Ovarian_cancer
Zijie Zhang
2018-08-21
Count reads
library(RADAR)
samplenames <- paste0("OvCa",c("1850", "1917", "2005" ,"2013", "2053" ,"2064", "2072" ,"2186", "2221", "2261", "2270" ,"2343", "2380"))
RADAR <- countReads(samplenames = samplenames,
gtf = "~/Database/genome/hg38/hg38_UCSC.gtf",
bamFolder = "~/PoissonGamma_Method/Ovary_Cancer/bam_files",
outputDir = "~/PoissonGamma_Method/Ovary_Cancer",
modification = "m6A",
binSize = 50,
strandToKeep = "opposite",
paired = TRUE,
threads = 40
)
save(RADAR,file = "~/PoissonGamma_Method/Ovary_Cancer/radar.RData")Summary of read count
library(RADAR)
# load( "~/PoissonGamma_Method/Ovary_Cancer/radar.RData" )
load( "~/PoissonGamma_Method/Ovary_Cancer/RADAR_analysis.RData" ) # load work space with analysis result for quick plot update
countSummary <- rbind("Input"= colSums(RADAR$reads)[1:length(RADAR$samplenames)], "IP" =colSums(RADAR$reads)[(1:length(RADAR$samplenames))+length(RADAR$samplenames)] )
colnames(countSummary) <- RADAR$samplenames
knitr::kable(countSummary,format = "pandoc" )| OvCa1850 | OvCa1917 | OvCa2005 | OvCa2013 | OvCa2053 | OvCa2064 | OvCa2072 | OvCa2186 | OvCa2221 | OvCa2261 | OvCa2270 | OvCa2343 | OvCa2380 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Input | 5451646 | 6774484 | 6712499 | 7025161 | 7210783 | 4994751 | 8137234 | 4805675 | 3367101 | 7728379 | 6098250 | 6184088 | 4677515 |
| IP | 12670799 | 14874196 | 14193427 | 15088959 | 15193462 | 13503265 | 13991803 | 11733340 | 10180252 | 14590303 | 11039885 | 13541646 | 12181408 |
monster <- RADAR
X <- c( rep("Ctl",7),rep("Tumor",6) )
RADAR <- normalizeLibrary(RADAR,X)
RADAR <- adjustExprLevel(RADAR)
RADAR <- filterBins(RADAR,minCountsCutOff = 15)
RADAR_pos <- adjustExprLevel(RADAR[1:12], adjustBy = "pos")
RADAR_pos <- filterBins(RADAR_pos,minCountsCutOff = 15)
RADAR_bin100 <- normalizeLibrary(RADAR_bin100,X)
RADAR_bin100 <- adjustExprLevel(RADAR_bin100)
RADAR_bin100 <- filterBins(RADAR_bin100,minCountsCutOff = 15)Local Vs geneSum
Plot distribution of number of reads in each 50 bp bins
hist(log10(rowMeans(RADAR$reads[rowMeans(RADAR$reads[,grep("input",colnames(RADAR$reads))])>1,grep("input",colnames(RADAR$reads))]) ),xlab = "log10 read count",main = "Distribution of INPUT read count in bins",xlim = c(0,3), col =rgb(0.2,0.2,0.2,0.5),cex.main = 2,cex.axis =2,cex.lab=2)
axis(side = 1, lwd = 2,cex.axis =2)
axis(side = 2, lwd = 2,cex.axis =2)
local read count V.S. geneSum
Compute the within group variability of INPUT geneSum VS INPUT local read count.
