T2D
Zijie Zhang
2018-08-21
Count reads
library(RADAR)
samplenames = c("Ctl1","Ctl4","Ctl8","Ctl16","Ctl17","Ctl18","Ctl20","T2D1","T2D4","T2D5","T2D6","T2D7","T2D9","T2D11","T2D12")
RADAR <- countReads(samplenames = samplenames,
gtf = "~/Database/genome/hg38/hg38_UCSC.gtf",
bamFolder = "~/Rohit_T2D/bam_files",
outputDir = "~/Rohit_T2D",
modification = "IP",
threads = 25
)
monster <- RADARSummary of read count
library(RADAR)
library(qvalue)
#load("~/Rohit_T2D/RADAR.RData")
load("~/Rohit_T2D/RADAR_analysis.RData")
countSummary <- rbind("Input"= colSums(RADAR$reads)[1:length(RADAR$samplenames)], "IP" =colSums(RADAR$reads)[(1:length(RADAR$samplenames))+length(RADAR$samplenames)] )
colnames(countSummary) <- RADAR$samplenames
knitr::kable(countSummary,format = "pandoc" )| Ctl1 | Ctl4 | Ctl8 | Ctl16 | Ctl17 | Ctl18 | Ctl20 | T2D1 | T2D4 | T2D5 | T2D6 | T2D7 | T2D9 | T2D11 | T2D12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Input | 18320414 | 17749453 | 17923796 | 11508719 | 15118237 | 25923296 | 20302596 | 16803517 | 16105448 | 14146820 | 17202593 | 14336611 | 17758817 | 19782144 | 18779123 |
| IP | 30287782 | 25105375 | 21908869 | 23197918 | 21409901 | 31653712 | 24753657 | 27733641 | 26634012 | 28457739 | 22539106 | 36100747 | 27434039 | 22053013 | 19402847 |
X <- c(rep("Ctl",7),rep("T2D",8))
RADAR <- normalizeLibrary(RADAR,X)
RADAR <- RADAR::adjustExprLevel(RADAR)
RADAR <- filterBins(RADAR,minCountsCutOff = 15)
RADAR_pos <- RADAR::adjustExprLevel(RADAR[1:12], adjustBy = "pos")
RADAR_pos <- filterBins(RADAR_pos,minCountsCutOff = 15)INPUT read count
Local INPUT read count
Plot distribution of number of reads in each 50 bp bins
hist(log10(rowMeans(RADAR$reads[rowMeans(RADAR$reads[,grep("input",colnames(RADAR$reads))])>1,grep("input",colnames(RADAR$reads))]) ),xlab = "log10 read count",main = "Distribution of INPUT read count in bins",xlim = c(0,3), col =rgb(0.2,0.2,0.2,0.5),cex.main = 2,cex.axis =2,cex.lab=2)
axis(side = 1, lwd = 2,cex.axis =2)
axis(side = 2, lwd = 2,cex.axis =2)
local read count V.S. geneSum
Compute the within group variability of INPUT geneSum VS INPUT local read count.
var.coef <- function(x){sd(as.numeric(x))/mean(as.numeric(x))}
## filter expressed genes
geneSum <- RADAR$geneSum[rowSums(RADAR$geneSum)>16,]
## within group variability
relative.var <- t( apply(geneSum,1,tapply,RADAR$X,var.coef) )
geneSum.var <- c(relative.var)
## For each gene used above, random sample a 50bp bin within this gene
set.seed(1)
r.bin50 <- tapply(rownames(RADAR$norm.input)[which(RADAR$geneBins$gene %in% rownames(geneSum))],as.character( RADAR$geneBins$gene[which(RADAR$geneBins$gene %in% rownames(geneSum))]) ,function(x){
n <- sample(1:length(x),1)
return(x[n])
})
relative.var <- apply(RADAR$norm.input[r.bin50,],1,tapply,RADAR$X,var.coef)
bin50.var <- c(relative.var)
bin50.var <- bin50.var[!is.na(bin50.var)]
## 100bp bins
r.bin100 <- tapply(rownames(RADAR$norm.input)[which(RADAR$geneBins$gene %in% rownames(geneSum))],as.character( RADAR$geneBins$gene[which(RADAR$geneBins$gene %in% rownames(geneSum))]) ,function(x){
n <- sample(1:(length(x)-2),1)
return(x[n:(n+1)])
})
relative.var <- lapply(r.bin100, function(x){ tapply( colSums(RADAR$norm.input[x,]),RADAR$X,var.coef ) })
bin100.var <- unlist(relative.var)
bin100.var <- bin100.var[!is.na(bin100.var)]
## 200bp bins
r.bin200 <- tapply(rownames(RADAR$norm.input)[which(RADAR$geneBins$gene %in% rownames(geneSum))],as.character( RADAR$geneBins$gene[which(RADAR$geneBins$gene %in% rownames(geneSum))]) ,function(x){
if(length(x)>4){
n <- sample(1:(length(x)-3),1)
return(x[n:(n+3)])
}else{
return(NULL)
}
})
r.bin200 <- r.bin200[which(!unlist(lapply(r.bin200,is.null)) ) ]
relative.var <- lapply(r.bin200, function(x){ tapply( colSums(RADAR$norm.input[x,]),RADAR$X,var.coef ) })
bin200.var <- unlist(relative.var)
bin200.var <- bin200.var[!is.na(bin200.var)]
relative.var <- data.frame(group=c(rep("geneSum",length(geneSum.var)),rep("bin-50bp",length(bin50.var)),rep("bin-100bp",length(bin100.var)),rep("bin-200bp",length(bin200.var))), variance=c(geneSum.var,bin50.var,bin100.var,bin200.var))ggplot(relative.var,aes(variance,colour=group))+geom_density()+xlab("Relative variation")+
theme_bw() + theme(panel.border = element_blank(), panel.grid.major = element_blank(),
panel.grid.minor = element_blank(), axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=20, face="bold", hjust=0.5,family = "arial"),
axis.title.y=element_text(size=20, face="bold", vjust=0.4, angle=90,family = "arial"),
legend.position = c(0.88,0.88),legend.title=element_blank(),legend.text = element_text(size = 14, face = "bold",family = "arial"),
axis.text = element_text(size = 15,face = "bold",family = "arial",colour = "black") )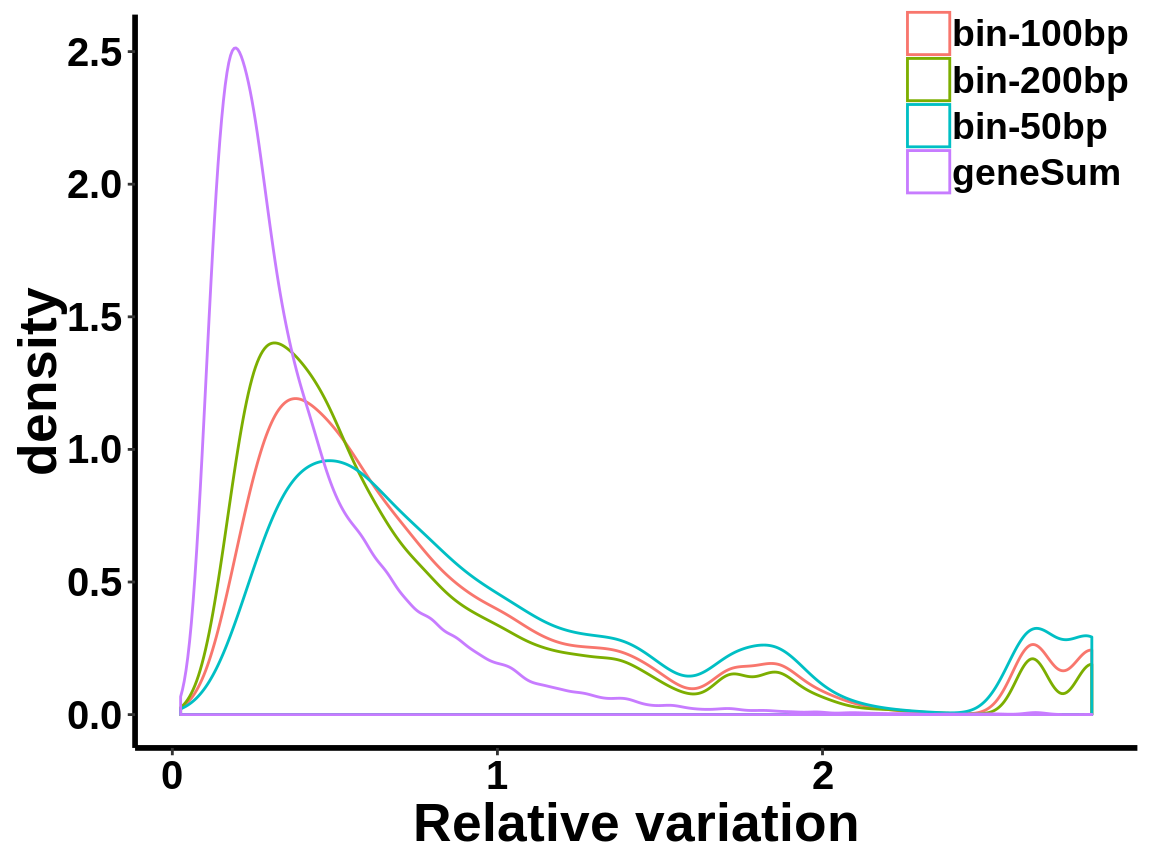
Compare MeRIP-seq IP data with regular RNA-seq data
var.coef <- function(x){sd(as.numeric(x))/mean(as.numeric(x))}
ip_coef_var <- t(apply(RADAR$ip_adjExpr_filtered,1,tapply,X,var.coef))
ip_coef_var <- ip_coef_var[!apply(ip_coef_var,1,function(x){return(any(is.na(x)))}),]
#hist(c(ip_coef_var),main = "M14KO mouse liver\n m6A-IP",xlab = "within group coefficient of variation",breaks = 50)
###
gene_coef_var <- t(apply(RADAR$geneSum,1,tapply,X,var.coef))
gene_coef_var <- gene_coef_var[!apply(gene_coef_var,1,function(x){return(any(is.na(x)))}),]
#hist(c(gene_coef_var),main = "M14KO mouse liver\n RNA-seq",xlab = "within group coefficient of variation",breaks = 50)
coef_var <- list('RNA-seq'=c(gene_coef_var),'m6A-IP'=c(ip_coef_var))
nn<-sapply(coef_var, length)
rs<-cumsum(nn)
re<-rs-nn+1
grp <- factor(rep(names(coef_var), nn), levels=names(coef_var))
coef_var.df <- data.frame(coefficient_var = c(c(gene_coef_var),c(ip_coef_var)),label = grp)ggplot(data = coef_var.df,aes(coefficient_var,colour = grp))+geom_density()+ theme_bw() + theme(panel.border = element_blank(), panel.grid.major = element_blank(),
panel.grid.minor = element_blank(), axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=20, face="bold", hjust=0.5,family = "arial"),
axis.title.y=element_text(size=20, face="bold", vjust=0.4, angle=90,family = "arial"),
legend.position = c(0.9,0.9),legend.title=element_blank(),legend.text = element_text(size = 18, face = "bold",family = "arial"),
axis.text = element_text(size = 15,face = "bold",family = "arial",colour = "black") )+
xlab("Coefficient of variation")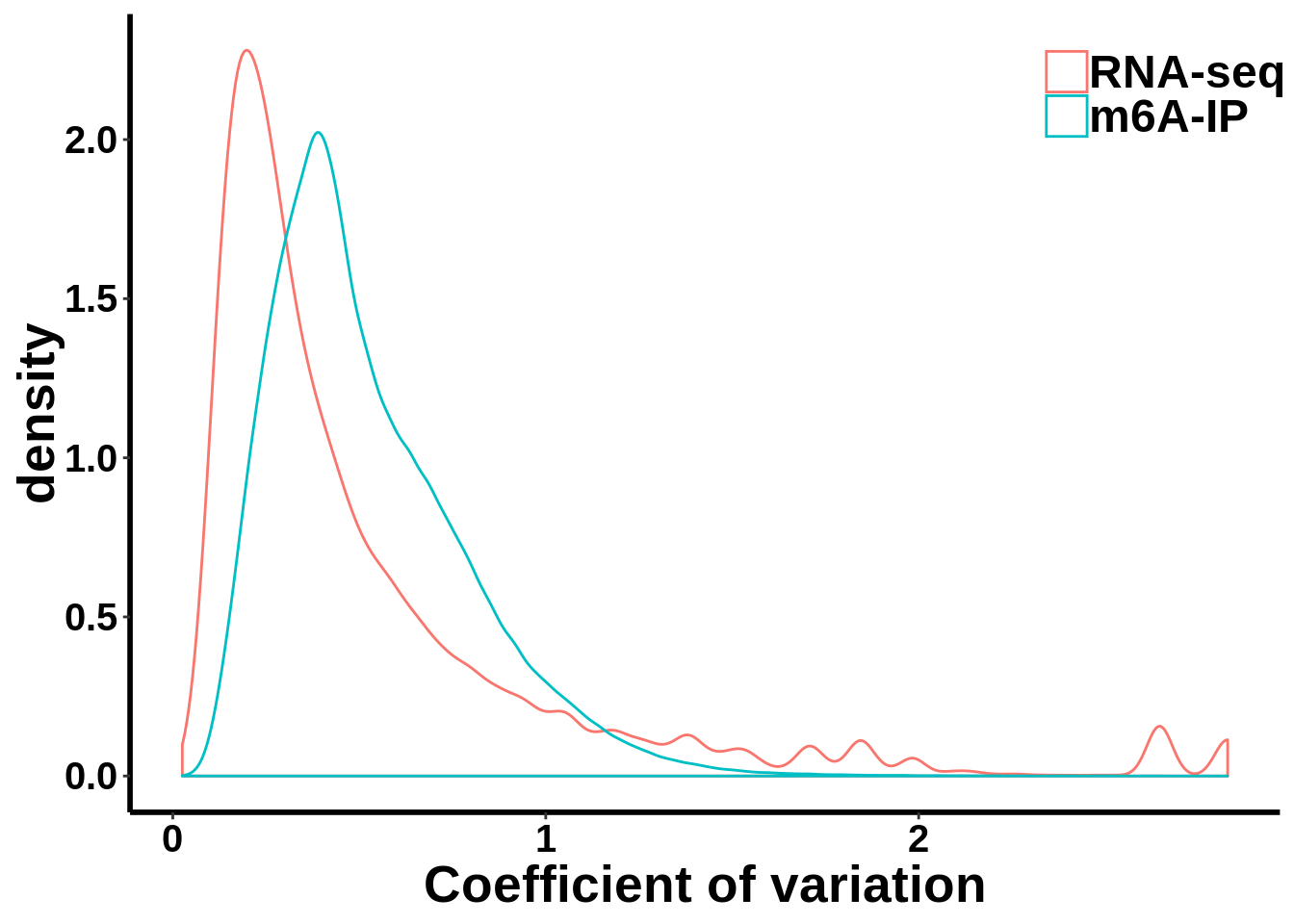
Check mean variance relationship of the data.
