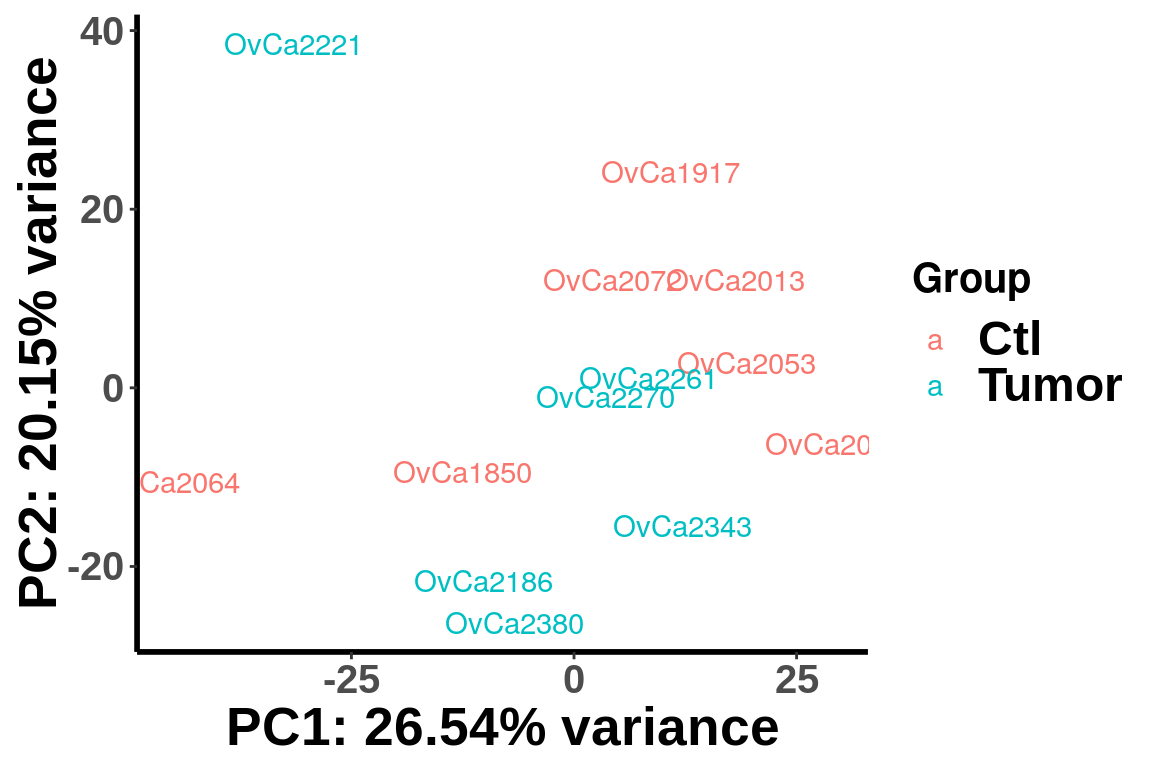
Run PoissonGamma test
PoissonGamma Test
RADAR_cov <- diffIP_parallel(RADAR,thread = 25 , fdrBy = "fdr")@test.est
RADAR_pos_cov <- diffIP_parallel(RADAR_pos,thread = 25 ,fdrBy = "fdr")@test.est
RADAR_nonCov <- RADAR
RADAR_pos_nonCov <- RADAR_pos
variable(RADAR_nonCov) <- variable(RADAR_pos_nonCov) <- variable(RADAR)[1]
RADAR_nonCov <- diffIP_parallel(RADAR_nonCov,thread = 25 , fdrBy = "fdr")
RADAR_pos_nonCov <- diffIP_parallel(RADAR_pos_nonCov,thread = 25 , fdrBy = "fdr")Other method
## fisher's exact test
fisherTest <- function( control_ip, treated_ip, control_input, treated_input, thread ){
registerDoParallel(cores = thread)
testResult <- foreach(i = 1:nrow(control_ip) , .combine = rbind )%dopar%{
tmpTest <- fisher.test( cbind( c( rowSums(treated_ip)[i], rowSums(treated_input)[i] ), c( rowSums(control_ip)[i], rowSums(control_input)[i] ) ), alternative = "two.sided" )
data.frame( logFC = log(tmpTest$estimate), pvalue = tmpTest$p.value )
}
rm(list=ls(name=foreach:::.foreachGlobals), pos=foreach:::.foreachGlobals)
testResult$fdr <- p.adjust(testResult$pvalue, method = "fdr")
return(testResult)
}
## wrapper for the MetDiff test
MeTDiffTest <- function( control_ip, treated_ip, control_input, treated_input, thread = 1 ){
registerDoParallel(cores = thread)
testResult <- foreach(i = 1:nrow(control_ip) , .combine = rbind )%dopar%{
x <- t( as.matrix(control_ip[i,]) )
y <- t( as.matrix(control_input[i,]) )
xx <- t( as.matrix(treated_ip[i,]) )
yy <- t( as.matrix(treated_input[i,]) )
xxx = cbind(x,xx)
yyy = cbind(y,yy)
logl1 <- MeTDiff:::.betabinomial.lh(x,y+1)
logl2 <- MeTDiff:::.betabinomial.lh(xx,yy+1)
logl3 <- MeTDiff:::.betabinomial.lh(xxx,yyy+1)
tst <- (logl1$logl+logl2$logl-logl3$logl)*2
pvalues <- 1 - pchisq(tst,2)
log.fc <- log( (sum(xx)+1)/(1+sum(yy)) * (1+sum(y))/(1+sum(x)) )
data.frame( logFC = log.fc, pvalue = pvalues )
}
rm(list=ls(name=foreach:::.foreachGlobals), pos=foreach:::.foreachGlobals)
testResult$fdr <- p.adjust(testResult$pvalue, method = "fdr")
return(testResult)
}
Bltest <- function(control_ip, treated_ip, control_input, treated_input){
control_ip_total <- sum(colSums(control_ip))
control_input_total <- sum(colSums(control_input))
treated_ip_total <- sum(colSums(treated_ip))
treated_input_total <- sum(colSums(treated_input))
tmpResult <- do.call(cbind.data.frame, exomePeak::bltest(rowSums(control_ip), rowSums(control_input),rowSums(treated_ip),rowSums(treated_input),control_ip_total, control_input_total, treated_ip_total, treated_input_total) )
return( data.frame(logFC = tmpResult[,"log.fc"], pvalue = exp(tmpResult[,"log.p"]), fdr = exp(tmpResult$log.fdr) ) )
}In order to compare performance of other method on this data set, we run other methods on default mode on this dataset.
cov <- matrix(age,ncol = 1)
colnames(cov) <- "age"
filteredBins <- rownames(RADAR@ip_adjExpr_filtered)
Metdiff.res <-MeTDiffTest(control_ip = round( RADAR@norm.ip[filteredBins,1:7] ) ,
treated_ip = round( RADAR@norm.ip[filteredBins,8:13] ),
control_input = round( RADAR@norm.input[filteredBins,1:7] ),
treated_input = round( RADAR@norm.input[filteredBins,8:13] ) , thread = 20)
QNB.res <- QNB::qnbtest(control_ip = round( RADAR@norm.ip[filteredBins,1:7] ) ,
treated_ip = round( RADAR@norm.ip[filteredBins,8:13] ),
control_input = round( RADAR@norm.input[filteredBins,1:7] ),
treated_input = round( RADAR@norm.input[filteredBins,8:13] ) ,plot.dispersion = FALSE )
fisher.res <- fisherTest(control_ip = round( RADAR@norm.ip[filteredBins,1:7] ) ,
treated_ip = round( RADAR@norm.ip[filteredBins,8:13] ),
control_input = round( RADAR@norm.input[filteredBins,1:7] ),
treated_input = round( RADAR@norm.input[filteredBins,8:13] ) )
exomePeak.res <- Bltest(control_ip = round( RADAR@norm.ip[filteredBins,1:7] ) ,
treated_ip = round( RADAR@norm.ip[filteredBins,8:13] ),
control_input = round( RADAR@norm.input[filteredBins,1:7] ),
treated_input = round( RADAR@norm.input[filteredBins,8:13] ) )Compare distribution of p value.
pvalues <- data.frame(pvalue = c(RADAR_cov[,"p_value"],Metdiff.res$pvalue,QNB.res$pvalue,fisher.res$pvalue, exomePeak.res$pvalue ),
method = factor(rep(c("RADAR","MeTDiff","QNB","Fisher","exomePeak"),c(length(RADAR_cov[,"p_value"]),
length(Metdiff.res$pvalue),
length(QNB.res$pvalue),
length(fisher.res$pvalue),
length(exomePeak.res$pvalue)
)
),levels =c("RADAR","MeTDiff","QNB","Fisher","exomePeak")
)
)
ggplot(pvalues, aes(x = pvalue))+geom_histogram(breaks = seq(0,1,0.04),col=I("black"))+facet_grid(.~method)+theme_bw()+xlab("p-value")+theme( axis.title = element_text(size = 22, face = "bold"),strip.text = element_text(size = 25, face = "bold"),axis.text.x = element_text(size = 18, face = "bold",colour = "black"),axis.text.y = element_text(size = 15, face = "bold",colour = "black",angle = 90),panel.grid = element_blank(), axis.line = element_line(size = 0.7 ,colour = "black"),axis.ticks = element_line(colour = "black"), panel.spacing = unit(0.4, "lines") )+ scale_x_continuous(breaks = seq(0,1,0.2),labels=function(x) sprintf("%.1f", x))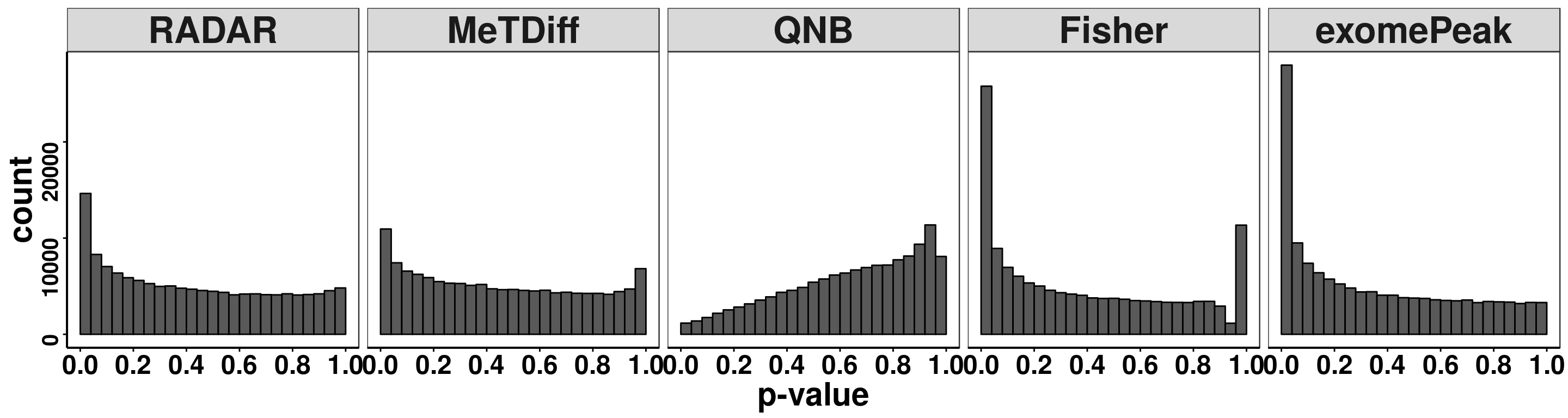
Compare adjusting expression level by geneSum vs. bin read counts.
tmp <- hist(c(RADAR_pos_cov[,"p_value"]),plot = F)
tmp$counts <- tmp$counts*(nrow(RADAR@ip_adjExpr_filtered)/nrow(RADAR_pos@ip_adjExpr_filtered))
hist(RADAR_cov[,"p_value"],main = "RADAR",xlab = "p value",col =rgb(1,0,0,0.4),cex.main = 2.5,cex.axis =2,cex.lab=2, ylim = c(0,max(hist(RADAR_cov[,"p_value"],plot = F)$counts,tmp$counts)) )
plot(tmp,col=rgb(0,0,1,0.5),add = T)
axis(side = 1, lwd = 1,cex.axis =2)
axis(side = 2, lwd = 1,cex.axis =2)
legend("topright", c("By geneSum", "By position"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 10,bty="n",text.font = 2)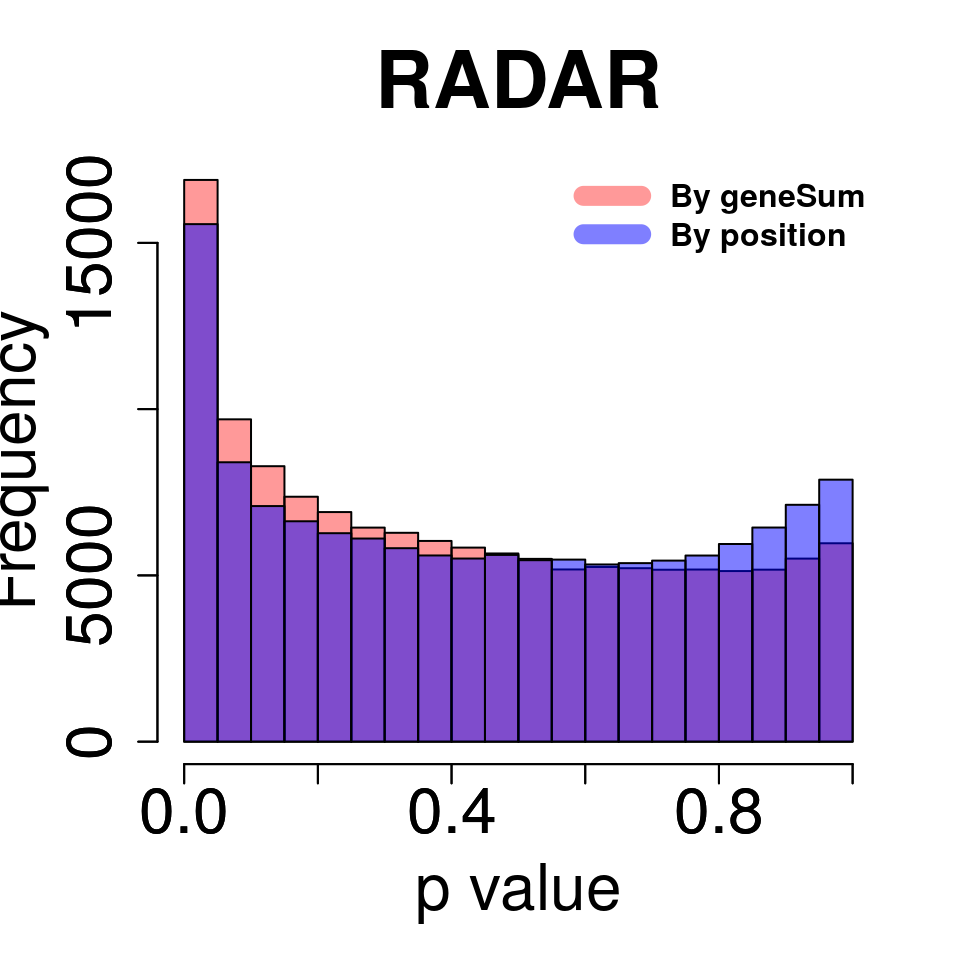
Compare the distibution of beta
par(mfrow = c(1,5))
hist(RADAR_cov[,"beta1"],main = "RADAR",xlab = "beta",breaks = 100,col =rgb(0.2,0.2,0.2,0.5),cex.main = 2.5,cex.axis =2,cex.lab=2)
#hist(radar_byPos$all.est[,"beta1"],main = "Norm by pos",xlab = "beta",breaks = 100)
hist(Metdiff.res$logFC, main = "MeTDiff",xlab = "beta",breaks = 100,col =rgb(0.2,0.2,0.2,0.5),cex.main = 2.5,cex.axis =2,cex.lab=2)
hist(QNB.res$log2.OR,main = "QNB", xlab = "beta",breaks = 100,col =rgb(0.2,0.2,0.2,0.5),cex.main = 2.5,cex.axis =2,cex.lab=2)
hist(fisher.res$logFC, main = "Fisher",xlab = "beta",breaks = 100,col =rgb(0.2,0.2,0.2,0.5),cex.main = 2.5,cex.axis =2,cex.lab=2)
hist(exomePeak.res$logFC, main = "exomePeak",xlab = "beta",breaks = 100,col =rgb(0.2,0.2,0.2,0.5),cex.main = 2.5,cex.axis =2,cex.lab=2)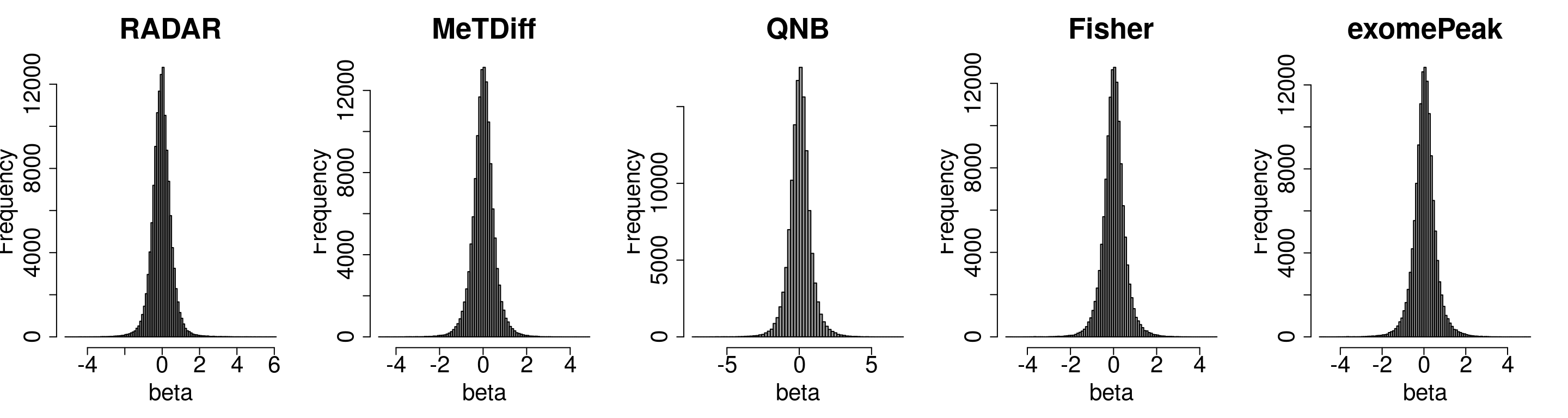
Number of significant bins detected at FDR < 0.1.