var.coef <- function(x){sd(as.numeric(x))/mean(as.numeric(x))}
## filter expressed genes
geneSum <- RADAR$geneSum[rowSums(RADAR$geneSum)>16,]
## within group variability
relative.var <- t( apply(geneSum,1,tapply,RADAR$X,var.coef) )
geneSum.var <- c(relative.var)
## For each gene used above, random sample a 50bp bin within this gene
set.seed(1)
r.bin50 <- tapply(rownames(RADAR$norm.input)[which(RADAR$geneBins$gene %in% rownames(geneSum))],as.character( RADAR$geneBins$gene[which(RADAR$geneBins$gene %in% rownames(geneSum))]) ,function(x){
n <- sample(1:length(x),1)
return(x[n])
})
relative.var <- apply(RADAR$norm.input[r.bin50,],1,tapply,RADAR$X,var.coef)
bin50.var <- c(relative.var)
bin50.var <- bin50.var[!is.na(bin50.var)]
## 100bp bins
r.bin100 <- tapply(rownames(RADAR$norm.input)[which(RADAR$geneBins$gene %in% rownames(geneSum))],as.character( RADAR$geneBins$gene[which(RADAR$geneBins$gene %in% rownames(geneSum))]) ,function(x){
n <- sample(1:(length(x)-2),1)
return(x[n:(n+1)])
})
relative.var <- lapply(r.bin100, function(x){ tapply( colSums(RADAR$norm.input[x,]),RADAR$X,var.coef ) })
bin100.var <- unlist(relative.var)
bin100.var <- bin100.var[!is.na(bin100.var)]
## 200bp bins
r.bin200 <- tapply(rownames(RADAR$norm.input)[which(RADAR$geneBins$gene %in% rownames(geneSum))],as.character( RADAR$geneBins$gene[which(RADAR$geneBins$gene %in% rownames(geneSum))]) ,function(x){
if(length(x)>4){