ip_var <- t(apply(RADAR$ip_adjExpr_filtered,1,tapply,X,var))
#ip_var <- ip_coef_var[!apply(ip_var,1,function(x){return(any(is.na(x)))}),]
ip_mean <- t(apply(RADAR$ip_adjExpr_filtered,1,tapply,X,mean))
###
gene_var <- t(apply(RADAR$geneSum,1,tapply,X,var))
#gene_var <- gene_var[!apply(gene_var,1,function(x){return(any(is.na(x)))}),]
gene_mean <- t(apply(RADAR$geneSum,1,tapply,X,mean))
all_var <- list('RNA-seq'=c(gene_var),'m6A-IP'=c(ip_var))
nn<-sapply(all_var, length)
rs<-cumsum(nn)
re<-rs-nn+1
group <- factor(rep(names(all_var), nn), levels=names(all_var))
all_var.df <- data.frame(variance = c(c(gene_var),c(ip_var)),mean= c(c(gene_mean),c(ip_mean)),label = group)ggplot(data = all_var.df,aes(x=mean,y=variance,colour = group,shape = group))+geom_point(size = I(0.2))+stat_smooth(se = T,show.legend = F)+stat_smooth(se = F)+theme_bw() +xlab("Mean")+ylab("Variance") +theme(panel.border = element_blank(), panel.grid.major = element_blank(),
panel.grid.minor = element_blank(), axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=22, face="bold", hjust=0.5,family = "arial"),
axis.title.y=element_text(size=22, face="bold", vjust=0.4, angle=90,family = "arial"),
legend.position = c(0.85,0.95),legend.title=element_blank(),legend.text = element_text(size = 20, face = "bold",family = "arial"),
axis.text = element_text(size = 15,face = "bold",family = "arial",colour = "black") ) +
scale_x_continuous(limits = c(0,5000))+scale_y_continuous(limits = c(0,4e6))
inputCount <- as.data.frame(RADAR$reads[,1:length(RADAR$samplenames)])
logInputCount <- as.data.frame(log( RADAR$geneSum ) )
ggplot(logInputCount, aes(x = `Ctl18` , y = `T2D6` ))+geom_point()+geom_abline(intercept = 0,slope = 1, lty = 2, colour = "red")+xlab(paste("Log counts Ctl18") )+ylab(paste("Log counts T2D6") )+theme_classic(base_line_size = 0.8)+theme(axis.title = element_text(size = 12,face = "bold"), axis.text = element_text(size = 10,face = "bold",colour = "black") )
ggplot(logInputCount, aes(x = `Ctl18` , y = `T2D5` ))+geom_point()+geom_abline(intercept = 0,slope = 1, lty = 2, colour = "red")+xlab(paste("Log counts Ctl18") )+ylab(paste("Log counts T2D5") )+theme_classic(base_line_size = 0.8)+theme(axis.title = element_text(size = 12,face = "bold"), axis.text = element_text(size = 10,face = "bold",colour = "black") )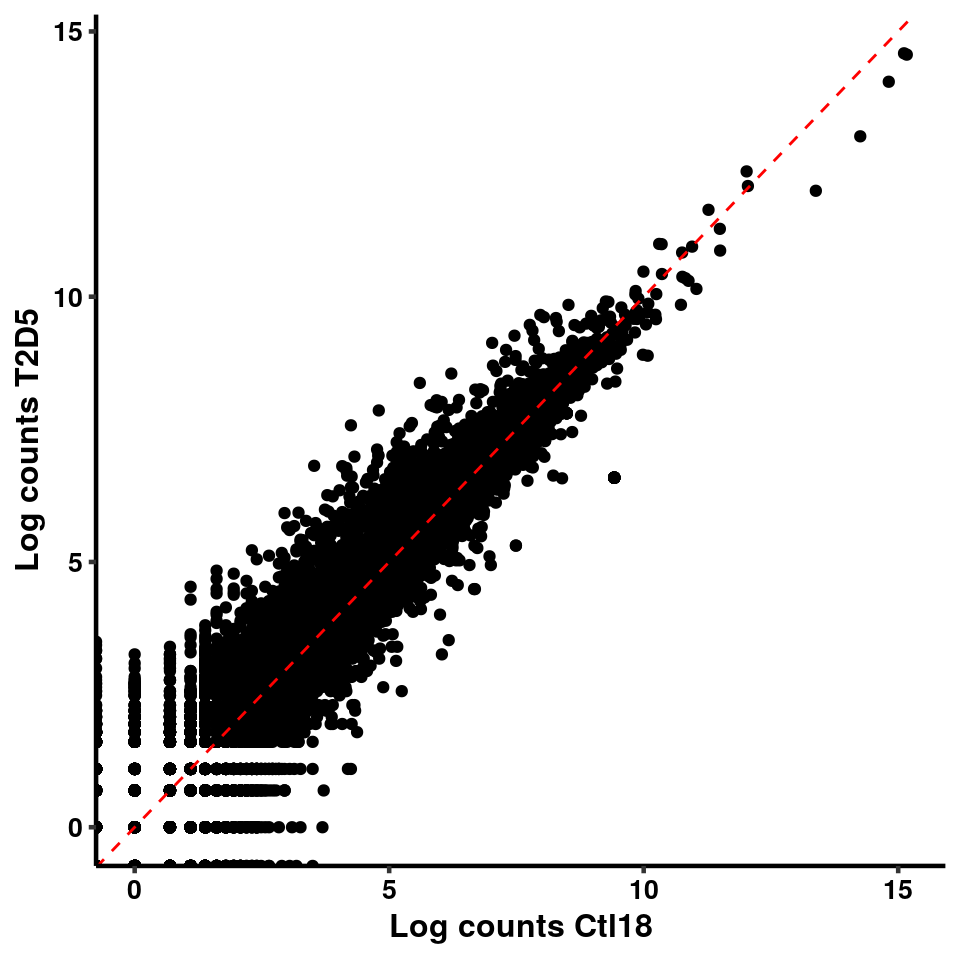
PCA analysis
After geting the filtered and normalized IP read count for testing, we can use PCA analysis to check unkown variation.
top_bins <- RADAR$ip_adjExpr_filtered[order(rowMeans(RADAR$ip_adjExpr_filtered))[1:15000],]We plot PCA colored by Ctl vs T2D
plotPCAfromMatrix(top_bins,group = X)+scale_color_discrete(name = "Group")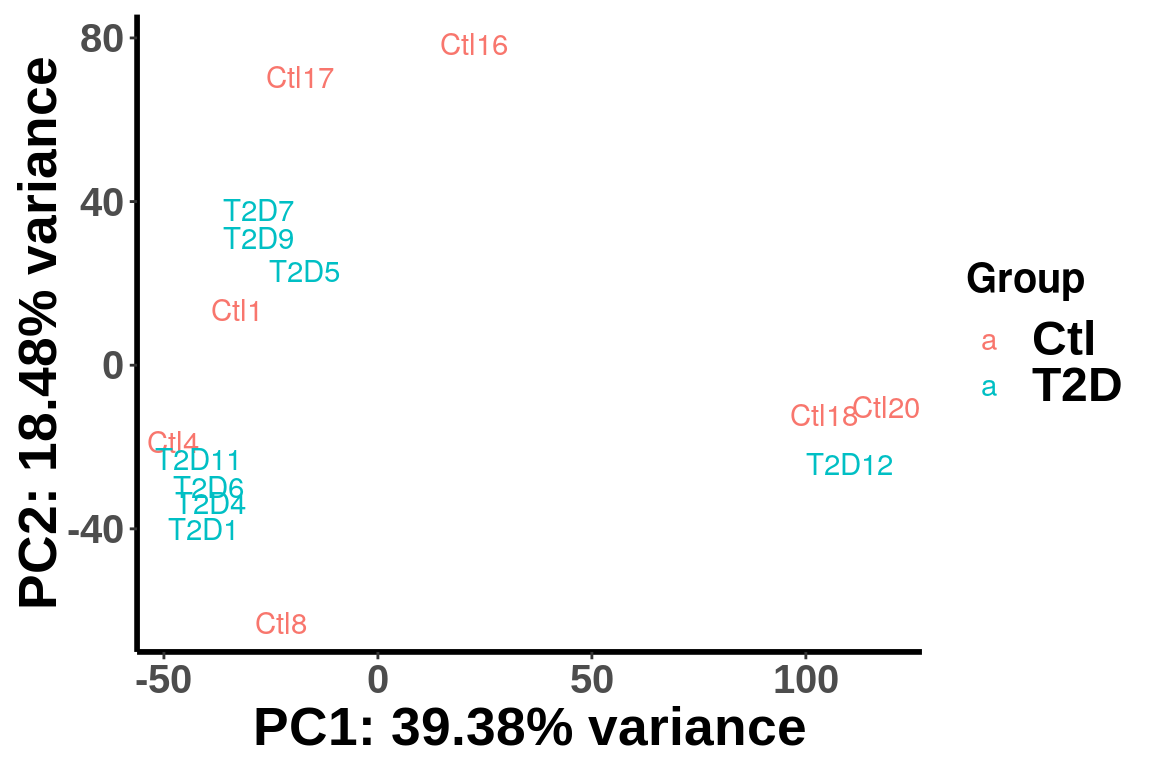
(Norm by position)
plotPCAfromMatrix(RADAR_pos$ip_adjExpr_filtered[order(rowMeans(RADAR_pos$ip_adjExpr_filtered))[1:15000],],group = X )+scale_color_discrete(name = "Group")
To check if samples are separated by different sequencing lanes, label the samples by lane batch.
batch <- c("A","A","A","B","B","C","C","A","A","B","A","B","B","A","C")
plotPCAfromMatrix(top_bins,group = paste0(batch))+scale_color_discrete(name = "Batch")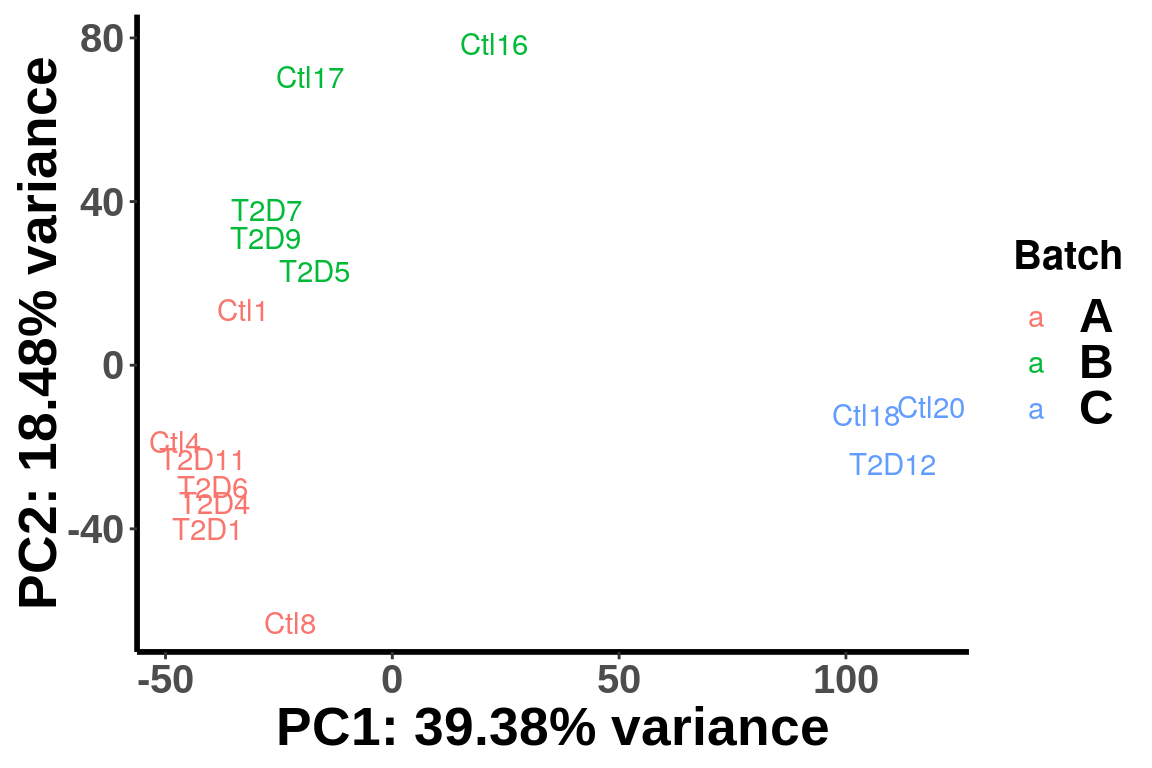
We can see that the samples are clearly separated by batches and we need to account for batch effect in the test. Thus we incorporate batch and gender as known covariates in the test.