sigBins <- apply(cbind("RADAR"=RADAR_cov[,"p_value"],"MeTDiff"=Metdiff.res$pvalue,"QNB"=QNB.res$pvalue,"Fisher"=fisher.res$pvalue,"exomePeak" = exomePeak.res$pvalue),2, function(x){
length( which( p.adjust(x,method = 'fdr') < 0.1 ) )
})
print(sigBins)## RADAR MeTDiff QNB Fisher exomePeak
## 2436 752 22 15956 18303RADAR_res <- reportResult(RADAR,cutoff = 0.1,Beta_cutoff = 0.5, threads = 20)Permutation Test
Because covariate “age” is continuous variable, which cannot be balanced when permuting diseases status. Therefore, permutation test is performed without covariate.
radar_permuted_P <-NULL
metdiff_permuted_P <- NULL
qnb_permuted_P <- NULL
fisher_permuted_P <- NULL
exomePeak_permute_P <- NULL
set.seed(2)
for(i in 1:15){
permute_X <- variable(RADAR)[,1]
permute_X[sample(1:7,4)] <- "Tumor"
permute_X[sample(8:13,3)] <- "Ctl"
RADAR_permute <- RADAR
variable(RADAR_permute) <- data.frame(condition = permute_X )
radar_permute <- diffIP_parallel( RADAR_permute )@test.est[,"p_value"]
metdiff_permute <-MeTDiffTest( control_ip = round( RADAR@norm.ip[filteredBins ,(which(permute_X == "Ctl" ) )] ),
treated_ip = round( RADAR@norm.ip[filteredBins,(which(permute_X == "Tumor" ) )] ),
control_input = round( RADAR@norm.input[filteredBins,which(permute_X == "Ctl" )] ),
treated_input = round( RADAR@norm.input[filteredBins,which(permute_X == "Tumor" )] ) ,
thread = 25)$pvalue
qnb_permute <- QNB::qnbtest(control_ip = round( RADAR@norm.ip[filteredBins ,(which(permute_X == "Ctl" ) )] ),
treated_ip = round( RADAR@norm.ip[filteredBins,(which(permute_X == "Tumor" ) )] ),
control_input = round( RADAR@norm.input[filteredBins,which(permute_X == "Ctl" )] ),
treated_input = round( RADAR@norm.input[filteredBins,which(permute_X == "Tumor" )] ),
plot.dispersion = FALSE)$pvalue
fisher_permute <- fisherTest( control_ip = round( RADAR@norm.ip[filteredBins ,(which(permute_X == "Ctl" ) )] ),
treated_ip = round( RADAR@norm.ip[filteredBins,(which(permute_X == "Tumor" ) )] ),
control_input = round( RADAR@norm.input[filteredBins,which(permute_X == "Ctl" )] ),
treated_input = round( RADAR@norm.input[filteredBins,which(permute_X == "Tumor" )] ) ,
thread = 25
)$pvalue
exomePeak_permute <- Bltest( control_ip = round( RADAR@norm.ip[filteredBins ,(which(permute_X == "Ctl" ) )] ),
treated_ip = round( RADAR@norm.ip[filteredBins,(which(permute_X == "Tumor" ) )] ),
control_input = round( RADAR@norm.input[filteredBins,which(permute_X == "Ctl" )] ),
treated_input = round( RADAR@norm.input[filteredBins,which(permute_X == "Tumor" )] )
)$pvalue
radar_permuted_P <- cbind(radar_permuted_P, radar_permute)
metdiff_permuted_P <- cbind(metdiff_permuted_P, metdiff_permute)
qnb_permuted_P <- cbind(qnb_permuted_P,qnb_permute)
fisher_permuted_P <- cbind(fisher_permuted_P, fisher_permute)
exomePeak_permute_P <- cbind(exomePeak_permute_P, exomePeak_permute)
}
for( i in 1:ncol(radar_permuted_P)){
par(mfrow=c(1,5))
hist(RADAR@test.est[,"p_value"], col =rgb(1,0,0,0.4),main = "RADAR", xlab = "p value",cex.main = 2.5,cex.axis =2,cex.lab=2)
hist(radar_permuted_P[,i],col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 10,bty="n")
hist(Metdiff.res$pvalue, col =rgb(1,0,0,0.4),main = "MeTdiff",xlab = "p value",cex.main = 2.5,cex.axis =2,cex.lab=2)
hist(metdiff_permuted_P[,i],col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 10,bty="n")
hist(QNB.res$pvalue, col =rgb(1,0,0,0.4),main = "QNB",xlab = "p value",cex.main = 2.5,cex.axis =2,cex.lab=2)
hist(qnb_permuted_P[,i],col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 10,bty="n")
hist(fisher.res$pvalue, col =rgb(1,0,0,0.4),main = "Fisher",xlab = "p value",cex.main = 2.5,cex.axis =2,cex.lab=2)
hist(fisher_permuted_P[,i],col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 10,bty="n")
hist(exomePeak.res$pvalue, col =rgb(1,0,0,0.4),main = "exomePeak",xlab = "p value",cex.main = 2.5,cex.axis =2,cex.lab=2)
hist(exomePeak_permute_P[,i],col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 10,bty="n")
}par(mfrow=c(1,5))
y.scale <- max(hist(exomePeak.res$pvalue,plot = F)$counts)
hist(RADAR_nonCov@test.est[,"p_value"], col =rgb(1,0,0,0.4),main = "RADAR", xlab = "p value",font=2, cex.lab=2.5, cex.axis = 1.5 , font.lab=2 ,cex.main = 3, lwd = 3,ylim = c(0,y.scale) )
tmp <- hist(c(radar_permuted_P),plot = F)
tmp$counts <- tmp$counts/15
plot(tmp,col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 12,bty="n",text.font = 2, cex = 1.5)
tmp <- hist(c(metdiff_permuted_P),plot = F)
tmp$counts <- tmp$counts/15
hist(Metdiff.res$pvalue, col =rgb(1,0,0,0.4),main = "MeTDiff",xlab = "p value" ,font=2, cex.lab=2.5, cex.axis = 1.5 , font.lab=2 ,cex.main = 3, lwd = 3, ylim = c(0,y.scale) )
plot(tmp,col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 12,bty="n",text.font = 2, cex = 1.5)
hist(QNB.res$pvalue, col =rgb(1,0,0,0.4),main = "QNB",xlab = "p value",font=2, cex.lab=2.5, cex.axis = 1.5 , font.lab=2 ,cex.main = 3,lwd = 3,ylim = c(0,y.scale))
tmp <- hist(c(qnb_permuted_P),plot = F)
tmp$counts <- tmp$counts/15
plot(tmp,col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 12,bty="n",text.font = 2, cex = 1.5)
hist(fisher.res$pvalue, col =rgb(1,0,0,0.4),main = "Fisher",xlab = "p value",font=2, cex.lab=2.5, cex.axis = 1.5 , font.lab=2 ,cex.main = 3,lwd = 3,ylim = c(0,y.scale))
tmp <- hist(c(fisher_permuted_P),plot = F)
tmp$counts <- tmp$counts/15
plot(tmp,col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 12,bty="n",text.font = 2, cex = 1.5)
hist(exomePeak.res$pvalue, col =rgb(1,0,0,0.4),main = "exomePeak",xlab = "p value",font=2, cex.lab=2.5, cex.axis = 1.5 , font.lab=2 ,cex.main = 3,lwd = 3,ylim = c(0,y.scale))
tmp <- hist(c(exomePeak_permute_P),plot = F)
tmp$counts <- tmp$counts/15
plot(tmp,col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 12,bty="n",text.font = 2, cex = 1.5)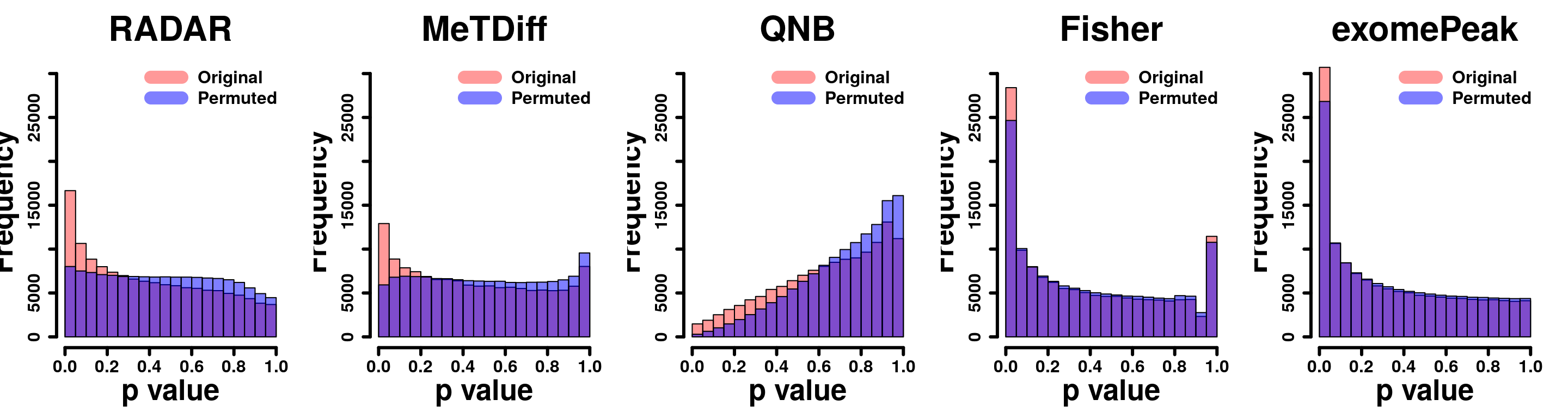
Permutation by position
radar_permuted_P_pos <-NULL
set.seed(2)
for(i in 1:15){
permute_X <- variable(RADAR)[,1]
permute_X[sample(1:7,4)] <- "Tumor"
permute_X[sample(8:13,3)] <- "Ctl"
RADAR_permute <- RADAR_pos
variable(RADAR_permute) <- data.frame(condition = permute_X )
radar_permute <- diffIP_parallel( RADAR_permute , thread = 20 )@test.est[,"p_value"]
radar_permuted_P_pos <- cbind(radar_permuted_P_pos, radar_permute)
} par(mfrow=c(1,2))
y.scale <- max(hist( RADAR_pos_nonCov@test.est[,"p_value"] ,plot = F)$counts)
hist(RADAR_nonCov@test.est[,"p_value"], col =rgb(1,0,0,0.4),main = "RADAR by geneSum", xlab = "p value",font=2, cex.lab=2.5, cex.axis = 1.5 , font.lab=2 ,cex.main = 3, lwd = 3, ylim = c(0,y.scale) )
tmp <- hist(c(radar_permuted_P),plot = F)
tmp$counts <- tmp$counts/15
plot(tmp,col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 12,bty="n",text.font = 2, cex = 1.5)
hist(RADAR_pos_nonCov@test.est[,"p_value"], col =rgb(1,0,0,0.4),main = "RADAR by pos", xlab = "p value",font=2, cex.lab=2.5, cex.axis = 1.5 , font.lab=2 ,cex.main = 3, lwd = 3, ylim = c(0,y.scale) )
tmp <- hist(c(radar_permuted_P_pos),plot = F)
tmp$counts <- tmp$counts/15
plot(tmp,col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 12,bty="n",text.font = 2, cex = 1.5)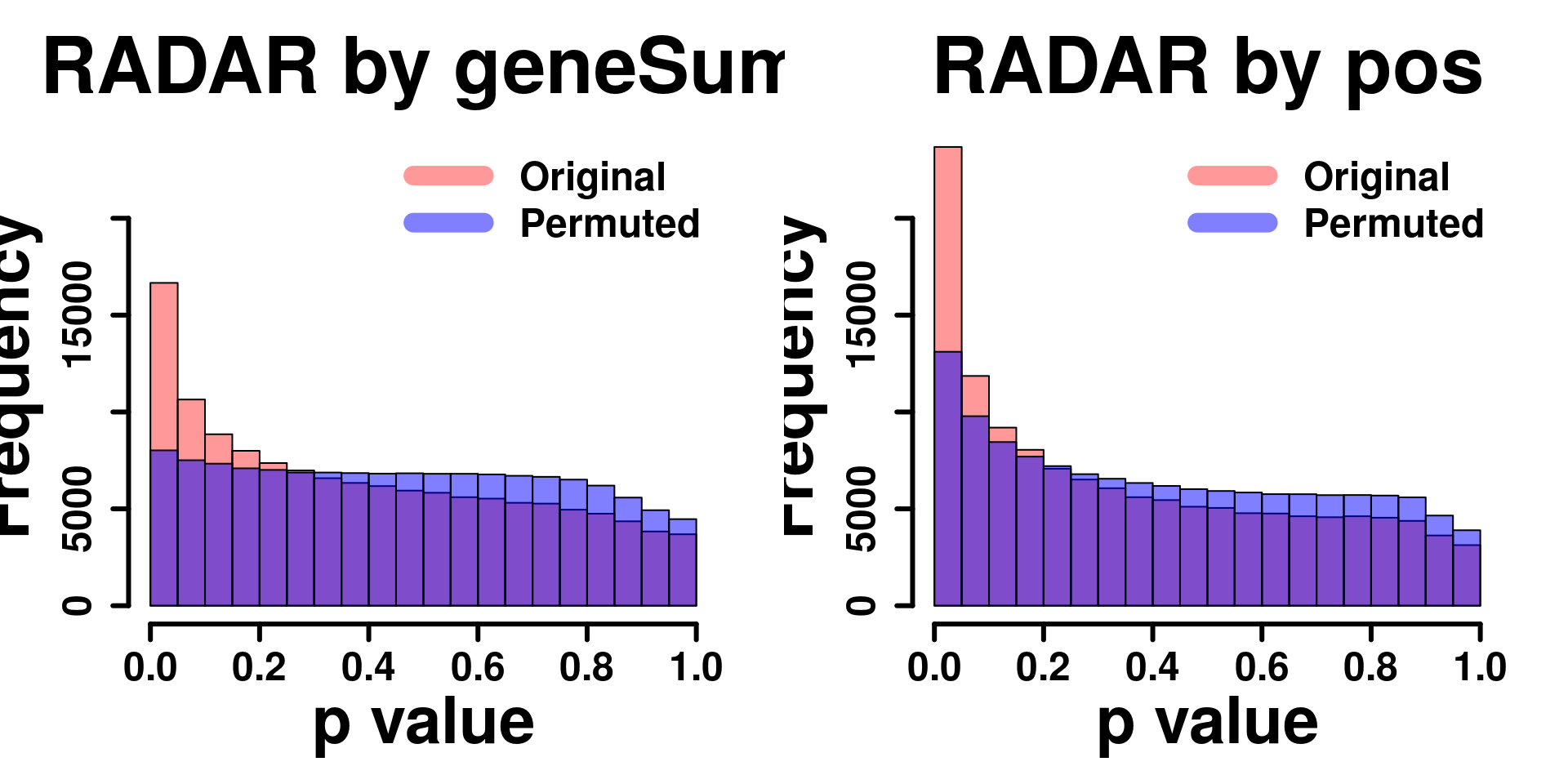
Downstream analysis of the differential peaks
Report differential peaks
RADAR <- reportResult(RADAR, cutoff = 0.1,threads = 20)
Diff_peaks_cov <- results(RADAR)
write.table(Diff_peaks_cov,"~/PoissonGamma_Method/Ovary_Cancer/Diff_peaks_ageCov.xls",sep = "\t", col.names = T,row.names = F,quote = F)
reportPeaks <- function(radar, est , cutoff = 0.1, Beta_cutoff = 0.5, threads = 1){
stats <- est
rownames(stats) <- rownames(radar@test.est)
colnames(stats) <- gsub("padj","fdr",colnames(stats))
colnames(stats) <- gsub("log2.OR","beta",colnames(stats))
colnames(stats) <- gsub("logFC","beta",colnames(stats))
colnames(stats) <- gsub("log2.OR","beta",colnames(stats))
colnames(stats) <- gsub("pvalue","p_value",colnames(stats))
num_bins <- length( which(stats[,"fdr"] < cutoff & abs(stats[,"beta"] )> Beta_cutoff) )
if(num_bins < 1 ){
stop("There is no bin passing the threshold...\n No differential peaks can be reported at current cutoff...")