n <- sample(1:(length(x)-3),1)
return(x[n:(n+3)])
}else{
return(NULL)
}
})
r.bin200 <- r.bin200[which(!unlist(lapply(r.bin200,is.null)) ) ]
relative.var <- lapply(r.bin200, function(x){ tapply( colSums(RADAR$norm.input[x,]),RADAR$X,var.coef ) })
bin200.var <- unlist(relative.var)
bin200.var <- bin200.var[!is.na(bin200.var)]
relative.var <- data.frame(group=c(rep("geneSum",length(geneSum.var)),rep("bin-50bp",length(bin50.var)),rep("bin-100bp",length(bin100.var)),rep("bin-200bp",length(bin200.var))), variance=c(geneSum.var,bin50.var,bin100.var,bin200.var))ggplot(relative.var,aes(variance,colour=group))+geom_density()+xlab("Relative variation")+
theme_bw() + theme(panel.border = element_blank(), panel.grid.major = element_blank(),
panel.grid.minor = element_blank(), axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=20, face="bold", hjust=0.5,family = "arial"),
axis.title.y=element_text(size=20, face="bold", vjust=0.4, angle=90,family = "arial"),
legend.position = c(0.88,0.88),legend.title=element_blank(),legend.text = element_text(size = 14, face = "bold",family = "arial"),
axis.text = element_text(size = 15,face = "bold",family = "arial",colour = "black") )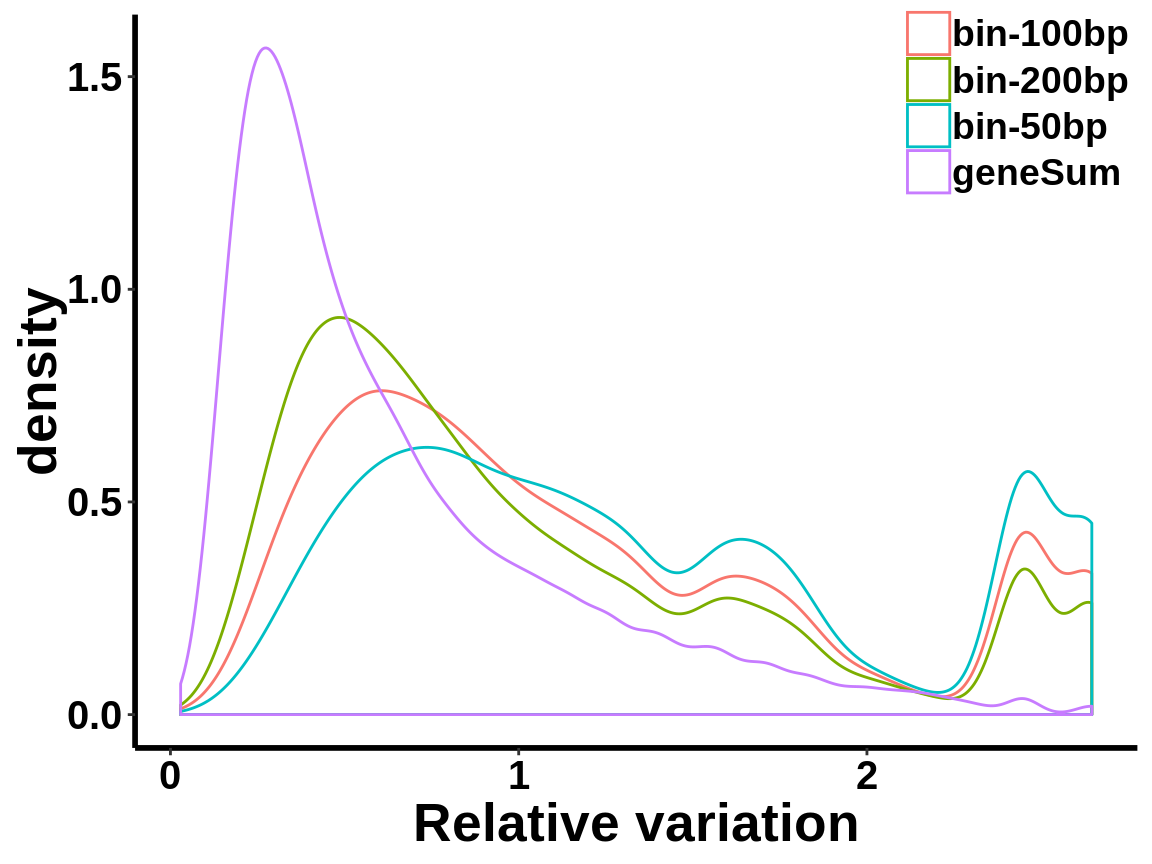
Check mean variance relationship of the data.
ip_var <- t(apply(RADAR$ip_adjExpr_filtered,1,tapply,X,var))
#ip_var <- ip_coef_var[!apply(ip_var,1,function(x){return(any(is.na(x)))}),]
ip_mean <- t(apply(RADAR$ip_adjExpr_filtered,1,tapply,X,mean))