Check the PCA after r egressing out the batch and gender.
library(rafalib)
X2 <- as.fumeric(c("A","A","A","B","B","A","A","A","A","B","A","B","B","A","A"))-1 # batch as covariates
X3 <- as.fumeric(c("A","A","A","A","A","C","C","A","A","A","A","A","A","A","C"))-1 # batch as covariates
X4 <- as.fumeric(c("M","F","M","M","M","F","M","M","M","F","M","M","M","M","F"))-1 # sex as covariates
registerDoParallel(cores = 10)
m6A.cov.out <- foreach(i = 1:nrow(top_bins), .combine = rbind) %dopar% {
Y = top_bins[i,]
resi <- residuals( lm(log(Y+1) ~ X2 + X3 + X4 ) )
resi
}
rm(list=ls(name=foreach:::.foreachGlobals), pos=foreach:::.foreachGlobals)
rownames(m6A.cov.out) <- rownames(top_bins)plotPCAfromMatrix(m6A.cov.out,X,loglink = FALSE)+scale_color_discrete(name = "Group")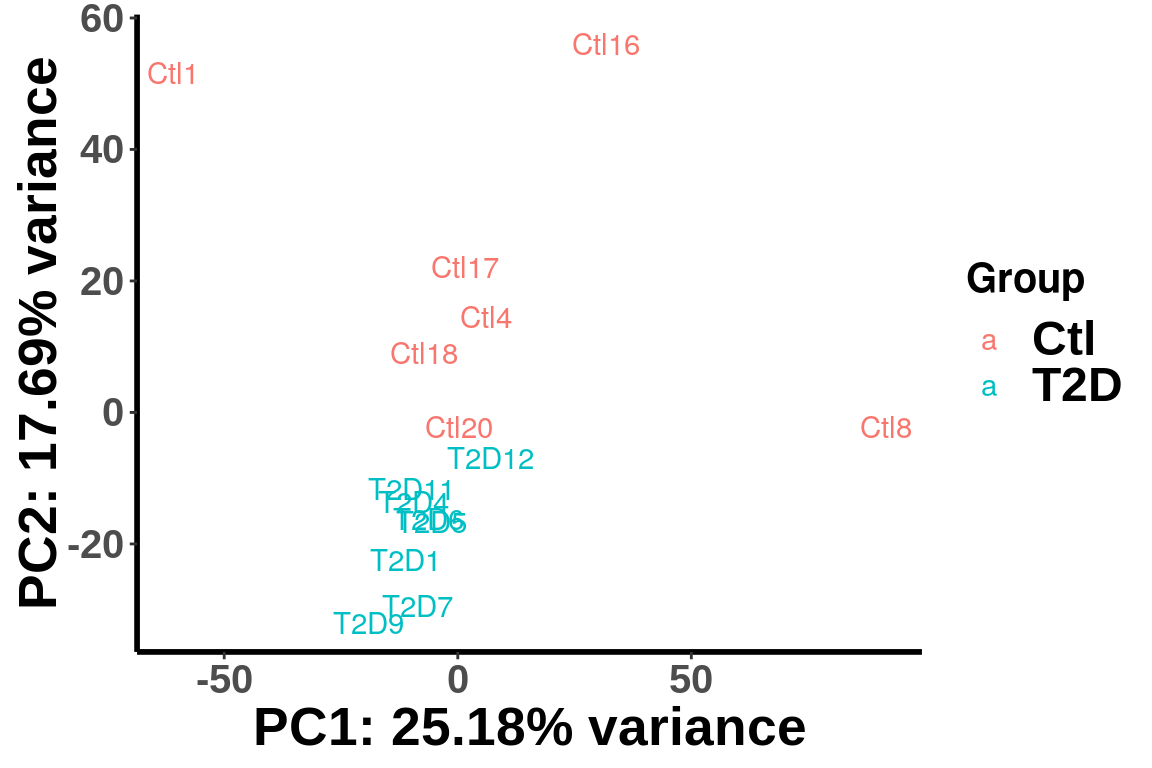
After regressing out the covariates, the samples are separated by diseases status in the PCA plot.
Run PoissonGamma test
cov <- cbind(X2,X3,X4)
RADAR <- diffIP_parallel(RADAR,Covariates = cov,thread = 20)
RADAR_pos <- diffIP_parallel(RADAR_pos[1:14],thread = 25, Covariates = cov,plotPvalue = F)Other method
In order to compare performance of other method on this data set, we run other three methods on default mode on this dataset.
deseq2.res <- DESeq2(countdata = RADAR$ip_adjExpr_filtered,pheno = matrix(c(rep(0,7),rep(1,8)),ncol = 1),covariates = cov)
edgeR.res <- edgeR(countdata = RADAR$ip_adjExpr_filtered,pheno = matrix(c(rep(0,7),rep(1,8)),ncol = 1),covariates = cov)
filteredBins <- rownames(RADAR$all.est)
QNB.res <- QNB::qnbtest(control_ip = RADAR$reads[filteredBins,16:22],
treated_ip = RADAR$reads[filteredBins,23:30],
control_input = RADAR$reads[filteredBins,1:7],
treated_input = RADAR$reads[filteredBins,8:15],plot.dispersion = FALSE)
deseq2.res_pos <- DESeq2(countdata = RADAR_pos$ip_adjExpr_filtered,pheno = matrix(X,ncol = 1),covariates = cov )
edgeR.res_pos <- edgeR(countdata = RADAR_pos$ip_adjExpr_filtered,pheno = matrix(X,ncol = 1),covariates = cov )Compare distribution of p value.
pvalues <- data.frame(pvalue = c(RADAR$all.est[,"p_value3"],deseq2.res$pvalue,edgeR.res$pvalue,QNB.res$pvalue),
method = factor(rep(c("RADAR","DESeq2","edgeR","QNB"),c(length(RADAR$all.est[,"p_value3"]),
length(deseq2.res$pvalue),
length(edgeR.res$pvalue),
length(QNB.res$pvalue)
)
),levels =c("RADAR","DESeq2","edgeR","QNB")
)
)
ggplot(pvalues, aes(x = pvalue))+geom_histogram(breaks = seq(0,1,0.04),col=I("black"))+facet_grid(.~method)+theme_bw()+xlab("p-value")+theme( axis.title = element_text(size = 22, face = "bold"),strip.text = element_text(size = 25, face = "bold"),axis.text.x = element_text(size = 18, face = "bold",colour = "black"),axis.text.y = element_text(size = 15, face = "bold",colour = "black",angle = 90),panel.grid = element_blank(), axis.line = element_line(size = 0.7 ,colour = "black"),axis.ticks = element_line(colour = "black"), panel.spacing = unit(0.4, "lines") )+ scale_x_continuous(breaks = seq(0,1,0.2),labels=function(x) sprintf("%.1f", x))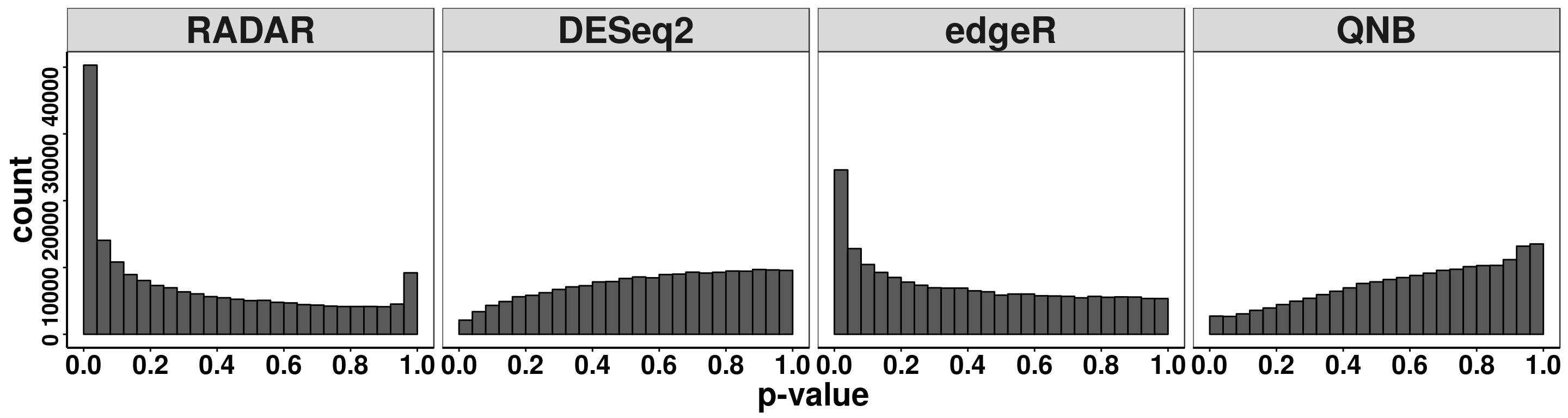
Compare the distibution of beta
par(mfrow = c(1,4))
hist(RADAR$all.est[,"beta1"],main = "RADAR",xlab = "beta",breaks = 50,col =rgb(0.2,0.2,0.2,0.5),cex.main = 2.5,cex.axis =2,cex.lab=2)
hist(deseq2.res$log2FC, main = "DESeq2",xlab = "beta",breaks = 100,col =rgb(0.2,0.2,0.2,0.5),cex.main = 2.5,cex.axis =2,cex.lab=2)
hist(edgeR.res$log2FC,main = "edgeR", xlab = "beta",breaks = 50,col =rgb(0.2,0.2,0.2,0.5),cex.main = 2.5,cex.axis =2,cex.lab=2)
hist(QNB.res$log2.OR, main = "QNB",xlab = "beta",breaks = 50,col =rgb(0.2,0.2,0.2,0.5),cex.main = 2.5,cex.axis =2,cex.lab=2)
Compare norm by geneSum with norm by position
par(mfrow = c(1,3))
tmp <- hist(c(RADAR_pos$all.est[,"p_value3"]),plot = F)
tmp$counts <- tmp$counts*(nrow(RADAR$ip_adjExpr_filtered)/nrow(RADAR_pos$ip_adjExpr_filtered))
hist(RADAR$all.est[,"p_value3"],main = "RADAR",xlab = "p value",col =rgb(1,0,0,0.4),cex.main = 2.5,cex.axis =2,cex.lab=2, ylim = c(0,max(hist(RADAR$all.est[,"p_value3"],plot = F)$counts,tmp$counts)) )
plot(tmp,col=rgb(0,0,1,0.5),add = T)