}else {
sig.bins <- rownames(stats)[which(stats[,"fdr"] < cutoff & abs(stats[,"beta"] )> Beta_cutoff)]
}
geneBins <- geneBins(radar)
ID <- (rownames(geneBins) %in% sig.bins)
num_lines <- length(ID)
# start ids of checkpoints
## find peak-starting checkpoints (either from nonpeak to peak, or peak in a different batch)
start_id <- which((ID[2:num_lines]-ID[1:num_lines-1]==1) |
((geneBins$gene[2:num_lines]!=geneBins$gene[1:num_lines-1]) & (ID[2:num_lines] == TRUE)) )
start_id <- start_id + 1 # add 1 since ID was counted from 2 to num_lines
if ( ID[1]==TRUE ) { start_id <- c(1,start_id) } # if the first checkpoint bin is peak
# end ids of checkpoints
## find peak-ending checkpoints (either from peak to nonpeak, or peak in a different batch)
end_id <- which((ID[1:num_lines-1]-ID[2:num_lines]==1) |
((geneBins$gene[1:num_lines-1]!=geneBins$gene[2:num_lines]) & (ID[1:num_lines-1] == TRUE)) )
if (ID[num_lines]==TRUE) {end_id <- c(end_id,num_lines)} # if the last checkpoint bin is peak
peak_id_pairs <- cbind(start_id, end_id)
num_peaks <- nrow(peak_id_pairs)
geneGRList <- radar@geneModel
peakGenes <- as.character(geneBins[peak_id_pairs[,1],"gene"])
cat(paste0("reporting ",num_peaks," of peaks\n"))
if (num_peaks == 0){return(data.frame())
}else{
start_time <- Sys.time()
registerDoParallel(cores = threads)
cat(paste("Hyper-thread registered:",getDoParRegistered(),"\n"))
cat(paste("Using",getDoParWorkers(),"thread(s) to report merged report...\n"))
merged.report<- foreach( p = 1:num_peaks, .combine = rbind)%dopar%{
peak_row_id <- peak_id_pairs[p,]
geneExons <- reduce ( geneGRList[peakGenes[p]][[1]] )
peak <- RADAR:::.getPeakBins(geneGRList,peakGenes[p],c(geneBins$bin[peak_row_id[1]],geneBins$bin[peak_row_id[2]]),radar@binSize )
peakE <- RADAR:::.peakExons(peak,as.data.frame(geneExons))
data.frame(chr=peak$chr,
start = peak$start,
end = peak$end,
name = peakGenes[p],
score = 0,
strand = as.character(strand(geneExons))[1],
thickStart = peak$start,
thickEnd = peak$end,
itemRgb=0,
blockCount = nrow(peakE),
blockSizes = paste(peakE$width,collapse=","),
blockStarts = paste(peakE$start - replicate(nrow(peakE),peakE$start[1]),collapse=","),
logFC = stats[rownames(geneBins[peak_row_id[1]:peak_row_id[2],]), "beta"][which.max(abs( stats[rownames(geneBins[peak_row_id[1]:peak_row_id[2],]), "beta"] ))],
p_value = RADAR:::.fishersMethod( stats[rownames(geneBins[peak_row_id[1]:peak_row_id[2],]), "p_value"] )
)
}
rm(list=ls(name=foreach:::.foreachGlobals), pos=foreach:::.foreachGlobals)
end_time <- Sys.time()
cat(paste("Time used to report peaks:",difftime(end_time, start_time, units = "mins"),"mins... \n"))
}
return(merged.report)
}
exomePeak_diff <- reportPeaks(RADAR, est = exomePeak.res, threads = 20)
QNB_diff <- reportPeaks(RADAR, est = QNB.res, threads = 20)
Metdiff_diff <- reportPeaks(RADAR, est = Metdiff.res, threads = 20)
fisher_diff <- reportPeaks(RADAR, est = fisher.res, threads = 20)library(MeRIPtools)
plotMetaGeneMulti(list("RADAR" = Diff_peaks_cov,"exomePeak"=exomePeak_diff, "QNB" = QNB_diff, "MeTDiff" = Metdiff_diff, "Fisher" = fisher_diff ),"~/Database/genome/hg38/hg38_UCSC.gtf")## [1] "Converting BED12 to GRangesList"