###
gene_var <- t(apply(RADAR$geneSum,1,tapply,X,var))
#gene_var <- gene_var[!apply(gene_var,1,function(x){return(any(is.na(x)))}),]
gene_mean <- t(apply(RADAR$geneSum,1,tapply,X,mean))
all_var <- list('RNA-seq'=c(gene_var),'m6A-IP'=c(ip_var))
nn<-sapply(all_var, length)
rs<-cumsum(nn)
re<-rs-nn+1
group <- factor(rep(names(all_var), nn), levels=names(all_var))
all_var.df <- data.frame(variance = c(c(gene_var),c(ip_var)),mean= c(c(gene_mean),c(ip_mean)),label = group)ggplot(data = all_var.df,aes(x=mean,y=variance,colour = group,shape = group))+geom_point(size = I(0.2))+stat_smooth(se = T,show.legend = F)+stat_smooth(se = F)+theme_bw() +xlab("Mean")+ ylab("Variance")+theme(panel.border = element_blank(), panel.grid.major = element_blank(),
panel.grid.minor = element_blank(), axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=22, face="bold", hjust=0.5,family = "arial"),
axis.title.y=element_text(size=22, face="bold", vjust=0.4, angle=90,family = "arial"),
legend.position = c(0.85,0.95),legend.title=element_blank(),legend.text = element_text(size = 20, face = "bold",family = "arial"),
axis.text = element_text(size = 15,face = "bold",family = "arial",colour = "black") ) +
scale_x_continuous(limits = c(0,7000))+scale_y_continuous(limits = c(0,5e6))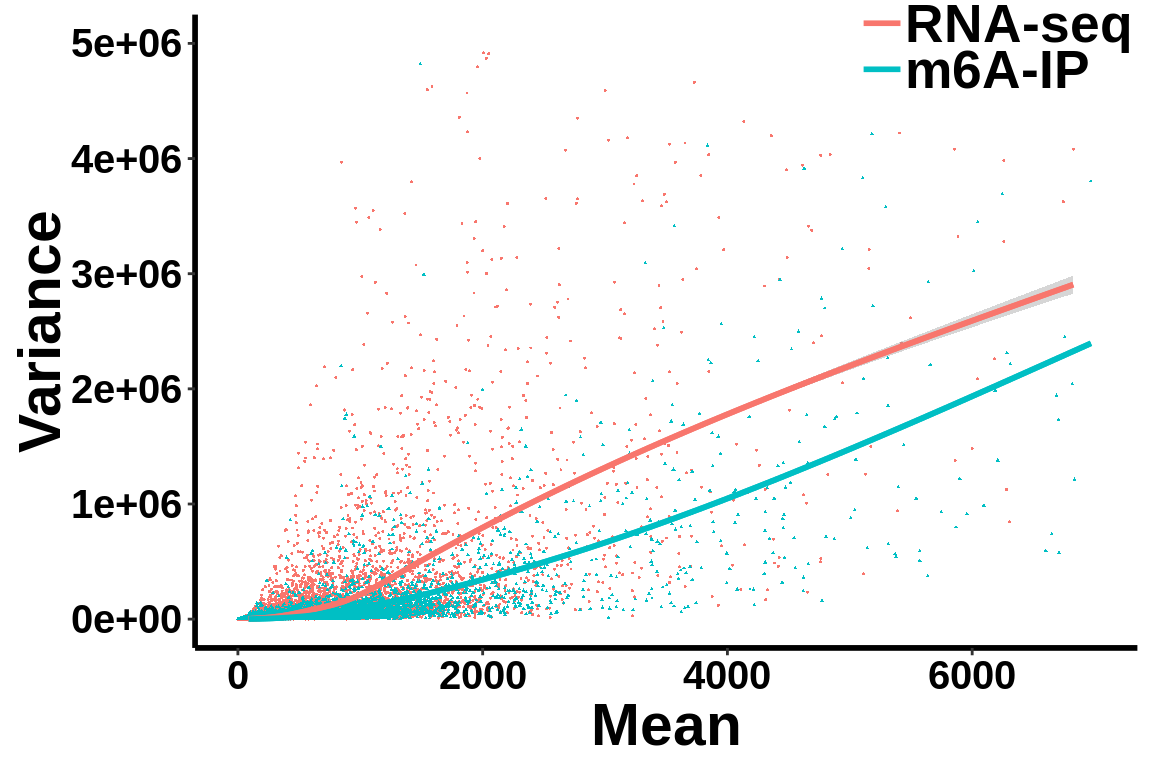
Compare MeRIP-seq IP data with regular RNA-seq data
var.coef <- function(x){sd(as.numeric(x))/mean(as.numeric(x))}