axis(side = 1, lwd = 1,cex.axis =2)
axis(side = 2, lwd = 1,cex.axis =2)
legend("topright", c("By geneSum", "By position"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 10,bty="n",text.font = 2)
y.scale <- max(hist(RADAR$all.est[,"p_value3"],plot = F)$counts)
tmp <- hist(c(deseq2.res_pos$pvalue),plot = F)
tmp$counts <- tmp$counts*(nrow(RADAR$ip_adjExpr_filtered)/nrow(RADAR_pos$ip_adjExpr_filtered))
hist(deseq2.res$pvalue,main = "DESeq2",xlab = "p value",col =rgb(1,0,0,0.4),cex.main = 2.5,cex.axis =2,cex.lab=2, ylim = c(0,y.scale) )
plot(tmp,col=rgb(0,0,1,0.5),add = T)
axis(side = 1, lwd = 1,cex.axis =2)
axis(side = 2, lwd = 1,cex.axis =2)
legend("topright", c("By geneSum", "By position"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 10,bty="n",text.font = 2)
tmp <- hist(c(edgeR.res_pos$pvalue),plot = F)
tmp$counts <- tmp$counts*(nrow(RADAR$ip_adjExpr_filtered)/nrow(RADAR_pos$ip_adjExpr_filtered))
hist(edgeR.res$pvalue,main = "edgeR",xlab = "p value",col =rgb(1,0,0,0.4),cex.main = 2.5,cex.axis =2,cex.lab=2, ylim = c(0,max(hist(edgeR.res$pvalue,plot = F)$counts,tmp$counts,y.scale)) )
plot(tmp,col=rgb(0,0,1,0.5),add = T)
axis(side = 1, lwd = 1,cex.axis =2)
axis(side = 2, lwd = 1,cex.axis =2)
legend("topright", c("By geneSum", "By position"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 10,bty="n",text.font = 2)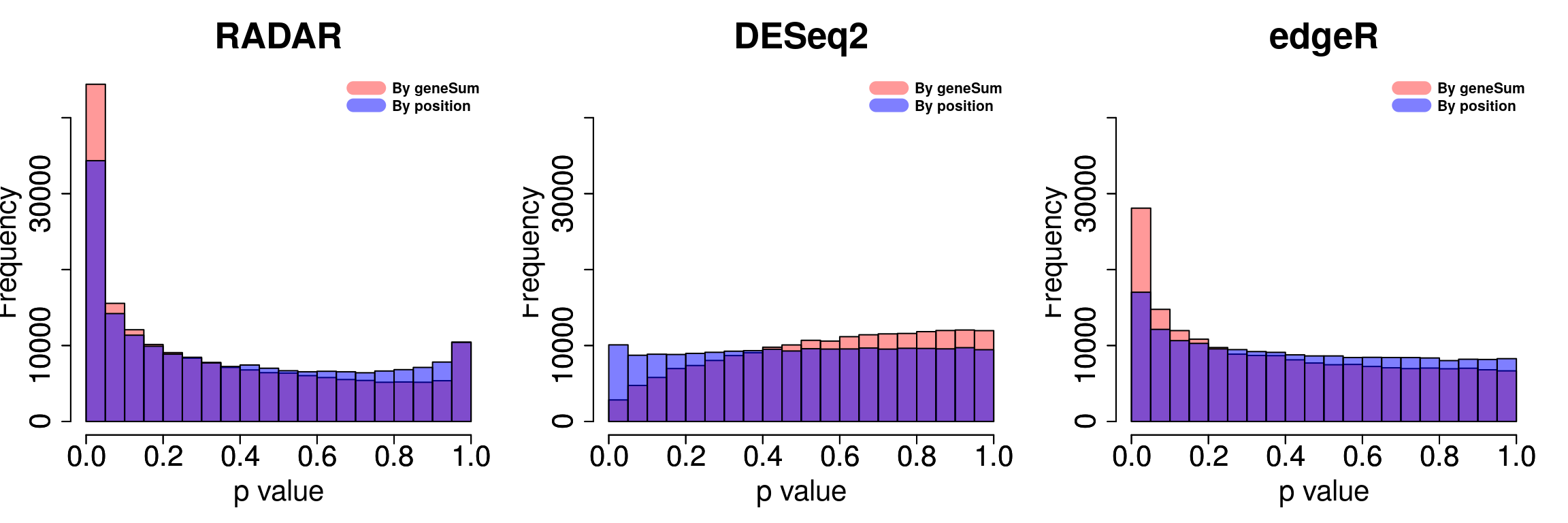
Number of significant bins detected at qvalue < 0.1
sigBins <- apply(cbind("RADAR"=RADAR$all.est[,"p_value3"],"DESeq2"=deseq2.res$pvalue,"edgeR"=edgeR.res$pvalue,"QNB"=QNB.res$pvalue),2, function(x){
length( which( qvalue::qvalue(x)$qvalue < 0.1 ) )
})
print(sigBins)## RADAR DESeq2 edgeR QNB
## 28031 17 7531 27Plot overlap of significant hits
plot.new()
MyTools::venn_diagram4( names(which( qvalue::qvalue(RADAR$all.est[,"p_value3"])$qvalue < 0.1 ) ),
rownames(RADAR$all.est)[which( qvalue::qvalue(deseq2.res$pvalue)$qvalue < 0.1 ) ],
rownames(RADAR$all.est)[which( qvalue::qvalue(edgeR.res$pvalue)$qvalue < 0.1 ) ],
rownames(RADAR$all.est)[which( qvalue::qvalue(QNB.res$pvalue)$qvalue < 0.1 ) ],
"RADAR","DESeq2","edgeR","QNB")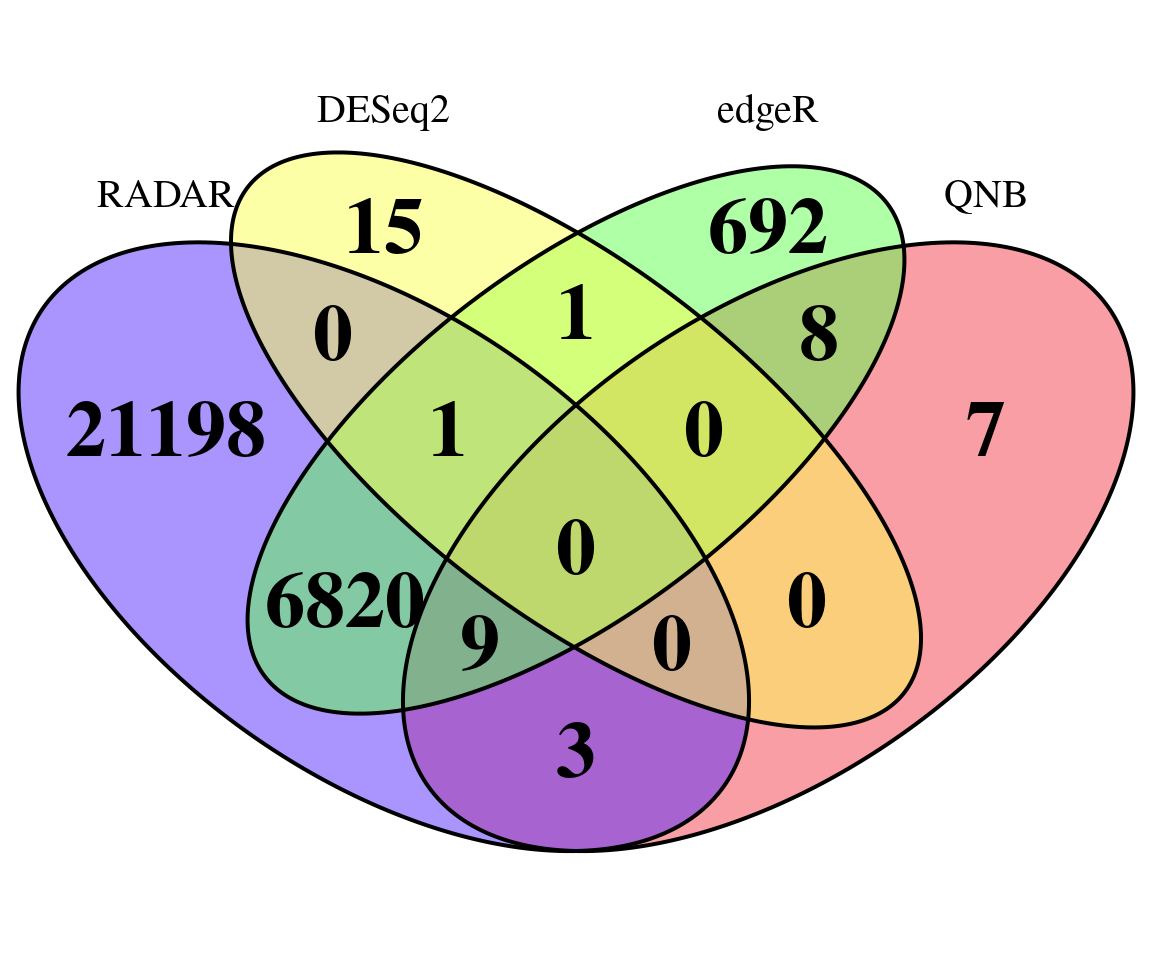
self-FDR test
load("~/PoissonGamma_Method/T2D_data/selfFDR.RData")
ggplot(data = selfFDR, aes(x=method, y= self_FDR, colour = qvalue_cutoff))+geom_boxplot()+theme_bw() +xlab("Method")+ylab("Self-FDR")+ theme(panel.border = element_blank(), panel.grid.major = element_blank(),
panel.grid.minor = element_blank(), axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=20, face="bold", hjust=0.5),
axis.title.y=element_text(size=20, face="bold", vjust=0.4, angle=90),
legend.title=element_text(size = 15,face = "bold"),legend.text = element_text(size = 18, face = "bold",family = "arial"),
axis.text = element_text(size = 15,face = "bold",colour = "black") , axis.ticks = element_line(colour = "black",size = 1) )+scale_color_discrete(name = "qvalue\ncutoff")
selfFDR_pos <- NULL
samplenames <- RADAR$samplenames
set.seed(3)
for(i in 1:20){
## Sample subset
sample_id <- c(sample(x = 1:7, size = 5,replace = F),7+sample(x = 1:8, size = 6,replace = F) )
Truth_id <- setdiff(1:12,sample_id)
if(length(unique(X2[sample_id])) == 1 ){ ## samples all from one batch
## run radar
radar_sample <- diffIP_parallel(RADAR_pos[1:14] ,Covariates = cov[,2:3],exclude = samplenames[setdiff(1:15,sample_id)],thread = 15,plotPvalue = F)$all.est
radar_sample <- cbind(radar_sample, "qvalue"= qvalue::qvalue(radar_sample[,"p_value3"])$qvalue )
radar_truth <- RADAR_pos$all.est
radar_truth <- cbind(radar_truth, "qvalue"= qvalue::qvalue(radar_truth[,"p_value3"])$qvalue )
radar_selfFDR <- sapply(c(0.1,0.05,0.01),function(x){length( which(! which(radar_sample[,"qvalue"]<x) %in% which(radar_truth[,"qvalue"]<0.1) ) )/length(which(radar_sample[,"qvalue"]<x))})
## run DESeq2
deseq_sample <- DESeq2(countdata = RADAR_pos$ip_adjExpr_filtered[,sample_id],pheno = matrix(c(rep(0,7),rep(1,8))[sample_id],ncol = 1),covariates = cov[sample_id,2:3] )
deseq_sample$qvalue <- qvalue::qvalue( deseq_sample$pvalue )$qvalue
deseq_truth <- deseq2.res_pos