## [1] "It may take a few minutes"
## [1] "Converting BED12 to GRangesList"
## [1] "It may take a few minutes"
## [1] "Converting BED12 to GRangesList"
## [1] "It may take a few minutes"
## [1] "Converting BED12 to GRangesList"
## [1] "It may take a few minutes"
## [1] "Converting BED12 to GRangesList"
## [1] "It may take a few minutes"
## [1] "total 58259 transcripts extracted ..."
## [1] "total 42543 transcripts left after ambiguity filter ..."
## [1] "total 21293 mRNAs left after component length filter ..."
## [1] "total 7986 ncRNAs left after ncRNA length filter ..."
## [1] "Building Guitar Coordinates. It may take a few minutes ..."
## [1] "Guitar Coordinates Built ..."
## [1] "Using provided Guitar Coordinates"
## [1] "resolving ambiguious features ..."
## [1] "no figure saved ..."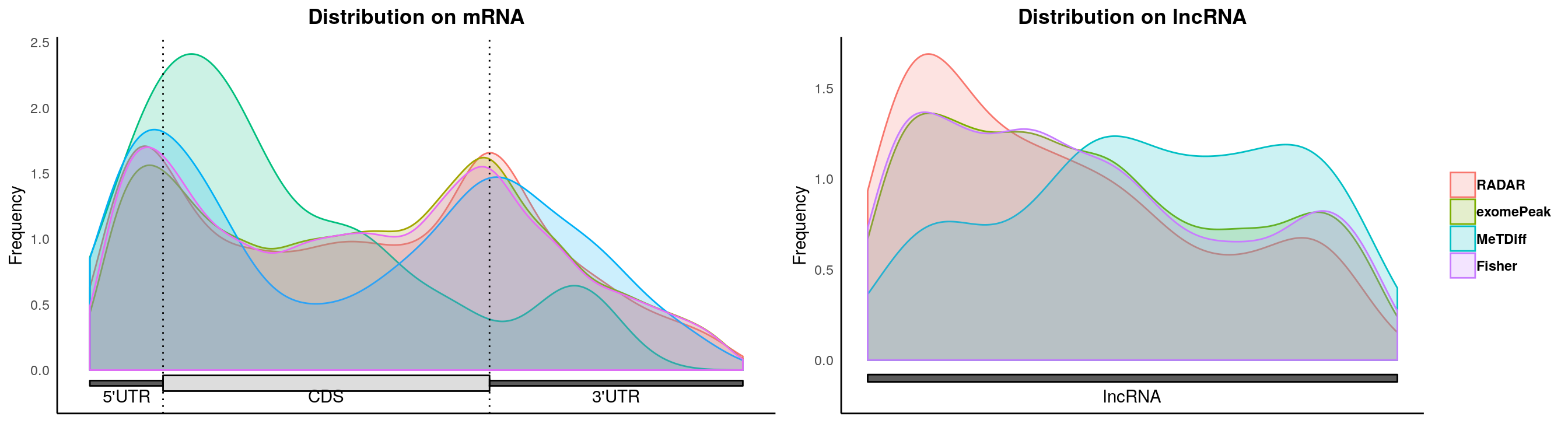
## NOTE this function is a wrapper for R package "Guitar".
## If you use the metaGene plot in publication, please cite the original reference:
## Cui et al 2016 BioMed Research Internationaldetach("package:MeRIPtools", unload = TRUE)extendPeak <- function(peak, extension = 50){
peak_extend <- peak
peak_extend$start <- peak$start-extension
peak_extend$end <- peak$end+extension
peak_extend$blockSizes <- unlist( lapply( strsplit(as.character(peak$blockSizes),split = ",") , function(x){
tmp = as.numeric(x)
tmp[1] = tmp[1]+extension
tmp[length(tmp)] = tmp[length(tmp)]+extension
paste(as.character(tmp),collapse = ",")
} ) )
return(peak_extend)
}
Diff_peaks_cov_extend <- extendPeak(Diff_peaks_cov)
exomePeak_diff_extend <- extendPeak(exomePeak_diff)
Metdiff_diff_extend <- extendPeak(Metdiff_diff)
fisher_diff_extend <- extendPeak(fisher_diff)
QNB_diff_extend <- extendPeak(QNB_diff)## analysis for RADAR detected signifiant bins
write.table(Diff_peaks_cov_extend[,1:12], file = paste0("~/Tools/RADARmannual/data/Ovary_Cancer_Diff_peaks_ageCov.bed"),sep = "\t",row.names = F,col.names = F,quote = F)
system2(command = "bedtools", args = paste0("getfasta -fi ~/Database/genome/hg38/hg38_UCSC.fa -bed ~/Tools/RADARmannual/data/Ovary_Cancer_Diff_peaks_ageCov.bed -s -split > ~/Tools/RADARmannual/data/Ovary_Cancer_Diff_peaks_ageCov.fa ") )
system2(command = "findMotifs.pl", args = paste0("~/Tools/RADARmannual/data/Ovary_Cancer_Diff_peaks_ageCov.fa fasta ~/Tools/RADARmannual/data/Ovary_Cancer_Diff_peaks_ageCov_Homer2 -fasta ~/Database/transcriptome/backgroud_peaks/hg38_90bpRandomPeak.fa -len 5,6 -rna -p 20 -S 5 -noknown"),wait = F )
## analysis for exomePeak detected significant bins
write.table(exomePeak_diff_extend[,1:12], file = paste0("~/Tools/RADARmannual/data/Ovary_Cancer_Diff_exomePeak.bed"),sep = "\t",row.names = F,col.names = F,quote = F)
system2(command = "bedtools", args = paste0("getfasta -fi ~/Database/genome/hg38/hg38_UCSC.fa -bed ~/Tools/RADARmannual/data/Ovary_Cancer_Diff_exomePeak.bed -s -split > ~/Tools/RADARmannual/data/Ovary_Cancer_Diff_exomePeak.fa ") )
system2(command = "findMotifs.pl", args = paste0("~/Tools/RADARmannual/data/Ovary_Cancer_Diff_exomePeak.fa fasta ~/Tools/RADARmannual/data/Ovary_Cancer_Diff_exomePeak_Homer2 -fasta ~/Database/transcriptome/backgroud_peaks/hg38_90bpRandomPeak.fa -len 5,6 -rna -p 20 -S 5 -noknown"),wait = F )
## analysis for MetDiff detected significant bins
write.table(Metdiff_diff_extend[,1:12], file = paste0("~/Tools/RADARmannual/data/Ovary_Cancer_Diff_MetDiff.bed"),sep = "\t",row.names = F,col.names = F,quote = F)
system2(command = "bedtools", args = paste0("getfasta -fi ~/Database/genome/hg38/hg38_UCSC.fa -bed ~/Tools/RADARmannual/data/Ovary_Cancer_Diff_MetDiff.bed -s -split > ~/Tools/RADARmannual/data/Ovary_Cancer_Diff_MetDiff.fa ") )
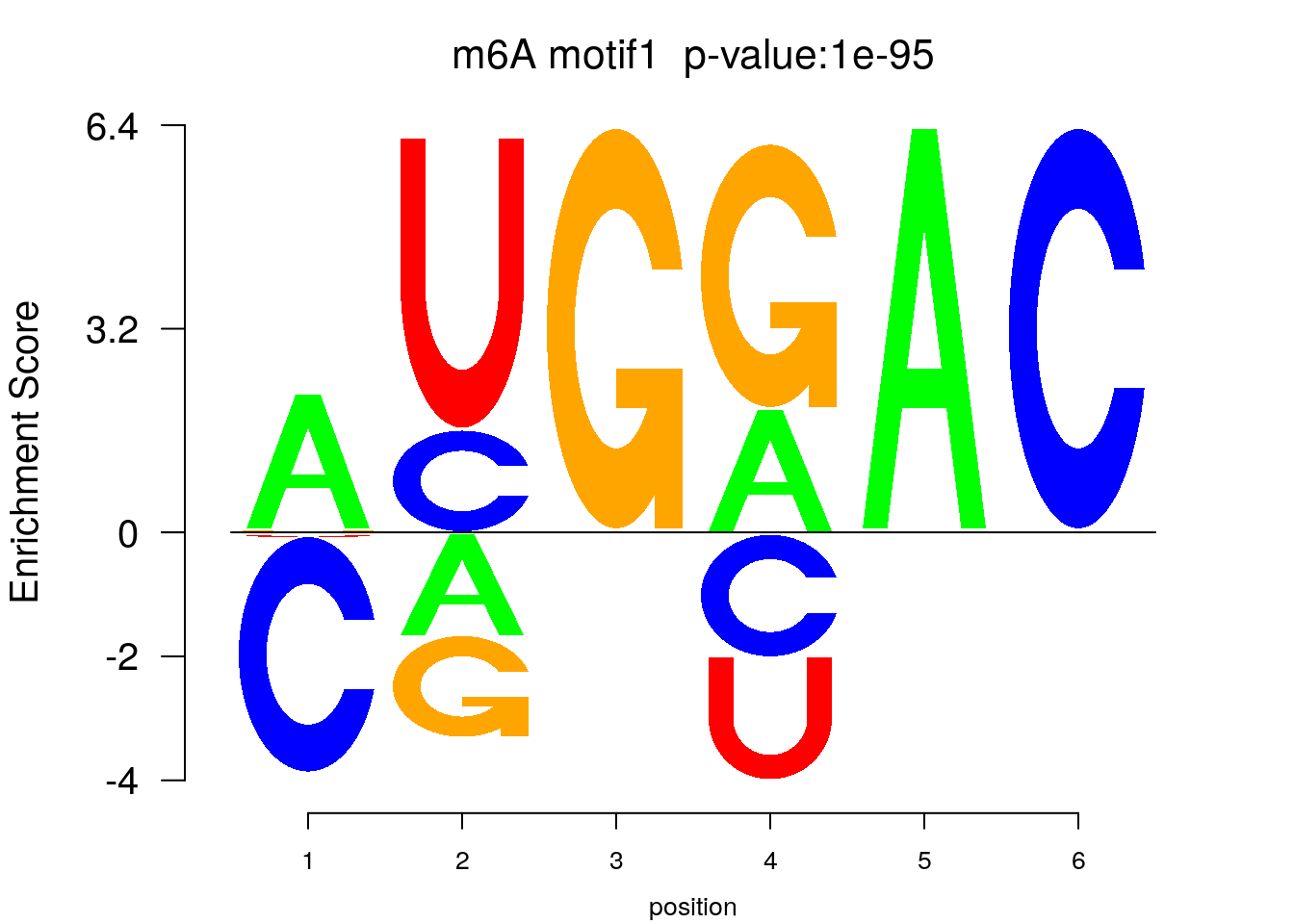
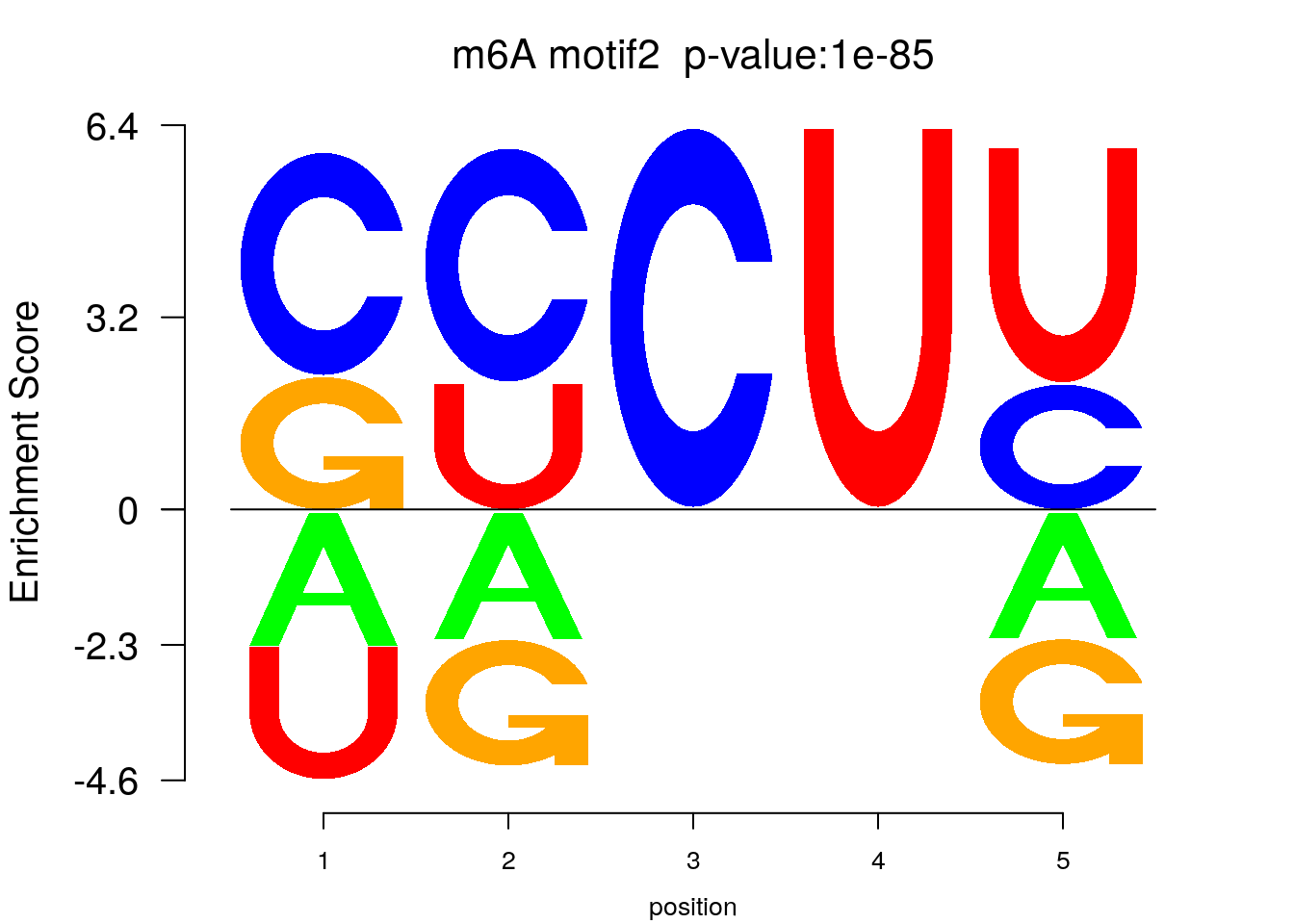
system2(command = "findMotifs.pl", args = paste0("~/Tools/RADARmannual/data/Ovary_Cancer_Diff_MetDiff.fa fasta ~/Tools/RADARmannual/data/Ovary_Cancer_Diff_MetDiff_Homer2 -fasta ~/Database/transcriptome/backgroud_peaks/hg38_90bpRandomPeak.fa -len 5,6 -rna -p 20 -S 5 -noknown"),wait = F )library(Logolas)