ip_coef_var <- t(apply(RADAR$ip_adjExpr_filtered,1,tapply,X,var.coef))
ip_coef_var <- ip_coef_var[!apply(ip_coef_var,1,function(x){return(any(is.na(x)))}),]
#hist(c(ip_coef_var),main = "M14KO mouse liver\n m6A-IP",xlab = "within group coefficient of variation",breaks = 50)
###
gene_coef_var <- t(apply(RADAR$geneSum,1,tapply,X,var.coef))
gene_coef_var <- gene_coef_var[!apply(gene_coef_var,1,function(x){return(any(is.na(x)))}),]
#hist(c(gene_coef_var),main = "M14KO mouse liver\n RNA-seq",xlab = "within group coefficient of variation",breaks = 50)
coef_var <- list('RNA-seq'=c(gene_coef_var),'m6A-IP'=c(ip_coef_var))
nn<-sapply(coef_var, length)
rs<-cumsum(nn)
re<-rs-nn+1
grp <- factor(rep(names(coef_var), nn), levels=names(coef_var))
coef_var.df <- data.frame(coefficient_var = c(c(gene_coef_var),c(ip_coef_var)),label = grp)ggplot(data = coef_var.df,aes(coefficient_var,colour = grp))+geom_density()+theme_bw() +theme(panel.border = element_blank(), panel.grid.major = element_blank(),
panel.grid.minor = element_blank(), axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=20, face="bold", hjust=0.5,family = "arial"),
axis.title.y=element_text(size=20, face="bold", vjust=0.4, angle=90,family = "arial"),
legend.position = c(0.9,0.9),legend.title=element_blank(),legend.text = element_text(size = 18, face = "bold",family = "arial"),
axis.text = element_text(size = 15,face = "bold",family = "arial",colour = "black") )+