deseq_truth$qvalue <- qvalue::qvalue( deseq_truth$pvalue )$qvalue
deseq_selfFDR <- sapply(c(0.1,0.05,0.01),function(x){length( which(! which(deseq_sample[,"qvalue"]<x) %in% which(deseq_truth[,"qvalue"]<0.1) ) )/length(which(deseq_sample[,"qvalue"]<x))})
## run edgeR
edgeR_sample <- edgeR(countdata = RADAR_pos$ip_adjExpr_filtered[,sample_id],pheno = matrix(c(rep(0,7),rep(1,8))[sample_id],ncol = 1),covariates = cov[sample_id,2:3] )
edgeR_sample$qvalue <- qvalue::qvalue( edgeR_sample$pvalue )$qvalue
edgeR_truth <- edgeR.res_pos
edgeR_truth$qvalue <- qvalue::qvalue( edgeR_truth$pvalue )$qvalue
edgeR_selfFDR <- sapply(c(0.1,0.05,0.01),function(x){length( which(! which(edgeR_sample[,"qvalue"]<x) %in% which(edgeR_truth[,"qvalue"]<0.1) ) )/length(which(edgeR_sample[,"qvalue"]<x))})
fdr <- rbind( data.frame(self_FDR = radar_selfFDR, qvalue_cutoff=c("0.1","0.05","0.01"), method = rep("RADAR",3) ),
data.frame(self_FDR = deseq_selfFDR, qvalue_cutoff=c("0.1","0.05","0.01"), method = rep("DESeq2",3) ),
data.frame(self_FDR = edgeR_selfFDR, qvalue_cutoff=c("0.1","0.05","0.01"), method = rep("edgeR",3) )
)
selfFDR_pos <- rbind(selfFDR_pos,fdr)
}else if(length(unique(X3[sample_id])) == 1 ){
## run radar
radar_sample <- diffIP_parallel(RADAR_pos[1:14] ,Covariates = cov[,c(1,3)],exclude = samplenames[setdiff(1:15,sample_id)],thread = 15,plotPvalue = F)$all.est
radar_sample <- cbind(radar_sample, "qvalue"= qvalue::qvalue(radar_sample[,"p_value3"])$qvalue )
radar_truth <- RADAR_pos$all.est
radar_truth <- cbind(radar_truth, "qvalue"= qvalue::qvalue(radar_truth[,"p_value3"])$qvalue )
radar_selfFDR <- sapply(c(0.1,0.05,0.01),function(x){length( which(! which(radar_sample[,"qvalue"]<x) %in% which(radar_truth[,"qvalue"]<0.1) ) )/length(which(radar_sample[,"qvalue"]<x))})
## run DESeq2
deseq_sample <- DESeq2(countdata = RADAR_pos$ip_adjExpr_filtered[,sample_id],pheno = matrix(c(rep(0,7),rep(1,8))[sample_id],ncol = 1),covariates = cov[sample_id,c(1,3)] )
deseq_sample$qvalue <- qvalue::qvalue( deseq_sample$pvalue )$qvalue
deseq_truth <- deseq2.res_pos
deseq_truth$qvalue <- qvalue::qvalue( deseq_truth$pvalue )$qvalue
deseq_selfFDR <- sapply(c(0.1,0.05,0.01),function(x){length( which(! which(deseq_sample[,"qvalue"]<x) %in% which(deseq_truth[,"qvalue"]<0.1) ) )/length(which(deseq_sample[,"qvalue"]<x))})
## run edgeR
edgeR_sample <- edgeR(countdata = RADAR_pos$ip_adjExpr_filtered[,sample_id],pheno = matrix(c(rep(0,7),rep(1,8))[sample_id],ncol = 1),covariates = cov[sample_id,c(1,3)] )
edgeR_sample$qvalue <- qvalue::qvalue( edgeR_sample$pvalue )$qvalue
edgeR_truth <- edgeR.res_pos
edgeR_truth$qvalue <- qvalue::qvalue( edgeR_truth$pvalue )$qvalue
edgeR_selfFDR <- sapply(c(0.1,0.05,0.01),function(x){length( which(! which(edgeR_sample[,"qvalue"]<x) %in% which(edgeR_truth[,"qvalue"]<0.1) ) )/length(which(edgeR_sample[,"qvalue"]<x))})
fdr <- rbind( data.frame(self_FDR = radar_selfFDR, qvalue_cutoff=c("0.1","0.05","0.01"), method = rep("RADAR",3) ),
data.frame(self_FDR = deseq_selfFDR, qvalue_cutoff=c("0.1","0.05","0.01"), method = rep("DESeq2",3) ),
data.frame(self_FDR = edgeR_selfFDR, qvalue_cutoff=c("0.1","0.05","0.01"), method = rep("edgeR",3) )
)
selfFDR_pos <- rbind(selfFDR_pos,fdr)
}else if(length(unique(X4[sample_id])) == 1){
## run radar
radar_sample <- diffIP_parallel(RADAR_pos[1:14] ,Covariates = cov[,c(1,2)],exclude = samplenames[setdiff(1:15,sample_id)],thread = 15,plotPvalue = F)$all.est
radar_sample <- cbind(radar_sample, "qvalue"= qvalue::qvalue(radar_sample[,"p_value3"])$qvalue )
radar_truth <- RADAR_pos$all.est
radar_truth <- cbind(radar_truth, "qvalue"= qvalue::qvalue(radar_truth[,"p_value3"])$qvalue )
radar_selfFDR <- sapply(c(0.1,0.05,0.01),function(x){length( which(! which(radar_sample[,"qvalue"]<x) %in% which(radar_truth[,"qvalue"]<0.1) ) )/length(which(radar_sample[,"qvalue"]<x))})
## run DESeq2
deseq_sample <- DESeq2(countdata = RADAR_pos$ip_adjExpr_filtered[,sample_id],pheno = matrix(c(rep(0,7),rep(1,8))[sample_id],ncol = 1),covariates = cov[sample_id,c(1,2)] )
deseq_sample$qvalue <- qvalue::qvalue( deseq_sample$pvalue )$qvalue