color_motif <- c( "orange", "blue", "red","green" )
for(i in 1:3){
pwm.m <- t( read.table(paste0("~/Tools/RADARmannual/data/Ovary_Cancer_Diff_peaks_ageCov_Homer2/homerResults/motif",i,".motif"), header = F, comment.char = ">",col.names = c("A","C","G","U")) )
motif_p <- sapply(1:3, function(x){
strsplit( as.character( read.table(paste0("~/Tools/RADARmannual/data/Ovary_Cancer_Diff_peaks_ageCov_Homer2/homerResults/motif",x,".motif"),comment.char = "", nrows = 1)[,12] ), split= ":")[[1]][4]
})
colnames(pwm.m) <- 1:ncol(pwm.m)
try(logomaker(pwm.m ,type = "EDLogo",colors = color_motif,color_type = "per_row" ,logo_control = list(pop_name = paste0("m6A motif",i," p-value:",motif_p[i])))
)
}
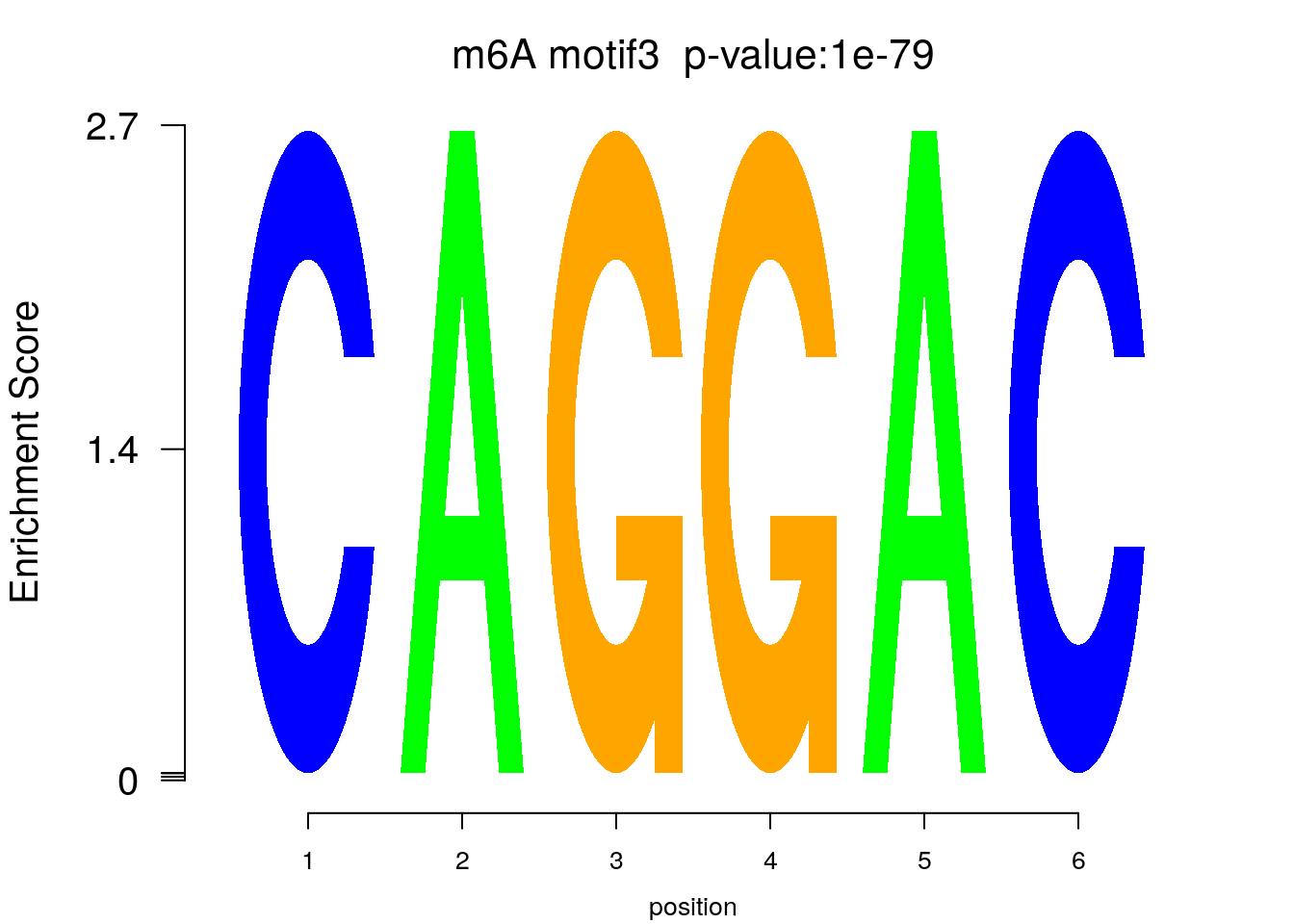
## Error in grid::unit(letters$x, "native") :
## 'x' and 'units' must have length > 0GO and KEGG enrichment analysis
library(clusterProfiler)
library(org.Hs.eg.db)eg_diffPeak_cov <- bitr(unique(Diff_peaks_cov$name), fromType="SYMBOL", toType="ENTREZID", OrgDb="org.Hs.eg.db")
KEGG_diffPeak_cov <- enrichKEGG(eg_diffPeak_cov$ENTREZID,organism = "hsa",pAdjustMethod = "none", pvalueCutoff = 0.05,minGSSize = 3)
ego_diffPeak_cov <- enrichGO(gene = eg_diffPeak_cov$ENTREZID,
universe = bitr(rownames(RADAR@geneSum), fromType="SYMBOL", toType="ENTREZID", OrgDb="org.Hs.eg.db")$ENTREZID,
OrgDb = org.Hs.eg.db,
ont = "BP",
pAdjustMethod = "BH",
pvalueCutoff = 0.01,
qvalueCutoff = 0.05,
readable = TRUE)Plot enriched pathways
emapplot(KEGG_diffPeak_cov,n = 20,vertex.label.font = 2)+theme(legend.text = element_text(face = "bold"),legend.title = element_text(face = "bold") )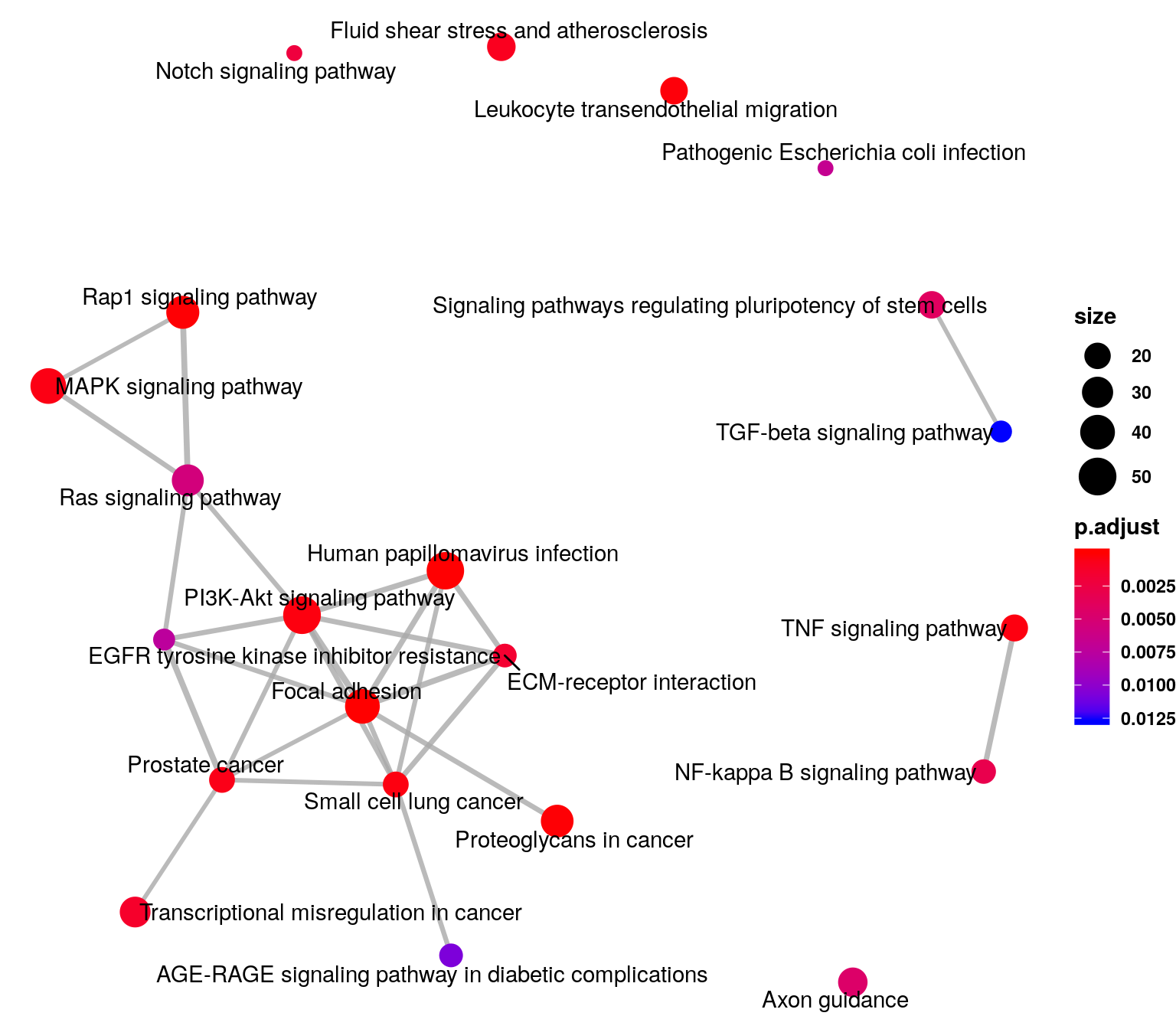
KEGG_diffPeak_cov_df <- as.data.frame(KEGG_diffPeak_cov)
KEGG_diffPeak_cov_df$geneID <- unlist( lapply(strsplit(KEGG_diffPeak_cov_df$geneID,"/"),function(x){ paste( bitr(x,fromType="ENTREZID", toType="SYMBOL",OrgDb="org.Hs.eg.db")$SYMBOL, collapse = "/") }) )
write.table(KEGG_diffPeak_cov_df, "~/PoissonGamma_Method/Ovary_Cancer/kegg_diffPeak_covariates.xls",sep = "\t", col.names = T, row.names = F,quote = F)
DT::datatable( KEGG_diffPeak_cov_df[KEGG_diffPeak_cov_df$Description %in% c("PI3K-Akt signaling pathway","MAPK signaling pathway","
Prostate cancer","Small cell lung cancer","Transcriptional misregulation in cancer","TNF signaling pathway"),c("ID","Description","pvalue","geneID")],options = list(scrollX = TRUE, keys = TRUE, pageLength = 10),rownames = F)Plot enriched biological process
emapplot(ego_diffPeak_cov,n = 25,vertex.label.font = 2)+theme(legend.text = element_text(face = "bold"),legend.title = element_text(face = "bold") )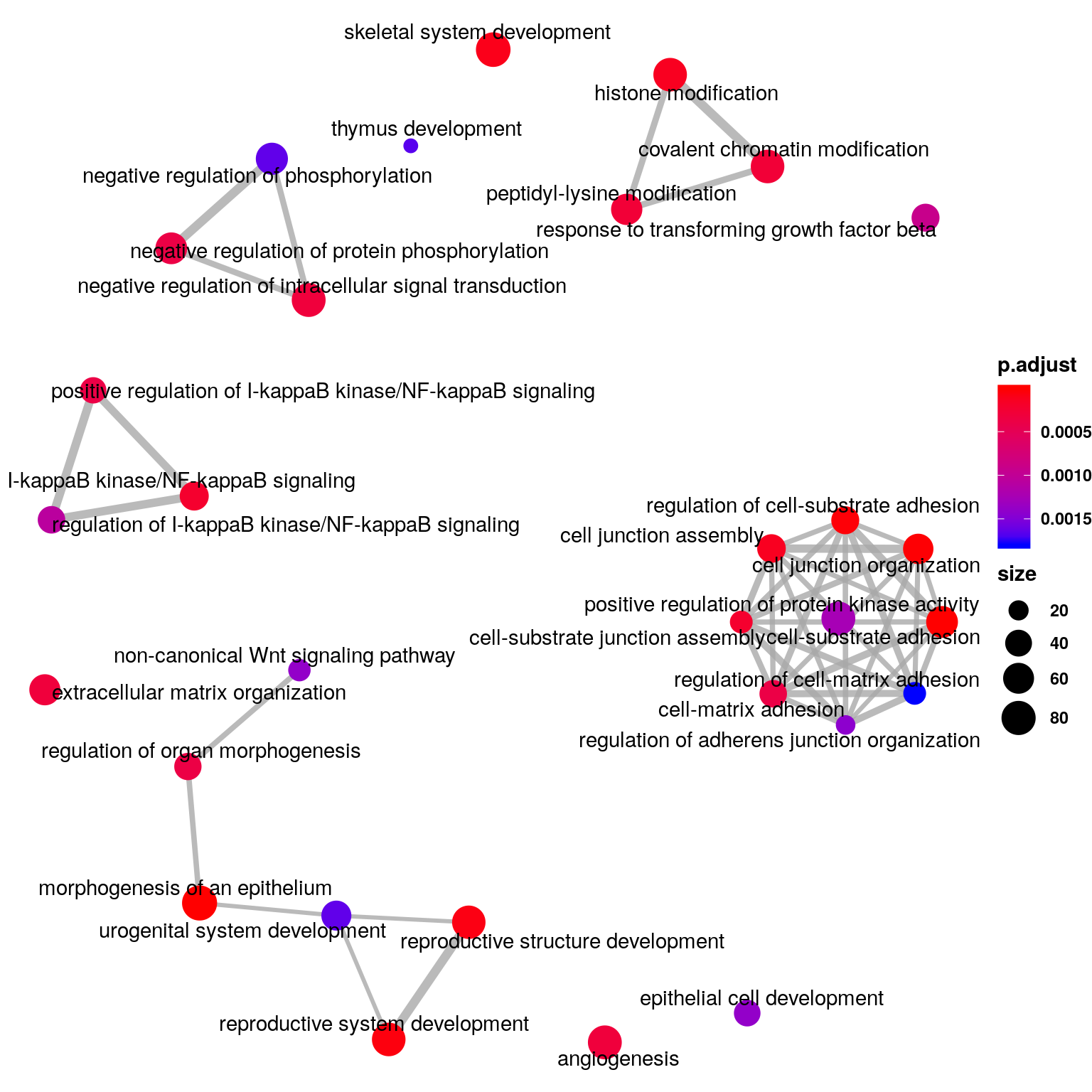
Enriched KEGG pathway for MeTDiff result
RADAR_metdiff <- RADAR
RADAR_metdiff@test.est[,"beta"] <- Metdiff.res$logFC
RADAR_metdiff@test.est[,"p_value"] <- Metdiff.res$pvalue
RADAR_metdiff@test.est[,"fdr"] <- Metdiff.res$fdr
RADAR_metdiff <- reportResult(RADAR_metdiff, cutoff = 0.1,threads = 20)
diffPeak_metidff <- results(RADAR_metdiff)
eg_diffPeak_metdiff <- bitr(unique(diffPeak_metidff$name), fromType="SYMBOL", toType="ENTREZID", OrgDb="org.Hs.eg.db")
KEGG_diffPeak_metdiff <- enrichKEGG(eg_diffPeak_metdiff$ENTREZID,organism = "hsa",pAdjustMethod = "none", pvalueCutoff = 0.05,minGSSize = 3)
ego_diffPeak_metdiff <- enrichGO(gene = eg_diffPeak_metdiff$ENTREZID,
universe = bitr(rownames(RADAR@geneSum), fromType="SYMBOL", toType="ENTREZID", OrgDb="org.Hs.eg.db")$ENTREZID,
OrgDb = org.Hs.eg.db,
ont = "BP",
pAdjustMethod = "BH",
pvalueCutoff = 0.01,
qvalueCutoff = 0.05,
readable = TRUE)emapplot(KEGG_diffPeak_metdiff,n = 20,vertex.label.font = 2)+theme(legend.text = element_text(face = "bold"),legend.title = element_text(face = "bold") )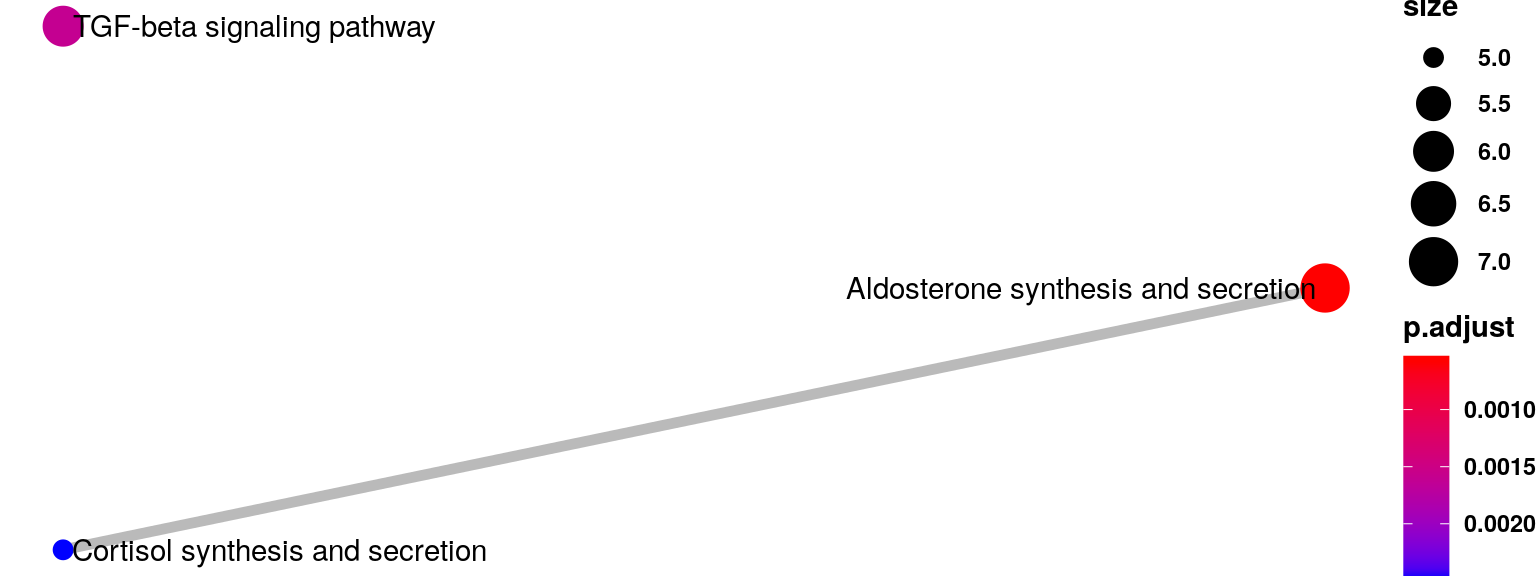
Plot differential peaks detected.