deseq_truth <- deseq2.res_pos
deseq_truth$qvalue <- qvalue::qvalue( deseq_truth$pvalue )$qvalue
deseq_selfFDR <- sapply(c(0.1,0.05,0.01),function(x){length( which(! which(deseq_sample[,"qvalue"]<x) %in% which(deseq_truth[,"qvalue"]<0.1) ) )/length(which(deseq_sample[,"qvalue"]<x))})
## run edgeR
edgeR_sample <- edgeR(countdata = RADAR_pos$ip_adjExpr_filtered[,sample_id],pheno = matrix(c(rep(0,7),rep(1,8))[sample_id],ncol = 1),covariates = cov[sample_id,c(1,2)] )
edgeR_sample$qvalue <- qvalue::qvalue( edgeR_sample$pvalue )$qvalue
edgeR_truth <- edgeR.res_pos
edgeR_truth$qvalue <- qvalue::qvalue( edgeR_truth$pvalue )$qvalue
edgeR_selfFDR <- sapply(c(0.1,0.05,0.01),function(x){length( which(! which(edgeR_sample[,"qvalue"]<x) %in% which(edgeR_truth[,"qvalue"]<0.1) ) )/length(which(edgeR_sample[,"qvalue"]<x))})
fdr <- rbind( data.frame(self_FDR = radar_selfFDR, qvalue_cuttoff=c("0.1","0.05","0.01"), method = rep("RADAR",3) ),
data.frame(self_FDR = deseq_selfFDR, qvalue_cuttoff=c("0.1","0.05","0.01"), method = rep("DESeq2",3) ),
data.frame(self_FDR = edgeR_selfFDR, qvalue_cuttoff=c("0.1","0.05","0.01"), method = rep("edgeR",3) )
)
selfFDR_pos <- rbind(selfFDR_pos,fdr)
}else{
## run radar
radar_sample <- diffIP_parallel(RADAR_pos[1:14],Covariates = cov ,exclude = samplenames[setdiff(1:15,sample_id)],thread = 15,plotPvalue = F)$all.est
radar_sample <- cbind(radar_sample, "qvalue"= qvalue::qvalue(radar_sample[,"p_value3"])$qvalue )
radar_truth <- RADAR_pos$all.est
radar_truth <- cbind(radar_truth, "qvalue"= qvalue::qvalue(radar_truth[,"p_value3"])$qvalue )
radar_selfFDR <- sapply(c(0.1,0.05,0.01),function(x){length( which(! which(radar_sample[,"qvalue"]<x) %in% which(radar_truth[,"qvalue"]<0.1) ) )/length(which(radar_sample[,"qvalue"]<x))})
## run DESeq2
deseq_sample <- DESeq2(countdata = RADAR_pos$ip_adjExpr_filtered[,sample_id],pheno = matrix(c(rep(0,7),rep(1,8))[sample_id],ncol = 1),covariates = cov[sample_id,] )
deseq_sample$qvalue <- qvalue::qvalue( deseq_sample$pvalue )$qvalue
deseq_truth <- deseq2.res_pos
deseq_truth$qvalue <- qvalue::qvalue( deseq_truth$pvalue )$qvalue
deseq_selfFDR <- sapply(c(0.1,0.05,0.01),function(x){length( which(! which(deseq_sample[,"qvalue"]<x) %in% which(deseq_truth[,"qvalue"]<0.1) ) )/length(which(deseq_sample[,"qvalue"]<x))})
## run edgeR
edgeR_sample <- edgeR(countdata = RADAR_pos$ip_adjExpr_filtered[,sample_id],pheno = matrix(c(rep(0,7),rep(1,8))[sample_id],ncol = 1),covariates = cov[sample_id,] )
edgeR_sample$qvalue <- qvalue::qvalue( edgeR_sample$pvalue )$qvalue
edgeR_truth <- edgeR.res_pos
edgeR_truth$qvalue <- qvalue::qvalue( edgeR_truth$pvalue )$qvalue
edgeR_selfFDR <- sapply(c(0.1,0.05,0.01),function(x){length( which(! which(edgeR_sample[,"qvalue"]<x) %in% which(edgeR_truth[,"qvalue"]<0.1) ) )/length(which(edgeR_sample[,"qvalue"]<x))})
fdr <- rbind( data.frame(self_FDR = radar_selfFDR, qvalue_cutoff=c("0.1","0.05","0.01"), method = rep("RADAR",3) ),
data.frame(self_FDR = deseq_selfFDR, qvalue_cutoff=c("0.1","0.05","0.01"), method = rep("DESeq2",3) ),
data.frame(self_FDR = edgeR_selfFDR, qvalue_cutoff=c("0.1","0.05","0.01"), method = rep("edgeR",3) )
)
selfFDR_pos <- rbind(selfFDR_pos,fdr)
}
}
selfFDR_pos$qvalue_cutoff <- factor(selfFDR_pos$qvalue_cutoff,levels = c("0.1","0.05","0.01"))
save(selfFDR_pos, file = "~/PoissonGamma_Method/T2D_data/selfFDR_pos.RData")Self-FDR norm by local bin read count
load("~/PoissonGamma_Method/T2D_data/selfFDR_pos.RData")
selfFDR_all <- cbind(rbind(selfFDR, selfFDR_pos), "NormBy"= c(rep("By gene-level count",nrow(selfFDR)),rep("By bin-level count",nrow(selfFDR_pos))) )
selfFDR_all$NormBy <- factor(selfFDR_all$NormBy,levels = c("By gene-level count","By bin-level count"))
ggplot(data = selfFDR_all, aes(x=method, y= self_FDR, colour = qvalue_cutoff))+geom_boxplot()+facet_grid(~NormBy)+theme_bw() +xlab("Method")+ylab("Self-FDR")+ theme( panel.grid.major = element_blank(),
axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=20, face="bold", hjust=0.5,family = "arial"),
axis.title.y=element_text(size=20, face="bold", vjust=0.4, angle=90,family = "arial"),
legend.title=element_text(size = 15,face = "bold"),legend.text = element_text(size = 18, face = "bold",family = "arial"),
axis.text = element_text(size = 18,face = "bold",family = "arial",colour = "black") ,strip.text.x = element_text(size = 20,face = "bold") )+scale_color_discrete(name = "qvalue\ncutoff")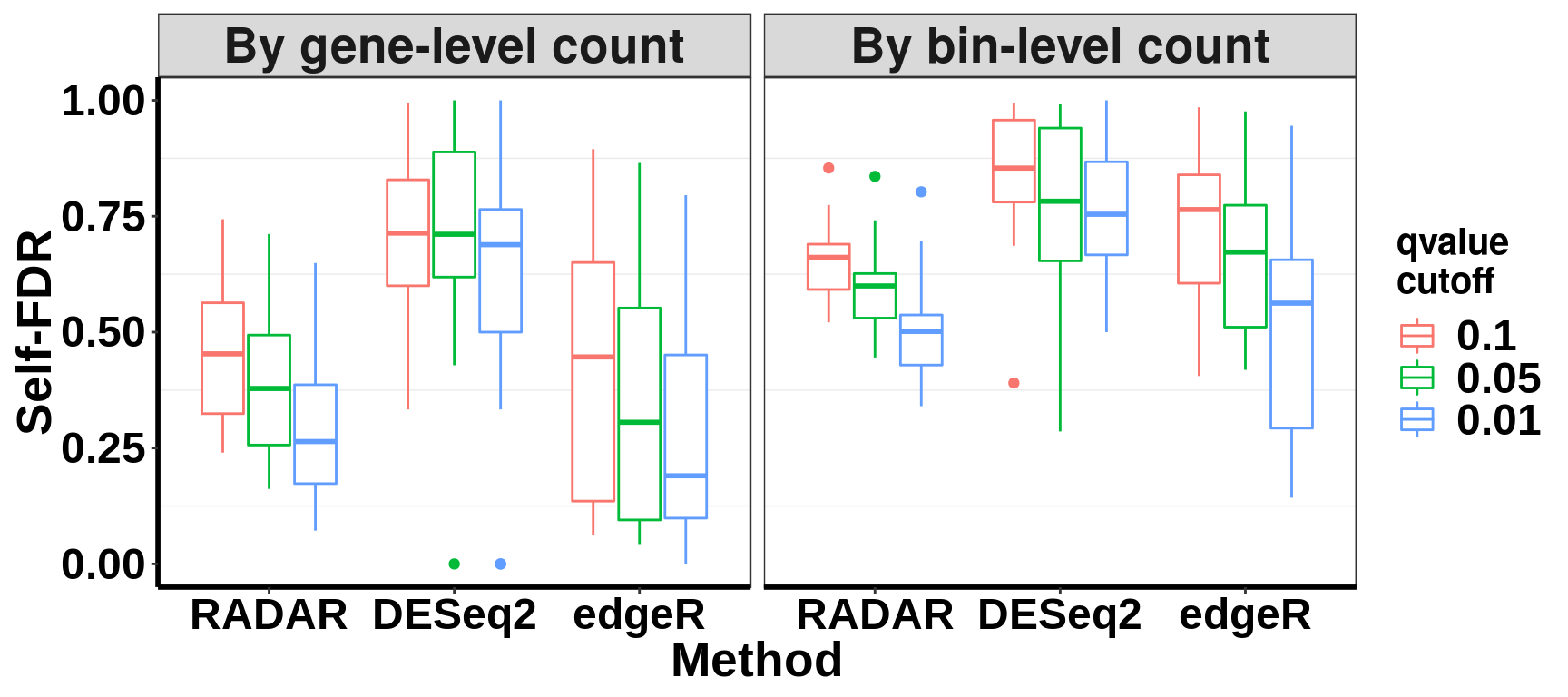
Permution
radar_permuted_P <-NULL
deseq_permuted_P <- NULL
edgeR_permuted_P <- NULL
set.seed(51)
for(i in 1:10){
permute_X <- unlist( tapply(RADAR$X,batch,function(x){sample(x,length(x))}) )[match(RADAR$samplenames,unlist( tapply(RADAR$samplenames,batch,function(x)return(x)) ))]
if ( all( permute_X == RADAR$X) | all( permute_X == c(rep("T2D",8),rep("Ctl",7))) ){
cat("Duplicated with original design...\n")
}else{
radar_permute <- diffIP_parallel(c(RADAR[1:11],list("X"=permute_X),RADAR[13:14]) ,thread = 15,Covariates = cov,plotPvalue = F)$all.est[,"p_value3"]
deseq_permute <- DESeq2(countdata = RADAR$ip_adjExpr_filtered,pheno = matrix(permute_X,ncol = 1),covariates = cov )$pvalue
edgeR_permute <- edgeR(countdata = RADAR$ip_adjExpr_filtered,pheno = matrix(permute_X,ncol = 1),covariates = cov )$pvalue
radar_permuted_P <- cbind(radar_permuted_P, radar_permute)
deseq_permuted_P <- cbind(deseq_permuted_P,deseq_permute)
edgeR_permuted_P <- cbind(edgeR_permuted_P,edgeR_permute)
}
}
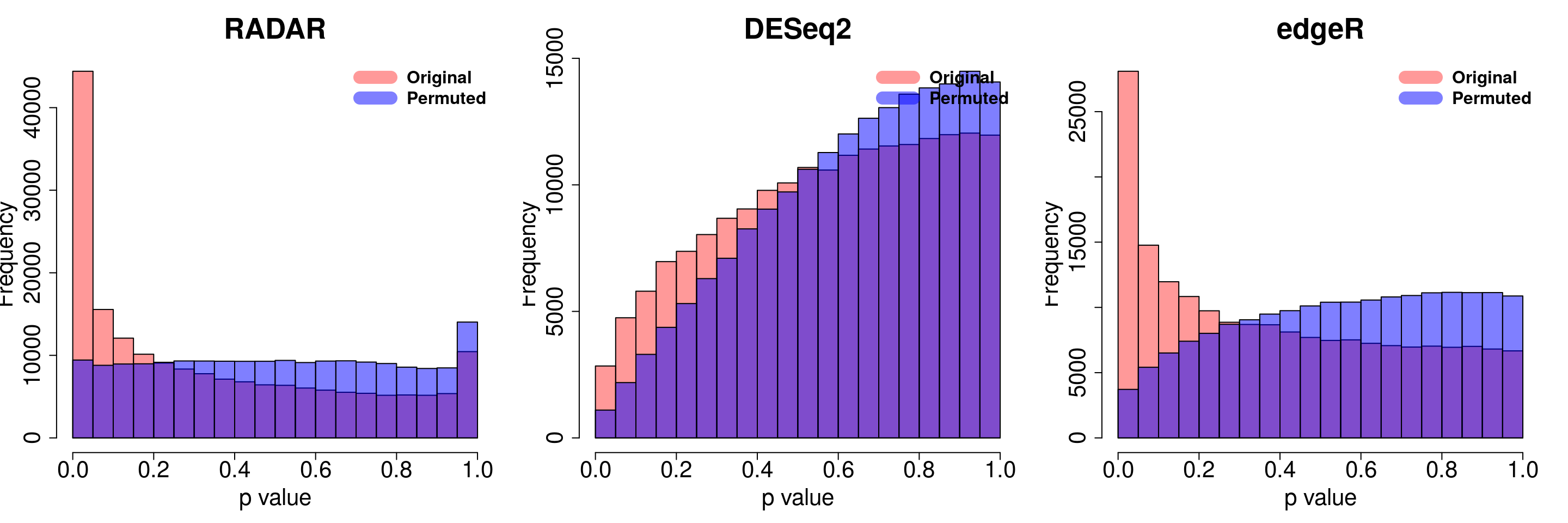
save(radar_permuted_P ,deseq_permuted_P ,edgeR_permuted_P, file= "~/PoissonGamma_Method/T2D_data/permutation.RData")load( "~/PoissonGamma_Method/T2D_data/permutation.RData")
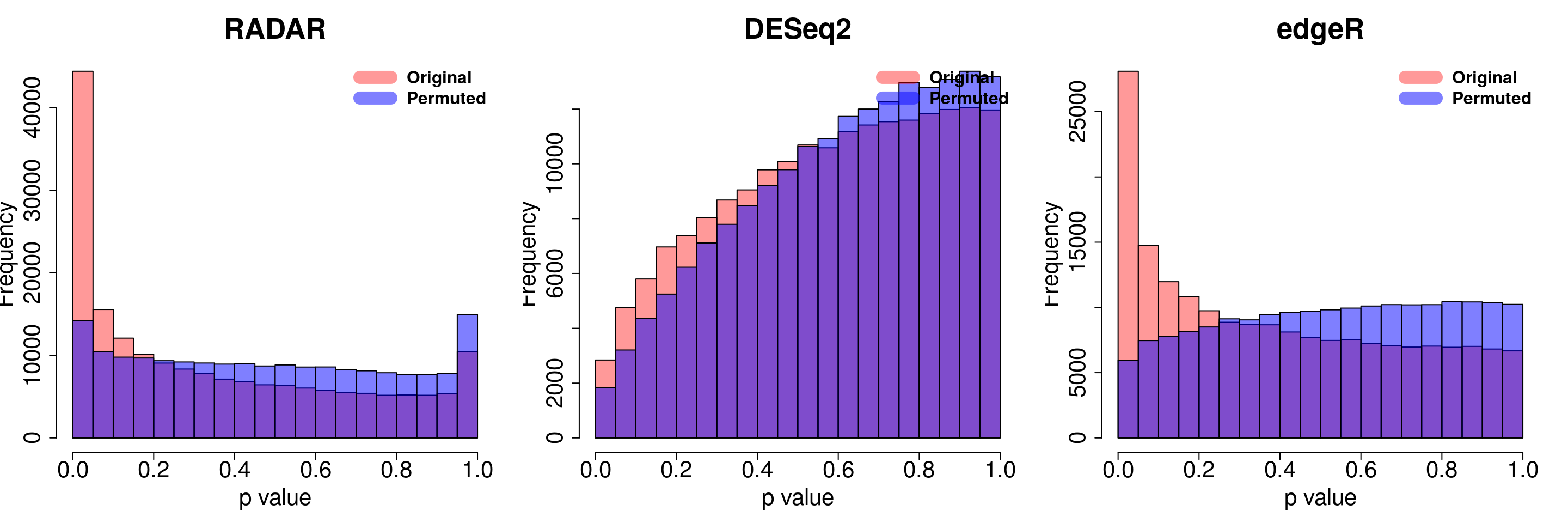
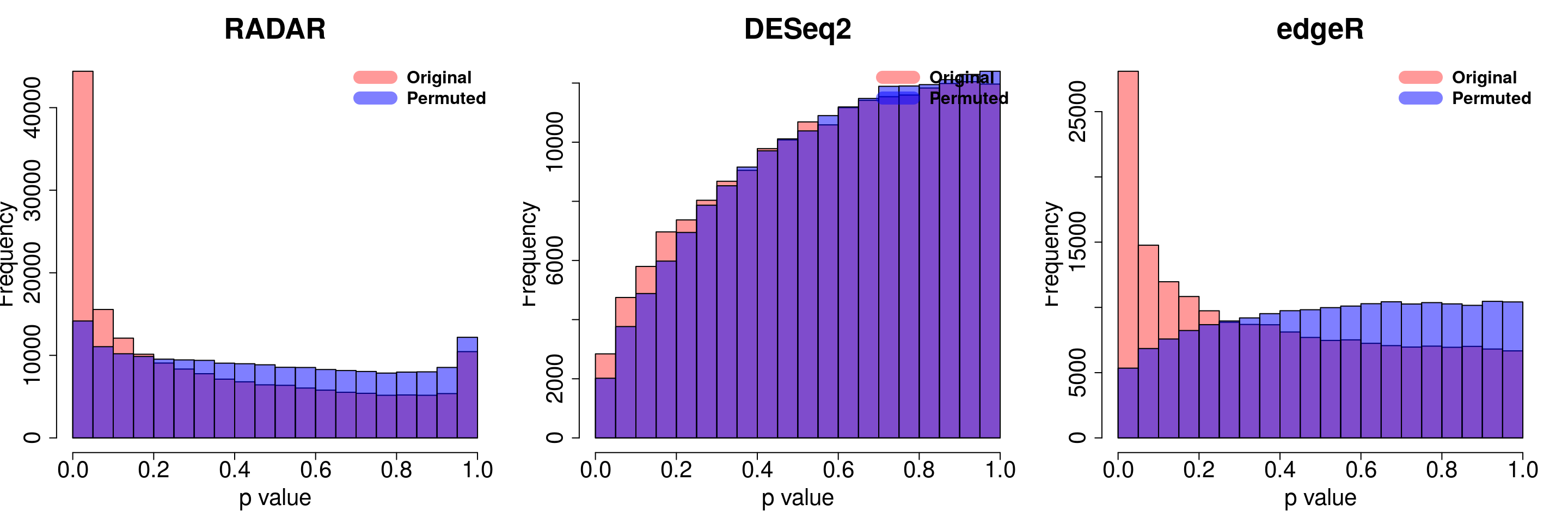
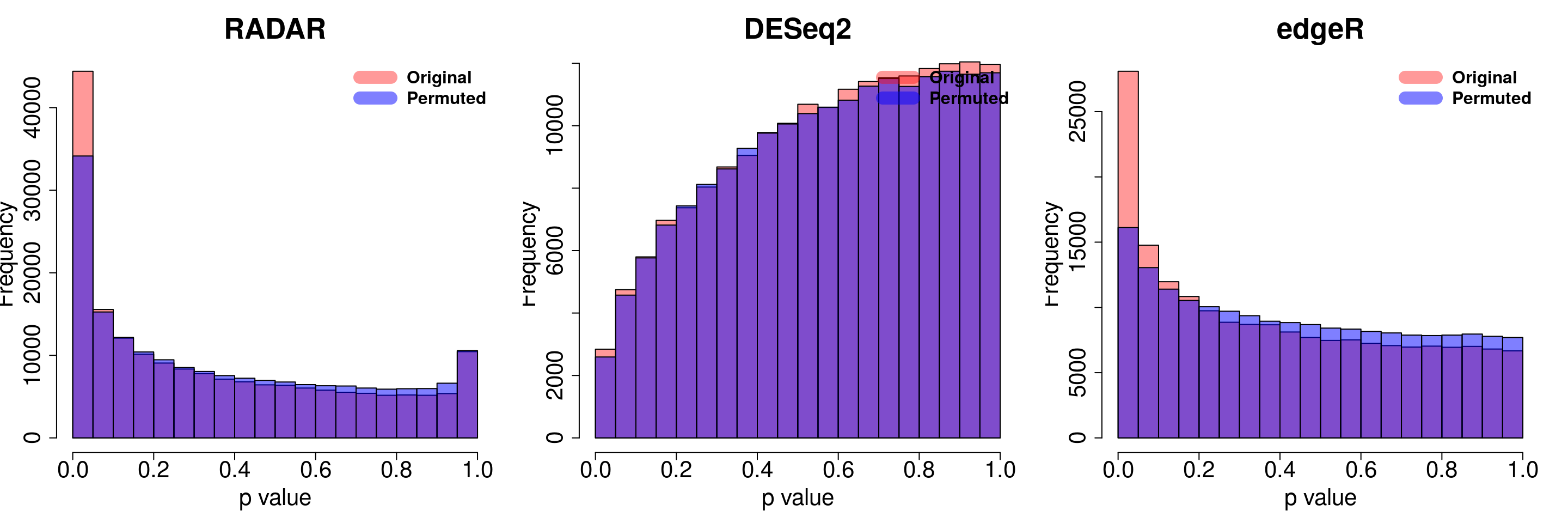
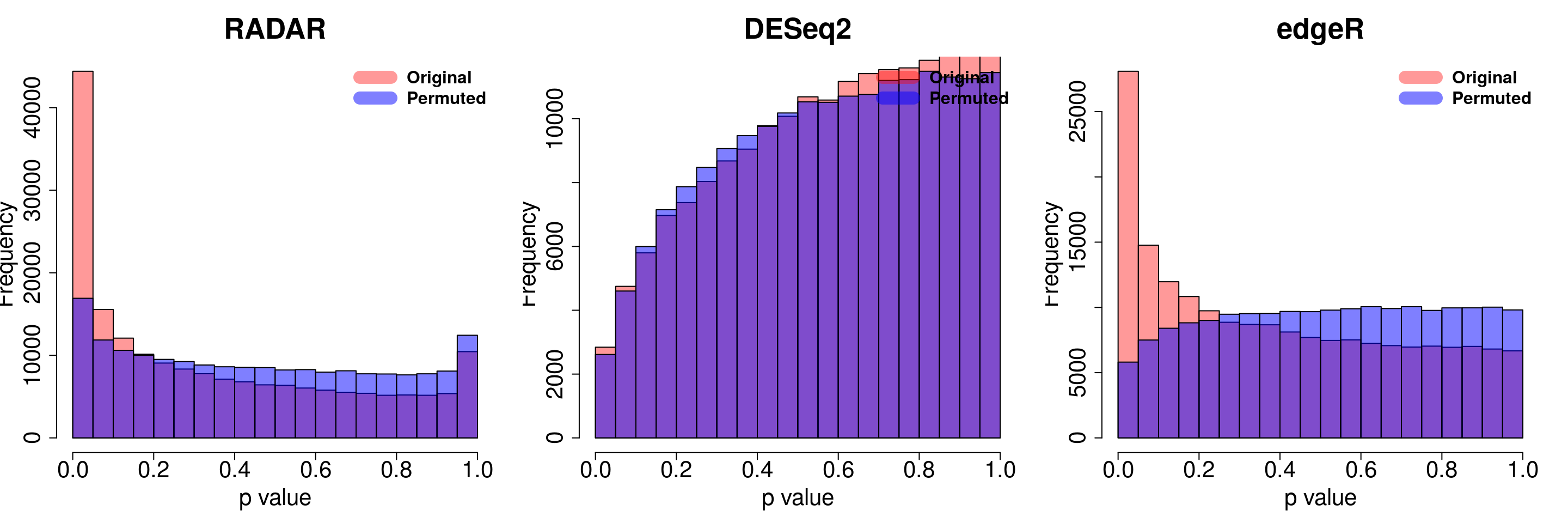
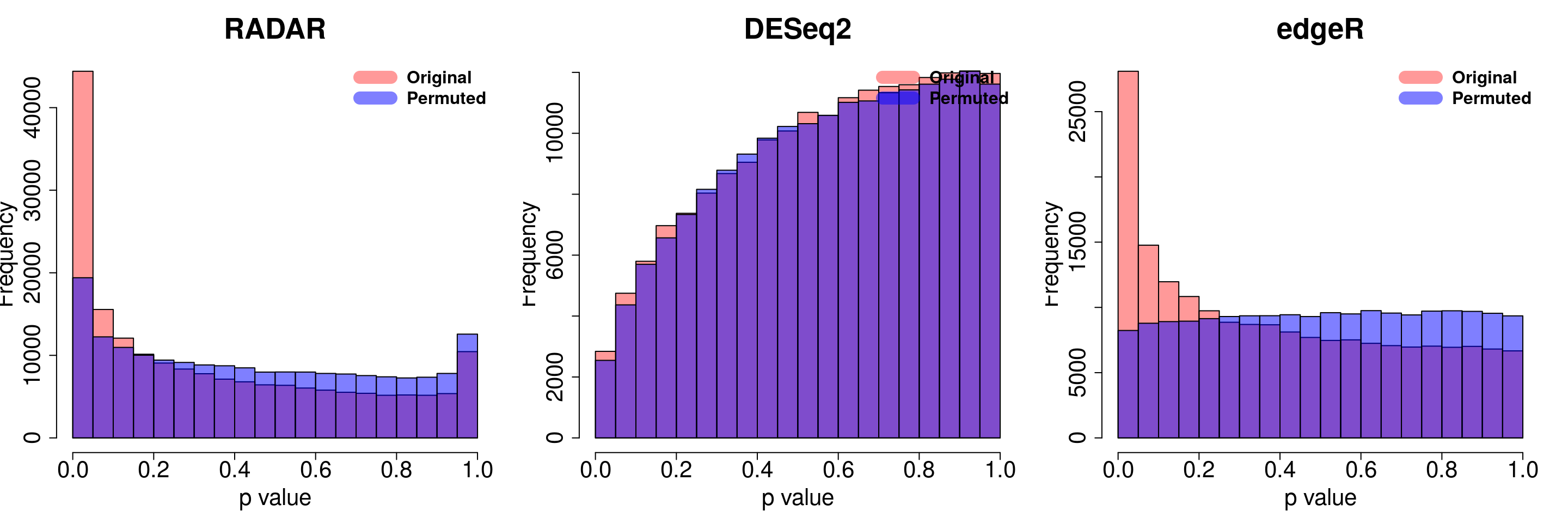
for( i in 1:ncol(radar_permuted_P)){
par(mfrow=c(1,3))
hist(RADAR$all.est[,"p_value3"], col =rgb(1,0,0,0.4),main = "RADAR", xlab = "p value",cex.main = 2.5,cex.axis =2,cex.lab=2)
hist(radar_permuted_P[,i],col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 12,bty="n",text.font = 2, cex = 1.5)
hist(deseq2.res$pvalue, col =rgb(1,0,0,0.4),main = "DESeq2",xlab = "p value",cex.main = 2.5,cex.axis =2,cex.lab=2, ylim =c(0,max(hist(deseq_permuted_P[,i],plot = F)$counts) ) )
hist(deseq_permuted_P[,i],col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 12,bty="n",text.font = 2, cex = 1.5)
hist(edgeR.res$pvalue, col =rgb(1,0,0,0.4),main = "edgeR",xlab = "p value",cex.main = 2.5,cex.axis =2,cex.lab=2)
hist(edgeR_permuted_P[,i],col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 12,bty="n",text.font = 2, cex = 1.5)
}
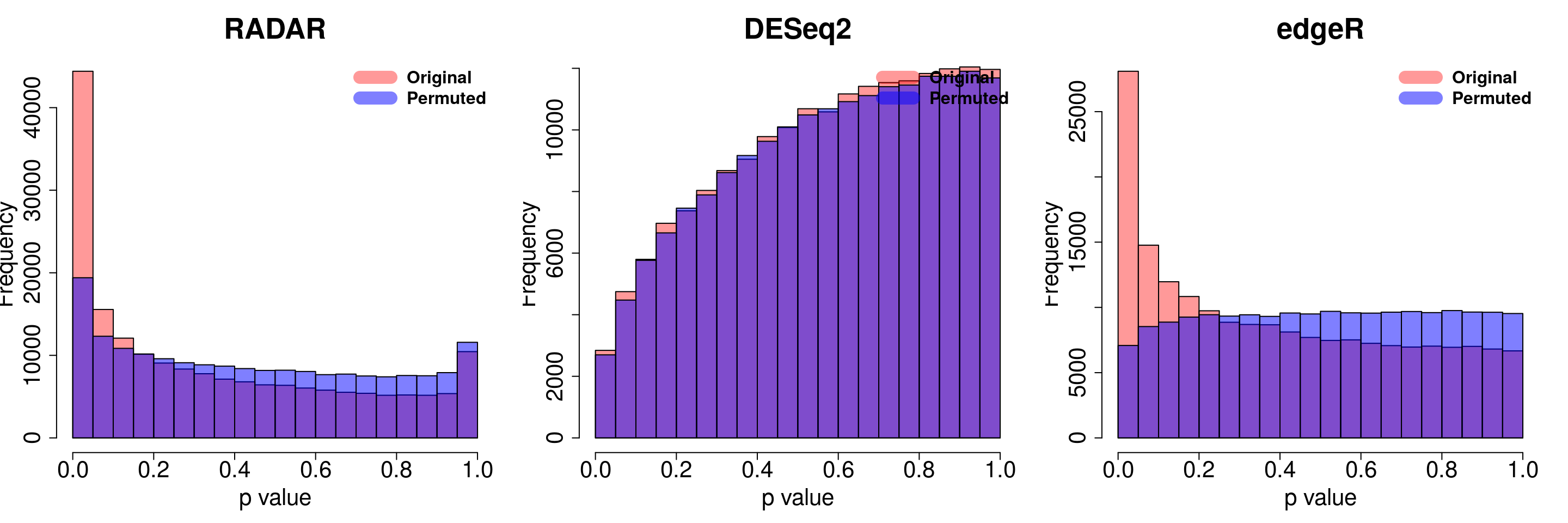


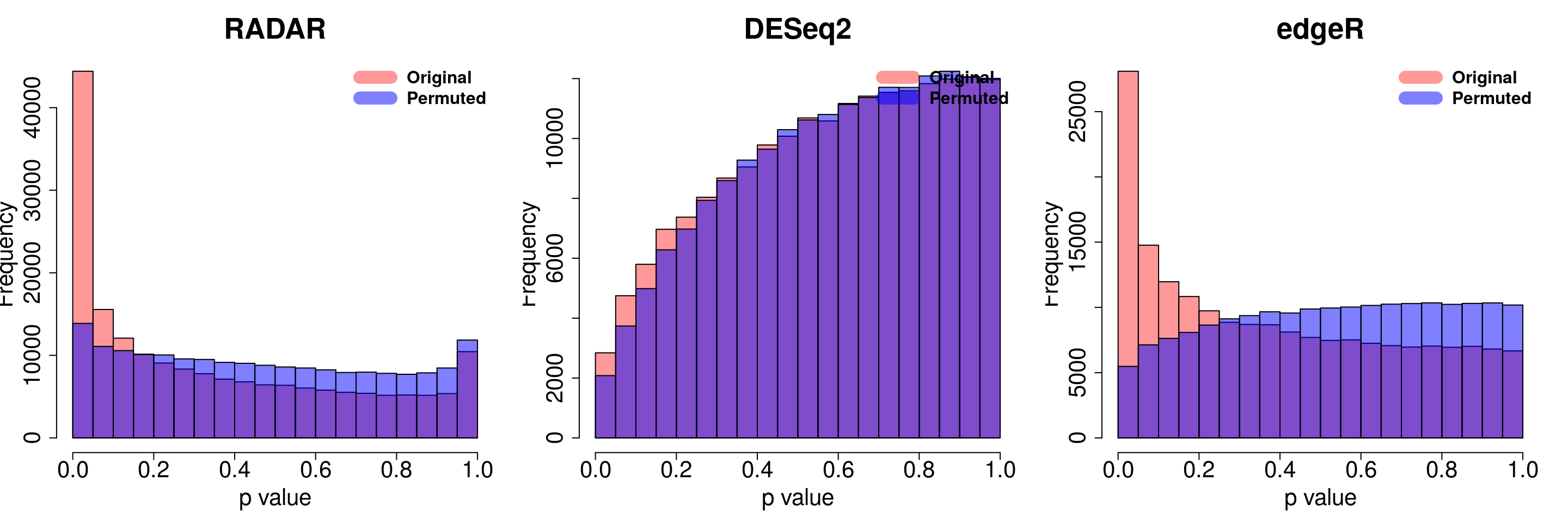
combine all sets of permuted p value in one
par(mfrow=c(1,2))
hist(RADAR$all.est[,"p_value3"], col =rgb(1,0,0,0.4),main = "RADAR", xlab = "p value",font=2, cex.lab=1.5, font.lab=2 ,cex.main = 2,cex.axis =1, lwd = 2.5)
tmp <- hist(c(radar_permuted_P),plot = F)
tmp$counts <- tmp$counts/10
plot(tmp,col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 12,bty="n",text.font = 2, cex = 1)
y.scale <- max(hist(RADAR$all.est[,"p_value3"],plot = F)$counts)
#hist(deseq2.res$pvalue, col =rgb(1,0,0,0.4),main = "DESeq2",xlab = "p value")
#tmp <- hist(c(deseq_permuted_P),plot = F)
#tmp$counts <- tmp$counts/10
#plot(tmp,col=rgb(0,0,1,0.5),add = T)
#legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 10,bty="n")
hist(edgeR.res$pvalue, col =rgb(1,0,0,0.4),main = "edgeR",xlab = "p value",font=2, cex.lab=1.5, font.lab=2 ,cex.main = 2 , cex.axis =1, ylim = c(0,y.scale), lwd = 2.5)
tmp <- hist(c(edgeR_permuted_P),plot = F)
tmp$counts <- tmp$counts/10
plot(tmp,col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 12,bty="n",text.font = 2, cex = 1)
Plot Heat map of significant bins by RADAR
topBins <- RADAR$ip_adjExpr_filtered[which(qvalue(RADAR$all.est[,"p_value3"])$qvalue<0.1),]
log_topBins <- log(topBins+1)
registerDoParallel(cores = 4)
cov.out.RADAR <- foreach(i = 1:nrow(log_topBins), .combine = rbind) %dopar% {
Y = log_topBins[i,]
tmp_data <- as.data.frame(cbind(Y,cov))