RADAR <- PrepCoveragePlot(RADAR)## GTF has already been imported as Granges, importing it again...PTEN
plotGeneCov(RADAR,"PTEN" ,adjustExprLevel = T)+scale_x_continuous(expand = c(0.02,0))+theme(legend.position = "none")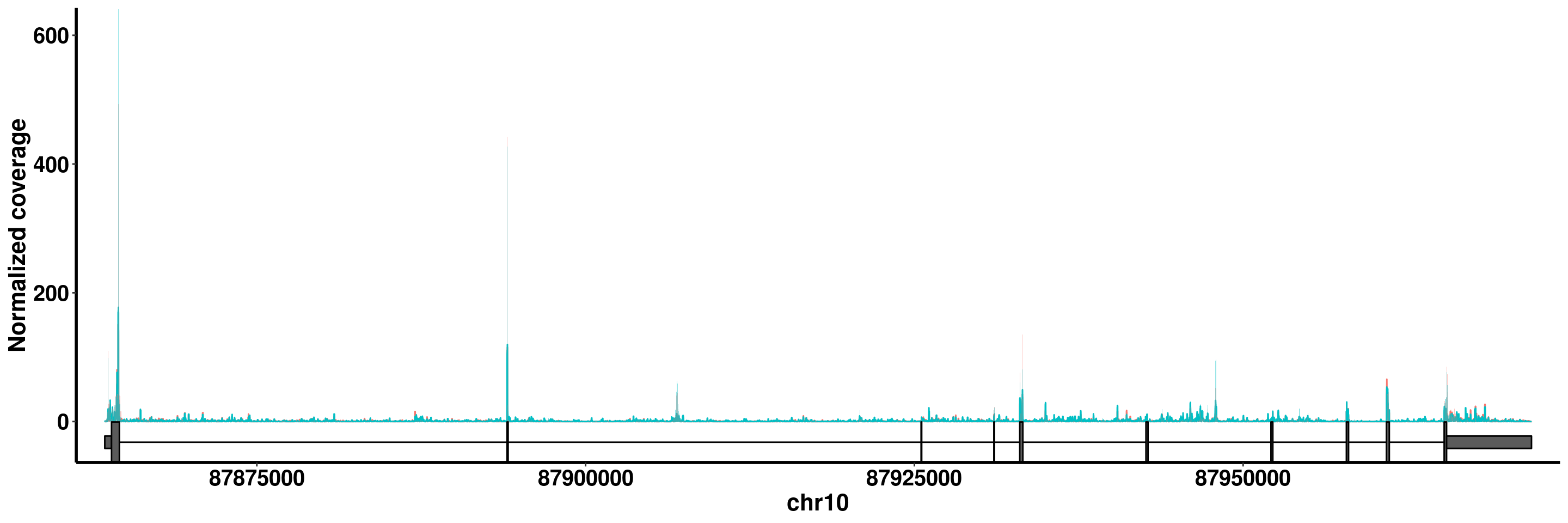
plotGeneCov(RADAR,"PTEN" ,ZoomIn = c(87965095,87966657),adjustExprLevel = T)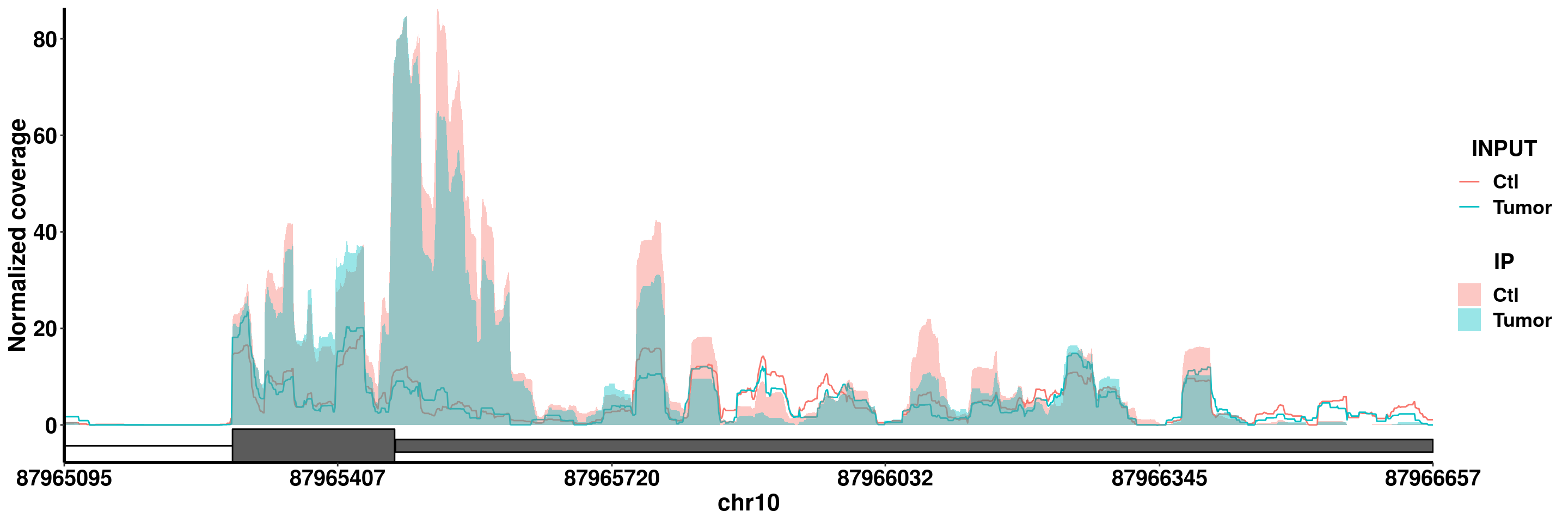
plotGeneCov(RADAR,"PTEN" ,ZoomIn = c(87929633,87936182),adjustExprLevel = T)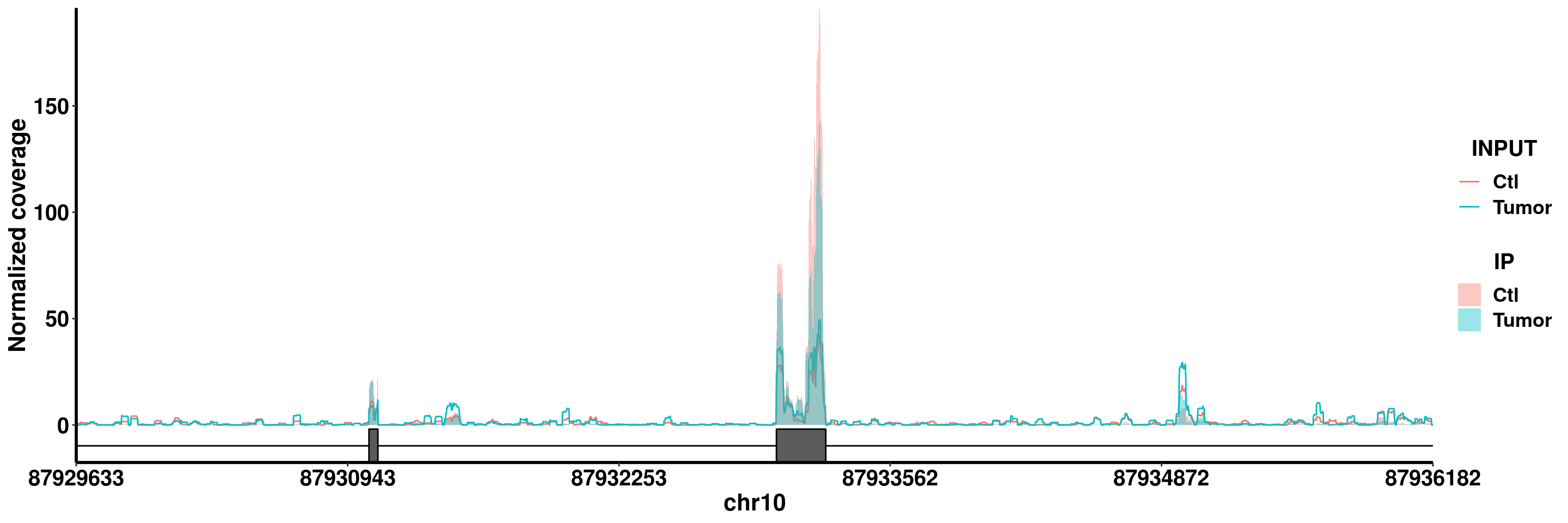
BCL2
plotGeneCov(RADAR,"BCL2",adjustExprLevel = T)+scale_x_continuous(expand = c(0.02,0) )+theme(legend.position = "none")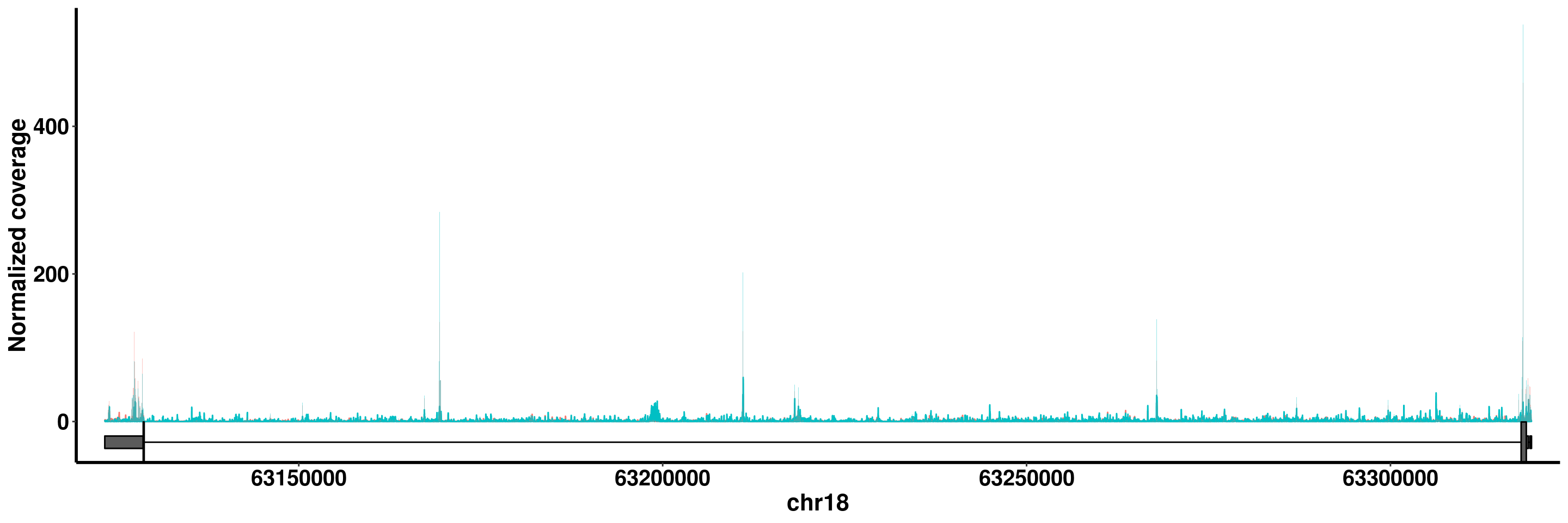
plotGeneCov(RADAR,"BCL2",ZoomIn = c(63317900,63318363),adjustExprLevel = T)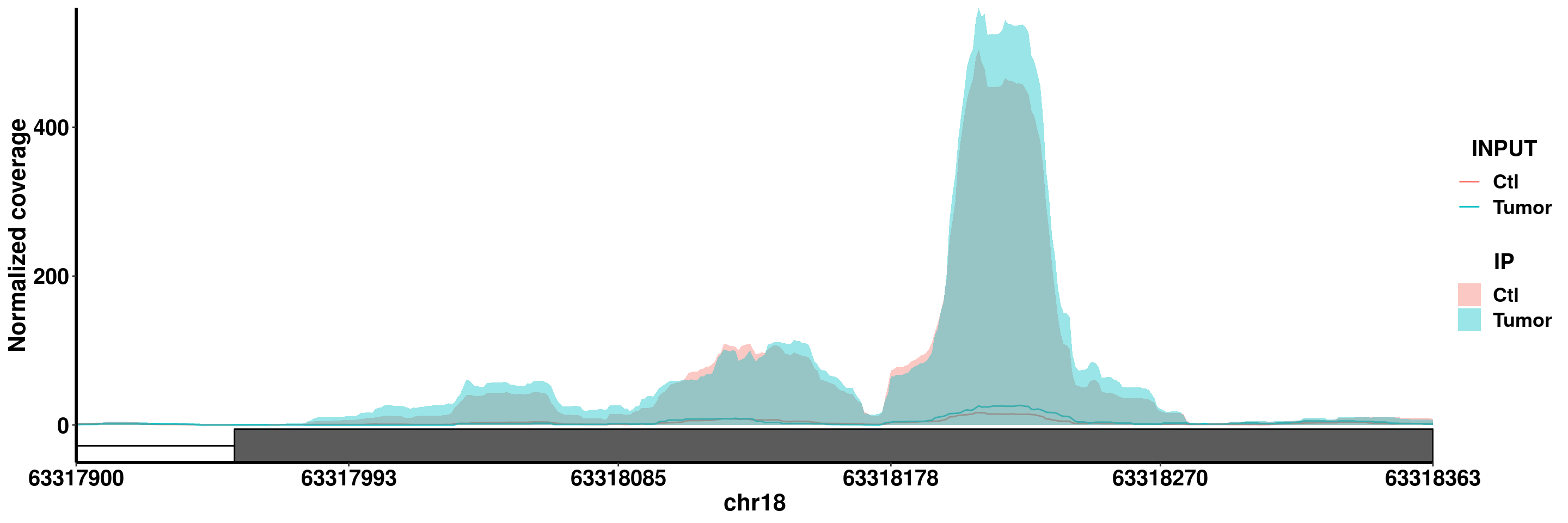
PIK3AP1
plotGeneCov(RADAR,"PIK3AP1",adjustExprLevel =T )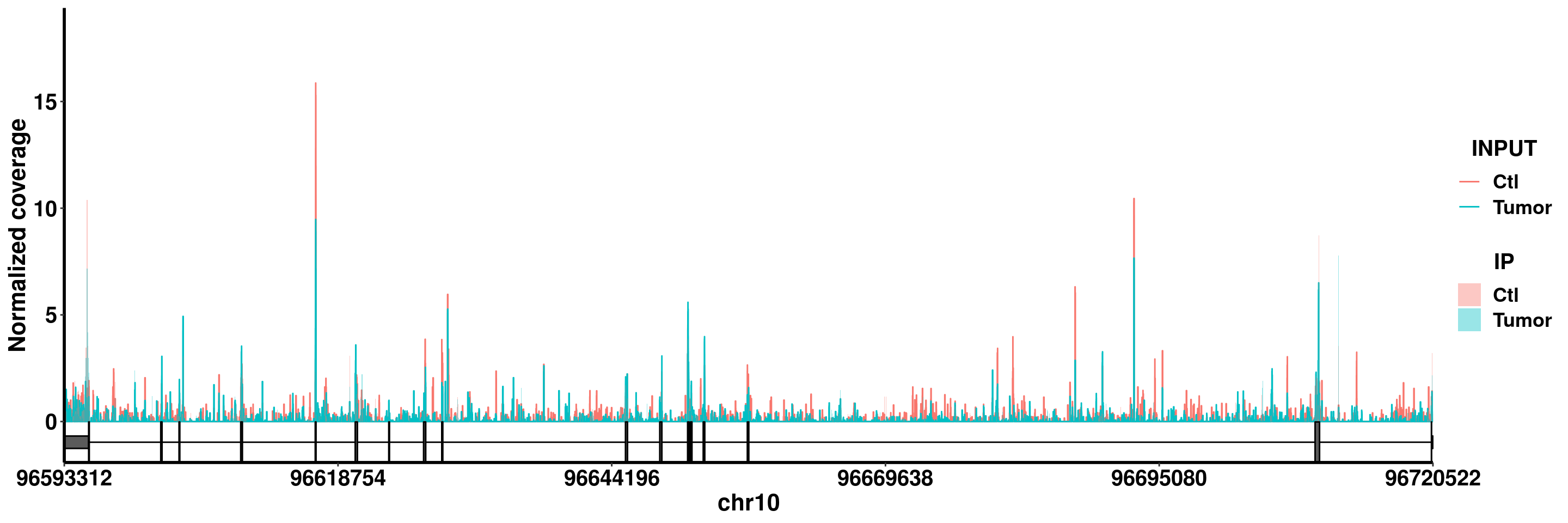
plotGeneCov(RADAR,"PIK3AP1",ZoomIn = c(96708976, 96710100),adjustExprLevel = TRUE)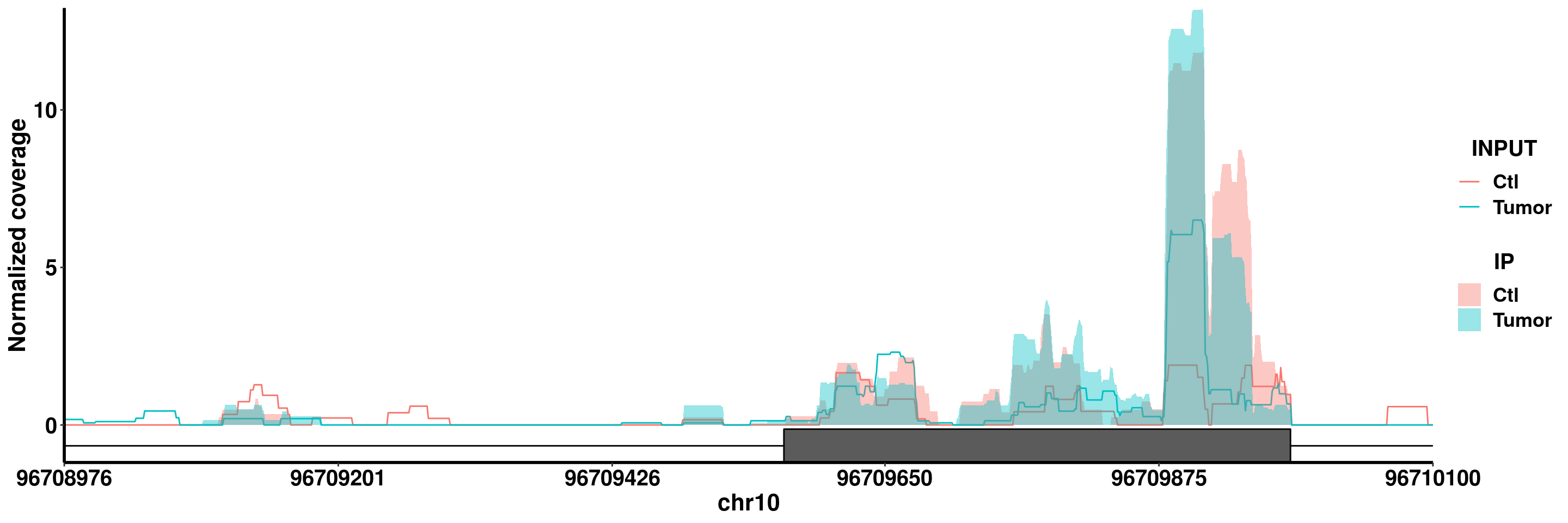
Heatmap for differential loci
topBins <- RADAR@ip_adjExpr_filtered[rownames(RADAR_cov[RADAR_cov[,"fdr"]<0.1,]),]
log_topBins <- log(topBins+1)
log_topBins_center <- t(apply(log_topBins,1,function(x){x-mean(x)}) )
colnames(log_topBins_center) <- unlist(variable(RADAR)[,1])
dist.pear <- function(x) as.dist(1-cor(t(x)))
hclust.ave <- function(x) hclust(x, method="average")
gplots::heatmap.2(log_topBins_center,scale="row",trace="none",labRow=NA,main = "Methylation level of significant bins",
distfun=dist.pear, hclustfun=hclust.ave,col=rev(RColorBrewer::brewer.pal(9,"RdBu")))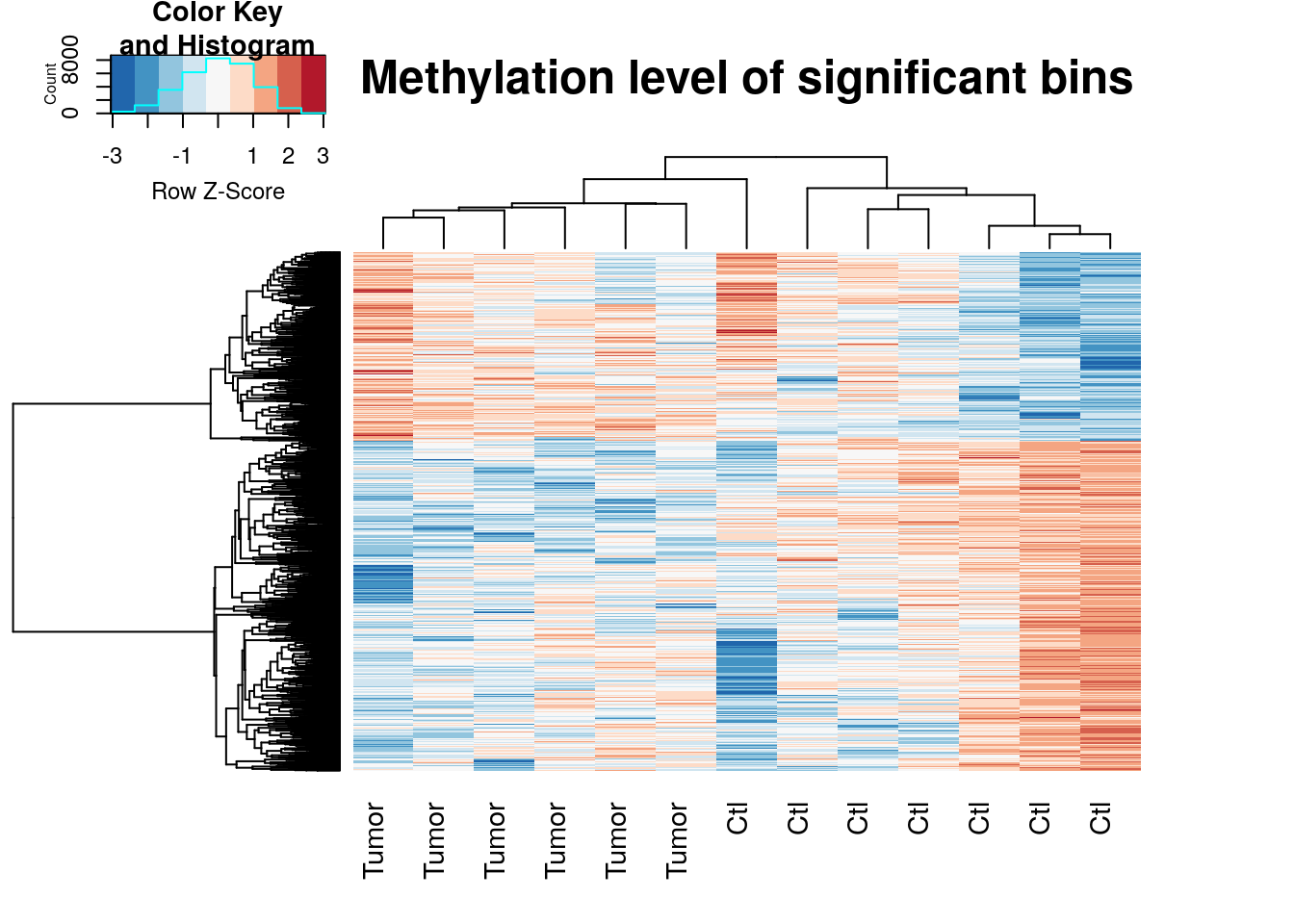
RADAR_diff_extend.gr <- makeGRangesFromDataFrame(Diff_peaks_cov_extend)
QNB_diff_extend.gr <- makeGRangesFromDataFrame(QNB_diff_extend)
exomePeak_diff_extend.gr <- makeGRangesFromDataFrame(exomePeak_diff_extend)
Metdiff_diff_extend.gr <- makeGRangesFromDataFrame(Metdiff_diff_extend)
fisher_diff_extend.gr <- makeGRangesFromDataFrame(fisher_diff_extend)
R_all_overlap <- findOverlaps( RADAR_diff_extend.gr , reduce(c(QNB_diff_extend.gr, Metdiff_diff_extend.gr,exomePeak_diff_extend.gr,fisher_diff_extend.gr)) )
RADAR_specific <- Diff_peaks_cov_extend[ ! 1:length(RADAR_diff_extend.gr)%in% queryHits(R_all_overlap),]
E_F_R_overlap <- findOverlaps( exomePeak_diff_extend.gr[unique( queryHits( findOverlaps(exomePeak_diff_extend.gr,fisher_diff_extend.gr) ) )], RADAR_diff_extend.gr)
bogus_specific <- exomePeak_diff_extend[ ! 1:length(exomePeak_diff_extend.gr) %in% queryHits(E_F_R_overlap) , ]RADAR specific DM sites
plotGeneCov(RADAR,"ARID3A", ZoomIn = c(972162,972360),adjustExprLevel =TRUE , split = T )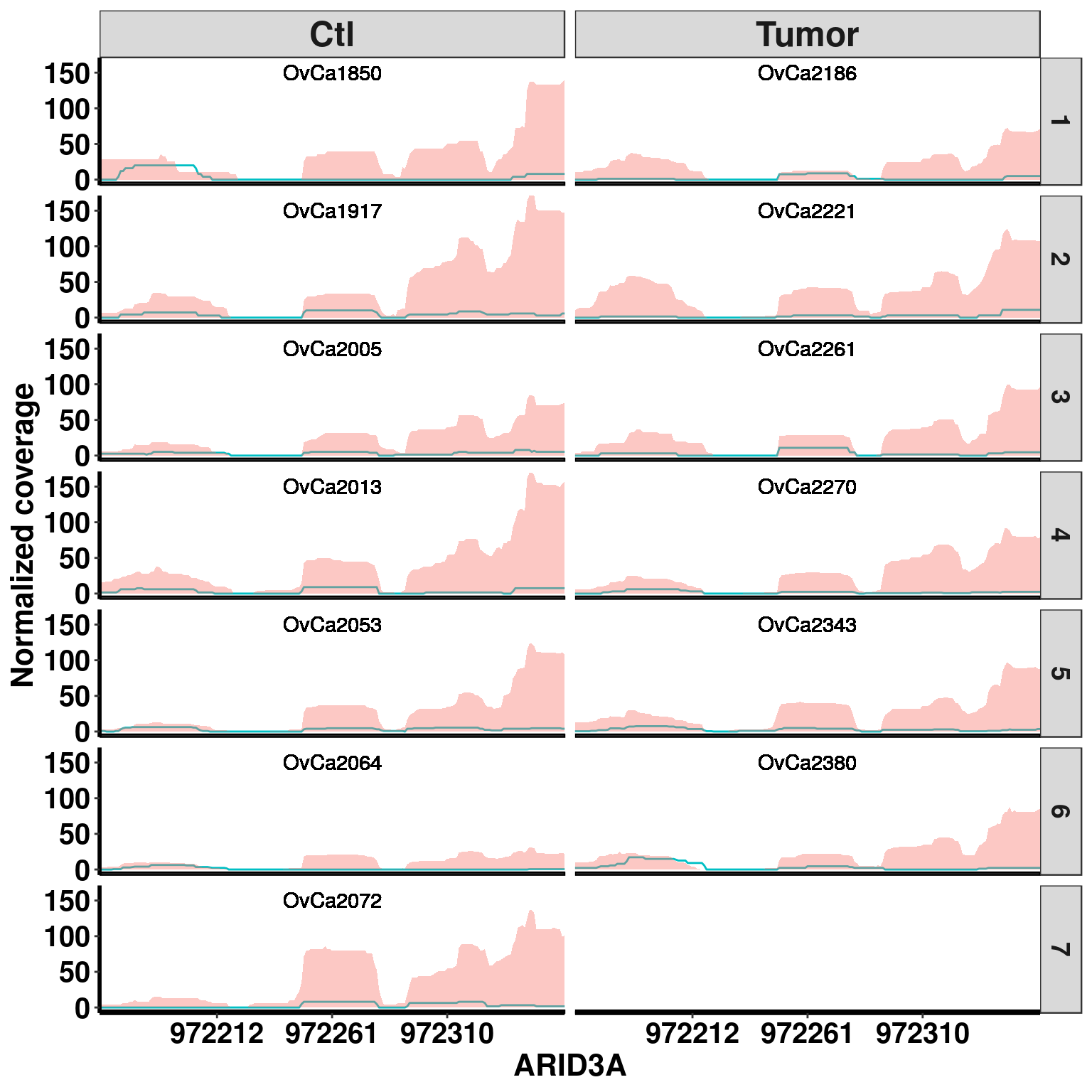
plotGeneCov(RADAR,"DPM1", ZoomIn = c(50958324,50958473),adjustExprLevel =TRUE , split = TRUE )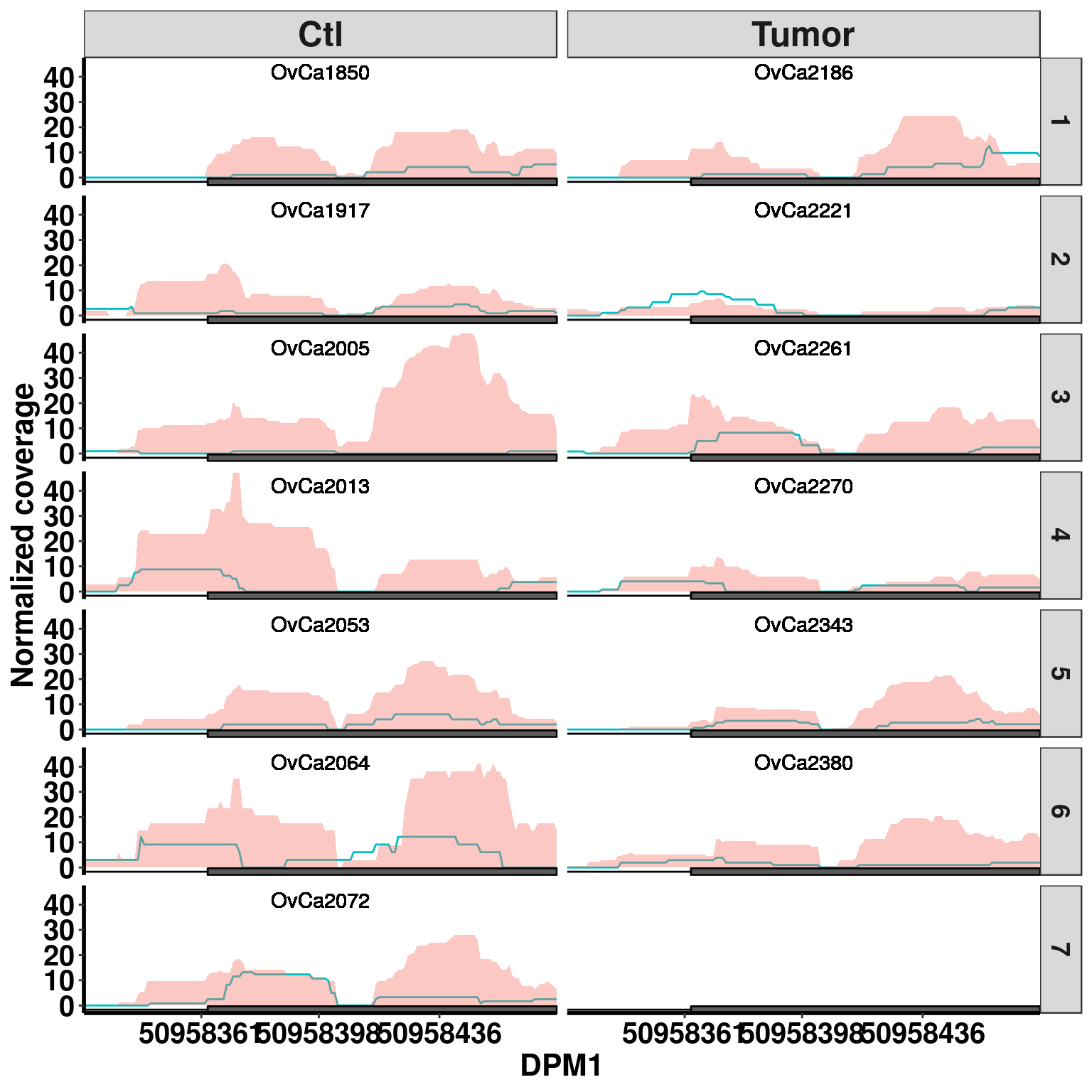
plotGeneCov(RADAR,"EID1", ZoomIn = c(48879586,48879785),adjustExprLevel =TRUE , split = TRUE )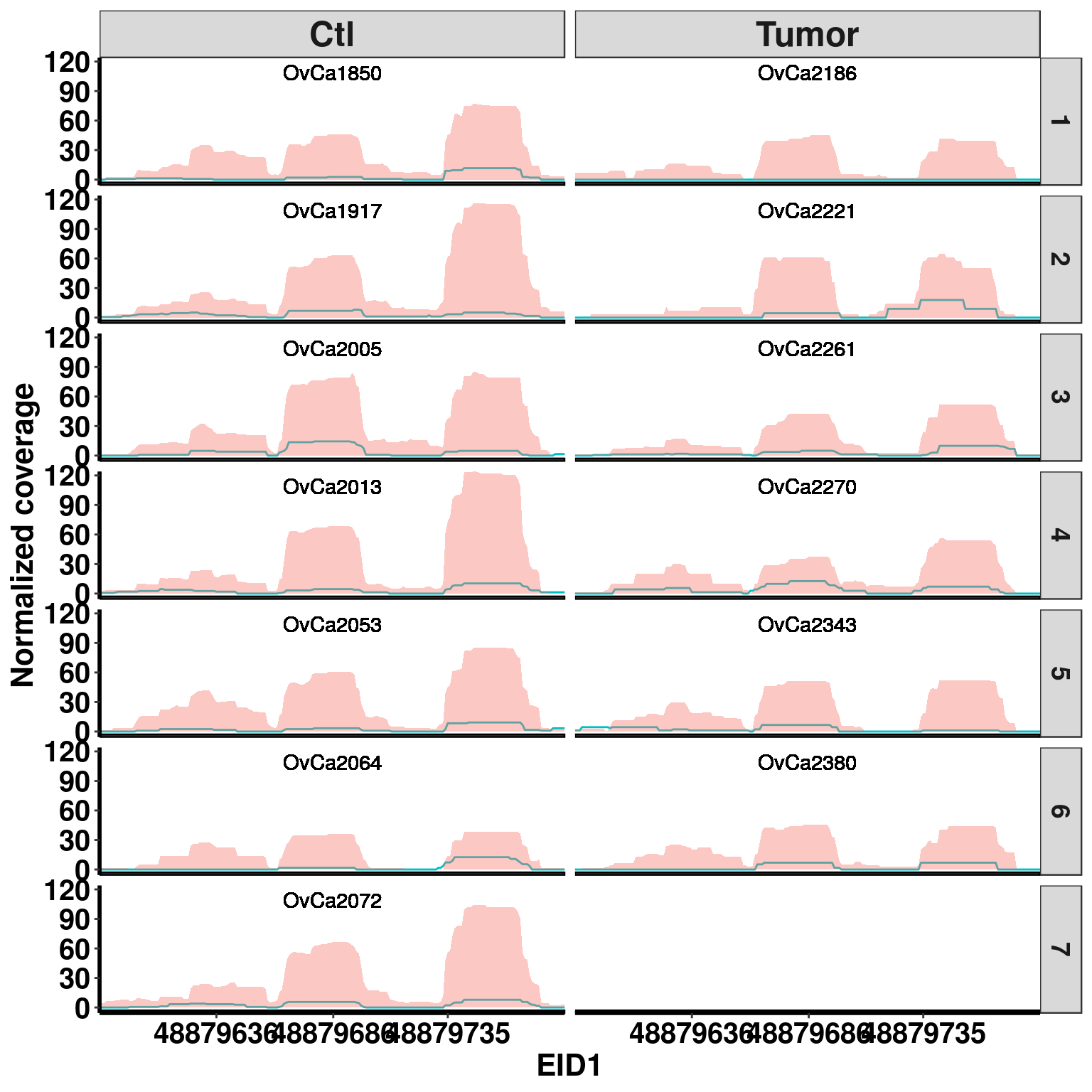
Bogus MD sites
plotGeneCov(RADAR,"ADIRF", ZoomIn = c(86970166,86970624),adjustExprLevel =TRUE , split = TRUE )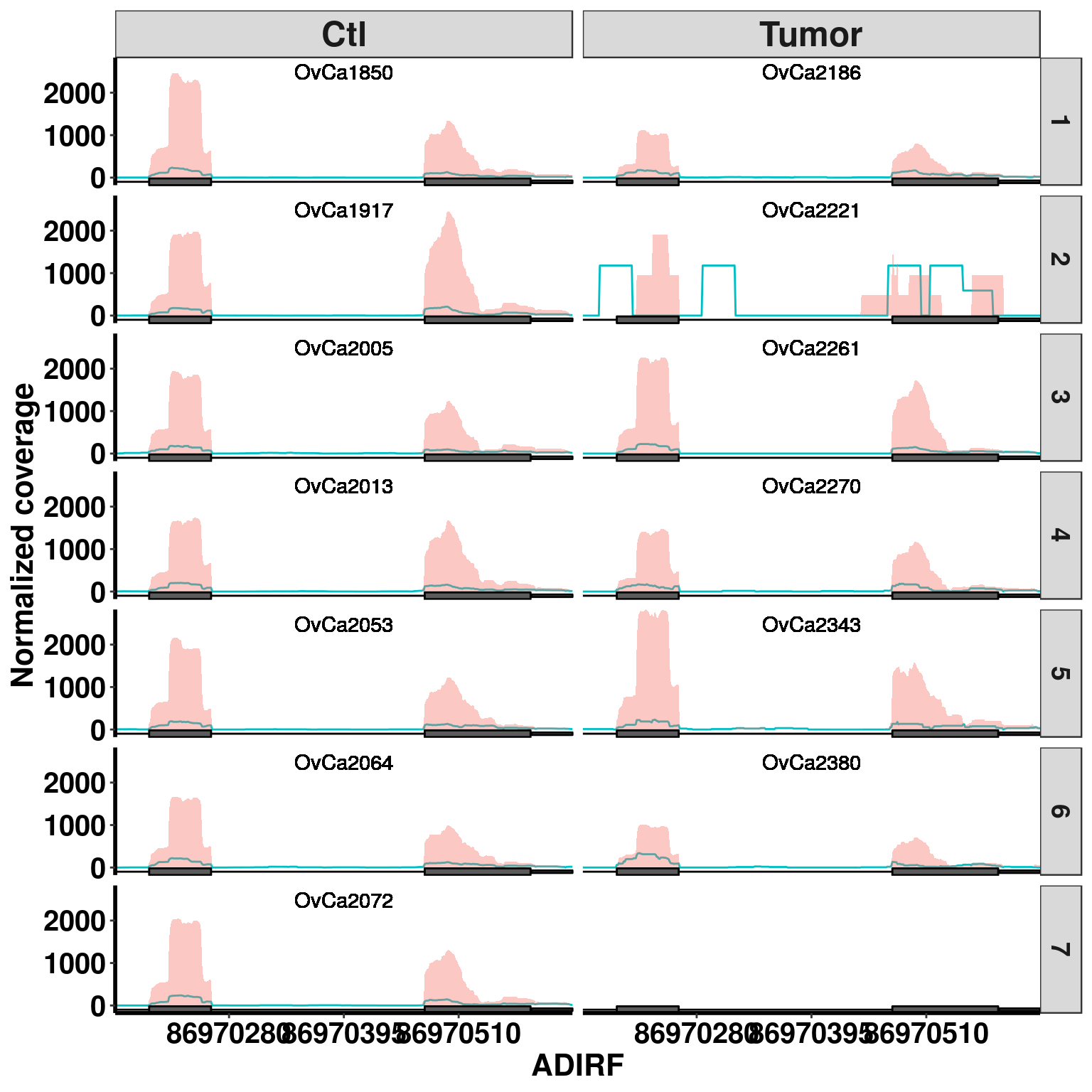
plotGeneCov(RADAR,"ADIRF", ZoomIn = c(86970166,86970624),adjustExprLevel =TRUE , split = TRUE )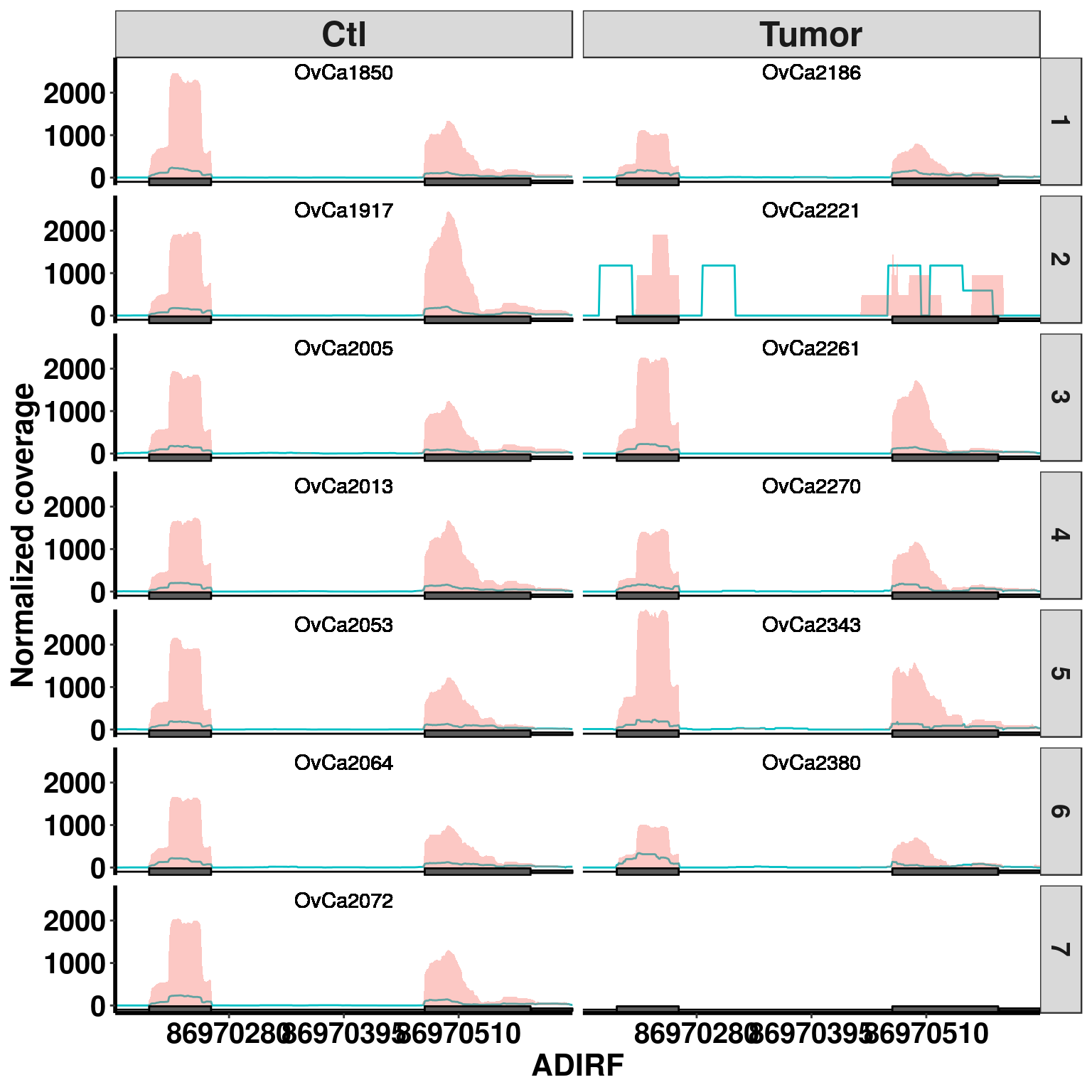
Session information
sessionInfo()## R version 3.5.3 (2019-03-11)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 17.10
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/libopenblasp-r0.2.20.so
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] grid stats4 parallel stats graphics grDevices utils
## [8] datasets methods base
##
## other attached packages:
## [1] org.Hs.eg.db_3.7.0 clusterProfiler_3.10.1
## [3] Logolas_1.6.0 reshape2_1.4.3
## [5] GenomicAlignments_1.18.1 SummarizedExperiment_1.12.0
## [7] DelayedArray_0.8.0 BiocParallel_1.16.6
## [9] matrixStats_0.54.0 rtracklayer_1.42.2
## [11] RADAR_0.2.1 qvalue_2.14.1
## [13] RcppArmadillo_0.9.400.2.0 Rcpp_1.0.1
## [15] RColorBrewer_1.1-2 gplots_3.0.1.1
## [17] doParallel_1.0.14 iterators_1.0.10
## [19] foreach_1.4.4 ggplot2_3.1.1
## [21] Rsamtools_1.34.1 Biostrings_2.50.2
## [23] XVector_0.22.0 GenomicFeatures_1.34.8
## [25] AnnotationDbi_1.44.0 Biobase_2.42.0
## [27] GenomicRanges_1.34.0 GenomeInfoDb_1.18.2
## [29] IRanges_2.16.0 S4Vectors_0.20.1
## [31] BiocGenerics_0.28.0
##
## loaded via a namespace (and not attached):
## [1] backports_1.1.4 Hmisc_4.2-0 fastmatch_1.1-0
## [4] plyr_1.8.4 igraph_1.2.4.1 