resi <- residuals( lm(Y~.,data=tmp_data ) )
resi
}
rm(list=ls(name=foreach:::.foreachGlobals), pos=foreach:::.foreachGlobals)
rownames(cov.out.RADAR) <- rownames(log_topBins)
colnames(cov.out.RADAR) <- colnames(log_topBins)
dist.pear <- function(x) as.dist(1-cor(t(x)))
hclust.ave <- function(x) hclust(x, method="average")
gplots::heatmap.2(cov.out.RADAR,scale="row",trace="none",labRow=NA,main = "Methylation level of significant bins\n(Covariates regressed out)",
distfun=dist.pear, hclustfun=hclust.ave,col=rev(RColorBrewer::brewer.pal(9,"RdBu")))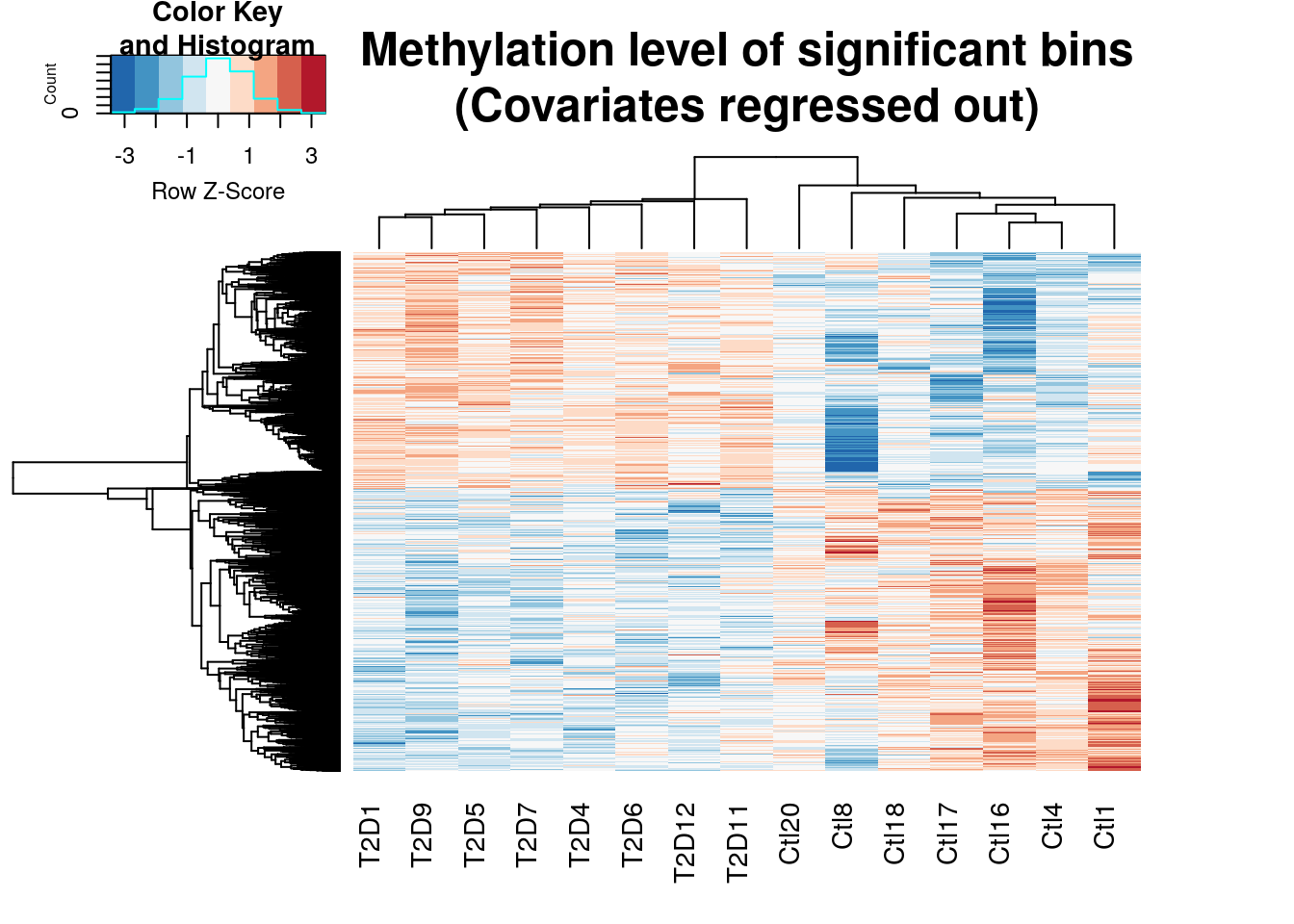
Plot heap map of significant bins by edgeR
topBins <- RADAR$ip_adjExpr_filtered[which(qvalue(edgeR.res$pvalue)$qvalue<0.1),]
log_topBins <- log(topBins+1)
registerDoParallel(cores = 4)
cov.out.edgeR <- foreach(i = 1:nrow(log_topBins), .combine = rbind) %dopar% {
Y = log_topBins[i,]
tmp_data <- as.data.frame(cbind(Y,cov))
resi <- residuals( lm(Y~.,data=tmp_data ) )
resi
}
rm(list=ls(name=foreach:::.foreachGlobals), pos=foreach:::.foreachGlobals)
rownames(cov.out.edgeR) <- rownames(log_topBins)
colnames(cov.out.edgeR) <- colnames(log_topBins)
dist.pear <- function(x) as.dist(1-cor(t(x)))
hclust.ave <- function(x) hclust(x, method="average")
gplots::heatmap.2(cov.out.edgeR,scale="row",trace="none",labRow=NA,main = "Methylation level of significant bins\n(Covariates regressed out)",
distfun=dist.pear, hclustfun=hclust.ave,col=rev(RColorBrewer::brewer.pal(9,"RdBu")))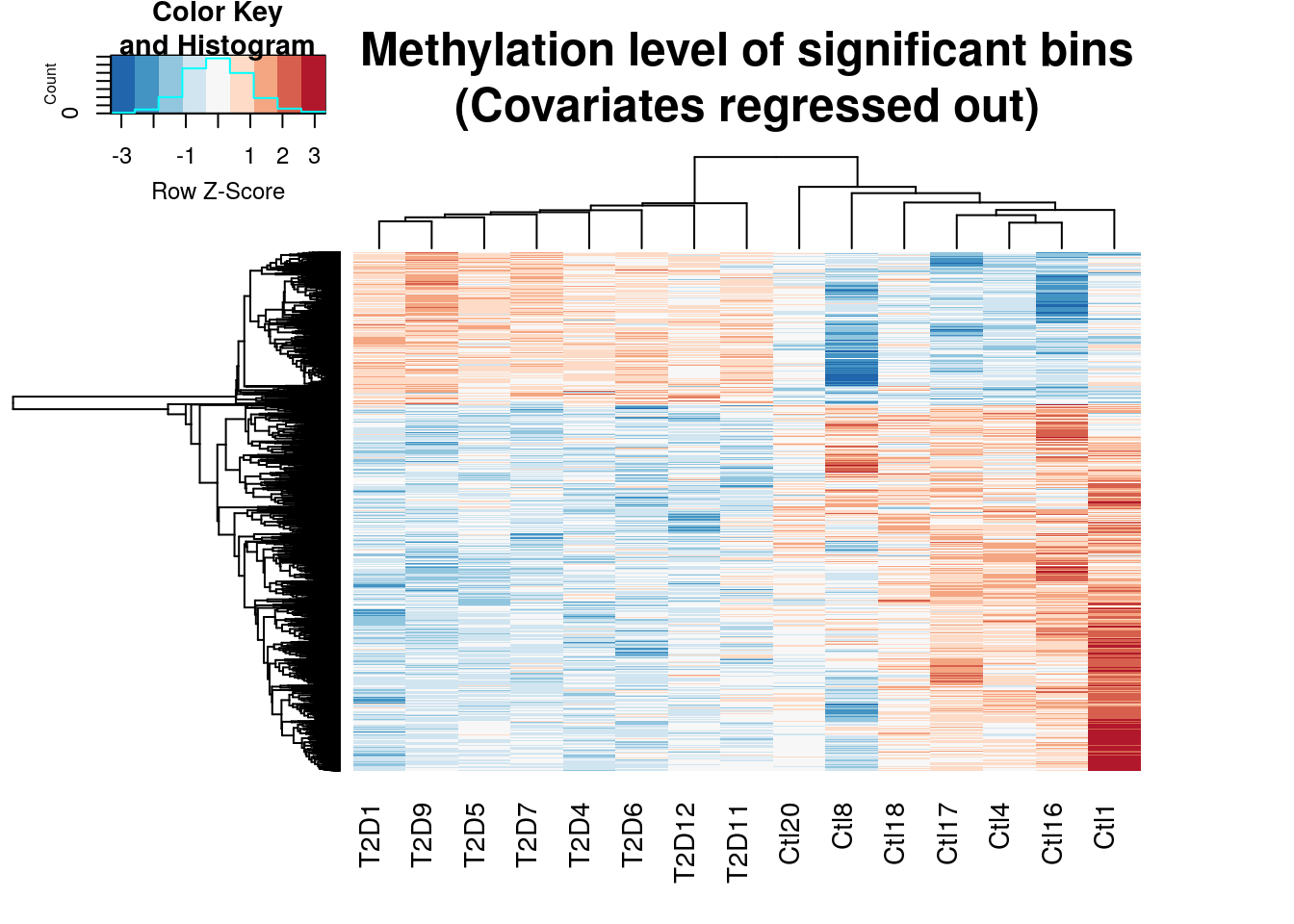
Session information
sessionInfo()## R version 3.4.4 (2018-03-15)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 17.10
##
## Matrix products: default
## BLAS: /usr/local/lib/R/lib/libRblas.so
## LAPACK: /usr/local/lib/R/lib/libRlapack.so
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] grid stats4 parallel stats graphics grDevices utils
## [8] datasets methods base
##
## other attached packages:
## [1] VennDiagram_1.6.20 futile.logger_1.4.3
## [3] qvalue_2.10.0 RADAR_0.1.5
## [5] RcppArmadillo_0.9.100.5.0 Rcpp_0.12.19
## [7] RColorBrewer_1.1-2 gplots_3.0.1
## [9] doParallel_1.0.14 iterators_1.0.10
## [11] foreach_1.4.4 ggplot2_3.0.0
## [13] Rsamtools_1.30.0 Biostrings_2.46.0
## [15] XVector_0.18.0 GenomicFeatures_1.30.3
## [17] AnnotationDbi_1.40.0 Biobase_2.38.0
## [19] GenomicRanges_1.30.3 GenomeInfoDb_1.14.0
## [21] IRanges_2.12.0 S4Vectors_0.16.0
## [23] BiocGenerics_0.24.0
##
## loaded via a namespace (and not attached):
## [1] nlme_3.1-131.1 bitops_1.0-6
## [3] matrixStats_0.54.0 bit64_0.9-7
## [5] progress_1.2.0 httr_1.3.1
## [7] rprojroot_1.3-2 tools_3.4.4
## [9] backports_1.1.2 R6_2.2.2
## [11] KernSmooth_2.23-15 mgcv_1.8-23
## [13] DBI_1.0.0 lazyeval_0.2.1
## [15] colorspace_1.3-2 withr_2.1.2
## [17] gridExtra_2.3 tidyselect_0.2.4
## [19] prettyunits_1.0.2 RMySQL_0.10.15
## [21] bit_1.1-14 compiler_3.4.4
## [23] formatR_1.5 DelayedArray_0.4.1
## [25] labeling_0.3 rtracklayer_1.38.3
## [27] caTools_1.17.1 scales_0.5.0
## [29] stringr_1.3.1 digest_0.6.15
## [31] rmarkdown_1.10 DOSE_3.4.0
## [33] pkgconfig_2.0.1 htmltools_0.3.6
## [35] highr_0.7 rlang_0.2.1
## [37] RSQLite_2.1.1 bindr_0.1.1
## [39] BiocParallel_1.12.0 gtools_3.8.1
## [41] GOSemSim_2.4.1 dplyr_0.7.6
## [43] RCurl_1.95-4.10 magrittr_1.5
## [45] GO.db_3.5.0 GenomeInfoDbData_1.0.0
## [47] Matrix_1.2-12 munsell_0.5.0
## [49] stringi_1.2.3 yaml_2.1.19
## [51] SummarizedExperiment_1.8.1 zlibbioc_1.24.0
## [53] plyr_1.8.4 blob_1.1.1
## [55] gdata_2.18.0 DO.db_2.9
## [57] crayon_1.3.4 lattice_0.20-35
## [59] splines_3.4.4 hms_0.4.2
## [61] knitr_1.20 pillar_1.2.3
## [63] igraph_1.2.1 fgsea_1.4.1
## [65] reshape2_1.4.3 codetools_0.2-15
## [67] biomaRt_2.34.2 futile.options_1.0.1
## [69] fastmatch_1.1-0 XML_3.98-1.11
## [71] glue_1.2.0 evaluate_0.10.1
## [73] lambda.r_1.2.3 data.table_1.11.4
## [75] tidyr_0.8.1 gtable_0.2.0
## [77] purrr_0.2.5 assertthat_0.2.0
## [79] tibble_1.4.2 clusterProfiler_3.6.0
## [81] rvcheck_0.1.0 GenomicAlignments_1.14.2
## [83] memoise_1.1.0 MyTools_0.0.0
## [85] bindrcpp_0.2.2This R Markdown site was created with workflowr