lazyeval_0.2.2
## [7] splines_3.5.3 crosstalk_1.0.0 gamlss_5.1-3
## [10] gridBase_0.4-7 urltools_1.7.3 digest_0.6.18
## [13] htmltools_0.3.6 GOSemSim_2.8.0 viridis_0.5.1
## [16] GO.db_3.7.0 SQUAREM_2017.10-1 gdata_2.18.0
## [19] magrittr_1.5 checkmate_1.9.1 memoise_1.1.0
## [22] BSgenome_1.50.0 cluster_2.0.7-1 annotate_1.60.1
## [25] enrichplot_1.2.0 prettyunits_1.0.2 colorspace_1.4-1
## [28] blob_1.1.1 ggrepel_0.8.0 gamlss.data_5.1-3
## [31] exomePeak_2.16.0 xfun_0.6 dplyr_0.8.0.1
## [34] crayon_1.3.4 RCurl_1.95-4.12 jsonlite_1.6
## [37] genefilter_1.64.0 ape_5.3 survival_2.44-1.1
## [40] glue_1.3.1 polyclip_1.10-0 gtable_0.3.0
## [43] zlibbioc_1.28.0 UpSetR_1.3.3 scales_1.0.0
## [46] DOSE_3.8.2 DBI_1.0.0 viridisLite_0.3.0
## [49] xtable_1.8-4 progress_1.2.0 htmlTable_1.13.1
## [52] gridGraphics_0.3-0 foreign_0.8-71 bit_1.1-14
## [55] europepmc_0.3 Formula_1.2-3 DT_0.5.1
## [58] htmlwidgets_1.3 httr_1.4.0 fgsea_1.8.0
## [61] acepack_1.4.1 pkgconfig_2.0.2 XML_3.98-1.19
## [64] farver_1.1.0 nnet_7.3-12 locfit_1.5-9.1
## [67] later_0.8.0 ggplotify_0.0.3 tidyselect_0.2.5
## [70] labeling_0.3 rlang_0.4.0 munsell_0.5.0
## [73] tools_3.5.3 generics_0.0.2 RSQLite_2.1.1
## [76] broom_0.5.2 ggridges_0.5.1 evaluate_0.13
## [79] stringr_1.4.0 yaml_2.2.0 knitr_1.22
## [82] bit64_0.9-7 fs_1.3.0 caTools_1.17.1.2
## [85] purrr_0.3.2 ggraph_1.0.2 nlme_3.1-137
## [88] mime_0.6 QNB_1.1.11 DO.db_2.9
## [91] xml2_1.2.0 biomaRt_2.38.0 compiler_3.5.3
## [94] rstudioapi_0.10 Guitar_1.20.1 tibble_2.1.1
## [97] tweenr_1.0.1 geneplotter_1.60.0 stringi_1.4.3
## [100] lattice_0.20-38 Matrix_1.2-17 permute_0.9-5
## [103] vegan_2.5-4 pillar_1.3.1 triebeard_0.3.0
## [106] data.table_1.12.2 cowplot_0.9.4 bitops_1.0-6
## [109] httpuv_1.5.1 R6_2.4.0 latticeExtra_0.6-28
## [112] promises_1.0.1 vcfR_1.8.0 KernSmooth_2.23-15
## [115] gridExtra_2.3 codetools_0.2-16 MASS_7.3-51.4
## [118] gtools_3.8.1 assertthat_0.2.1 DESeq2_1.22.2
## [121] pinfsc50_1.1.0 withr_2.1.2 GenomeInfoDbData_1.2.0
## [124] mgcv_1.8-28 hms_0.4.2 rpart_4.1-13
## [127] tidyr_0.8.3 rmarkdown_1.12 rvcheck_0.1.3
## [130] ggforce_0.2.2 shiny_1.3.2 gamlss.dist_5.1-3
## [133] base64enc_0.1-3This R Markdown site was created with workflowr