xlab("Coefficient of variation")
PCA analysis
We plot PCA colored by Ctl vs Tumor
plotPCAfromMatrix(RADAR$ip_adjExpr_filtered,group = X )+scale_color_discrete(name = "Group")
(Adjust expression level by local read count)
plotPCAfromMatrix(RADAR_pos$ip_adjExpr_filtered,group = X )+scale_color_discrete(name = "Group")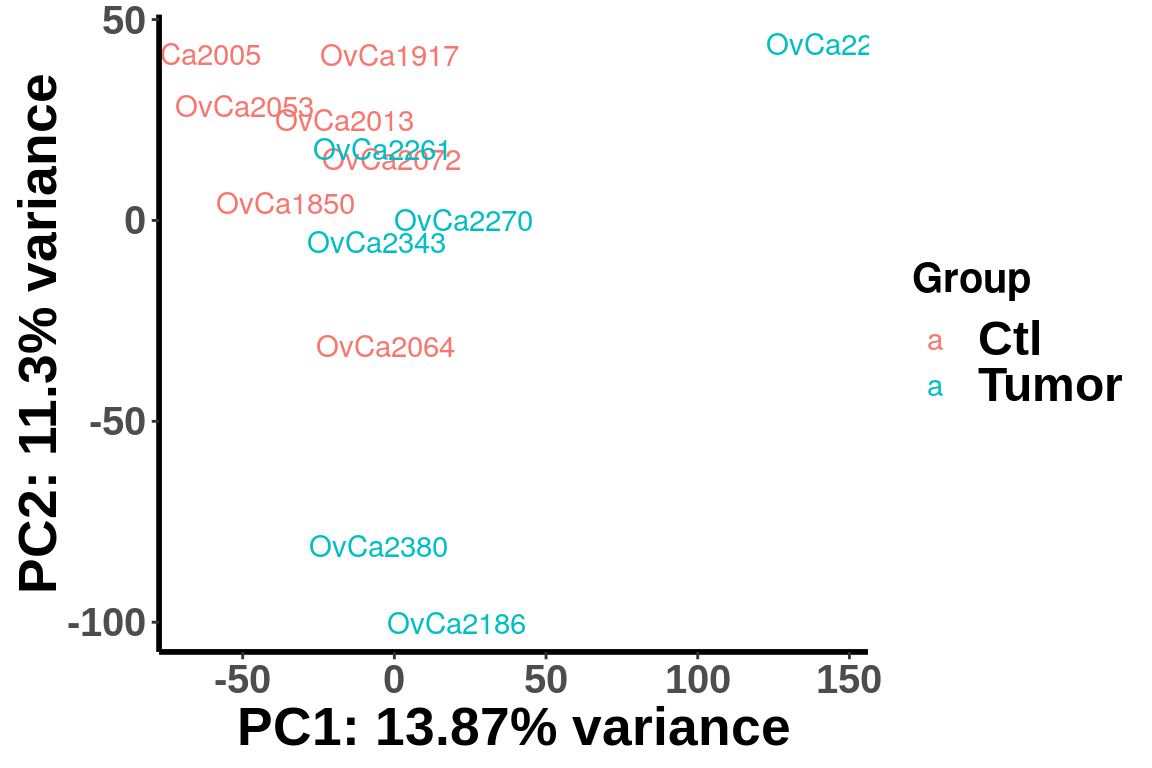
age <- c(45,54,45,59,35,36,51,73,68,64,70,68,62)
IP_counts <- RADAR$ip_adjExpr_filtered
colnames(IP_counts)<- c(paste0("Ctl",c("1850", "1917", "2005" ,"2013", "2053" ,"2064", "2072")), paste0("Tumor",c("2186", "2221", "2261", "2270" ,"2343", "2380")) )
plotPCAfromMatrix(IP_counts[order(rowSums(RADAR$ip_adjExpr_filtered),decreasing = T)[1:15000],], group = age )+scale_colour_continuous(name = "Age",low = "blue",high = "red")+scale_color_continuous(name = "Age")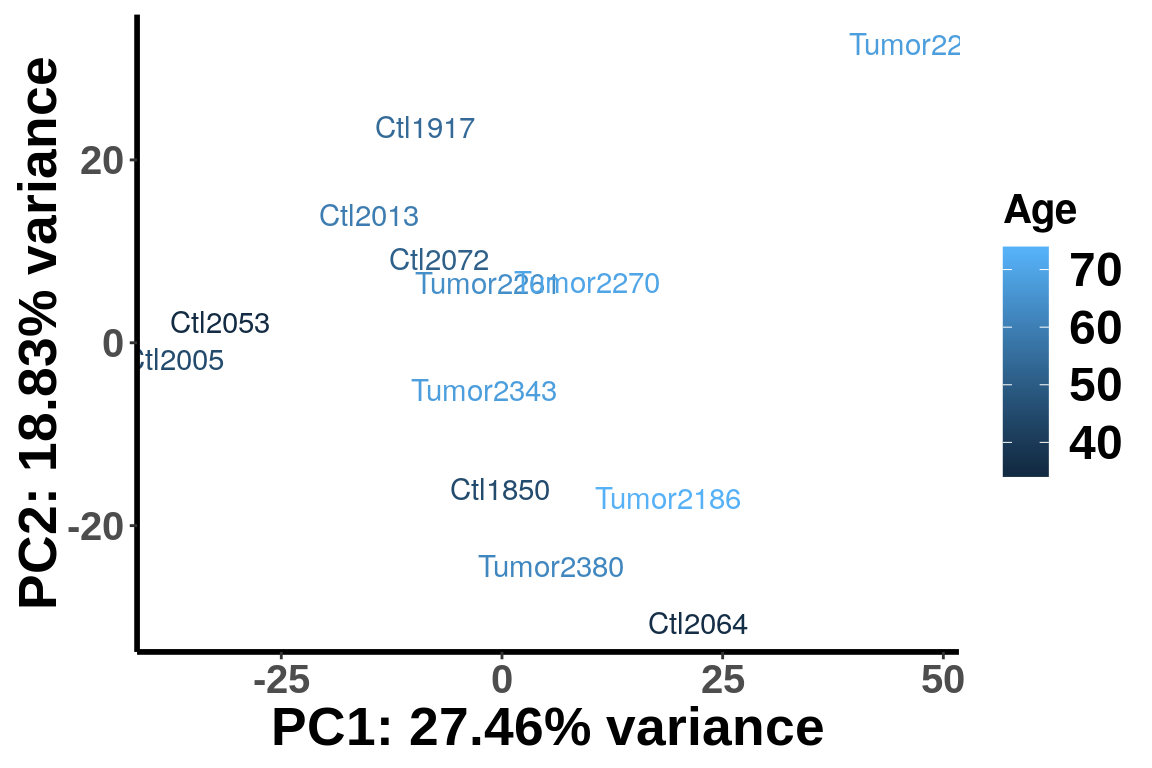
Regress out age and re-plot PCA
registerDoParallel(cores = 10)
m6A.cov.out <- foreach(i = 1:nrow(RADAR$ip_adjExpr_filtered), .combine = rbind) %dopar% {
Y = RADAR$ip_adjExpr_filtered[i,]
resi <- residuals( lm(log(Y+1) ~ age ) )
resi
}
rm(list=ls(name=foreach:::.foreachGlobals), pos=foreach:::.foreachGlobals)
rownames(m6A.cov.out) <- rownames(RADAR$ip_adjExpr_filtered)
plotPCAfromMatrix(m6A.cov.out[order(rowSums(RADAR$ip_adjExpr_filtered),decreasing = T)[1:15000],],X,loglink = FALSE)+scale_color_discrete(name = "Group")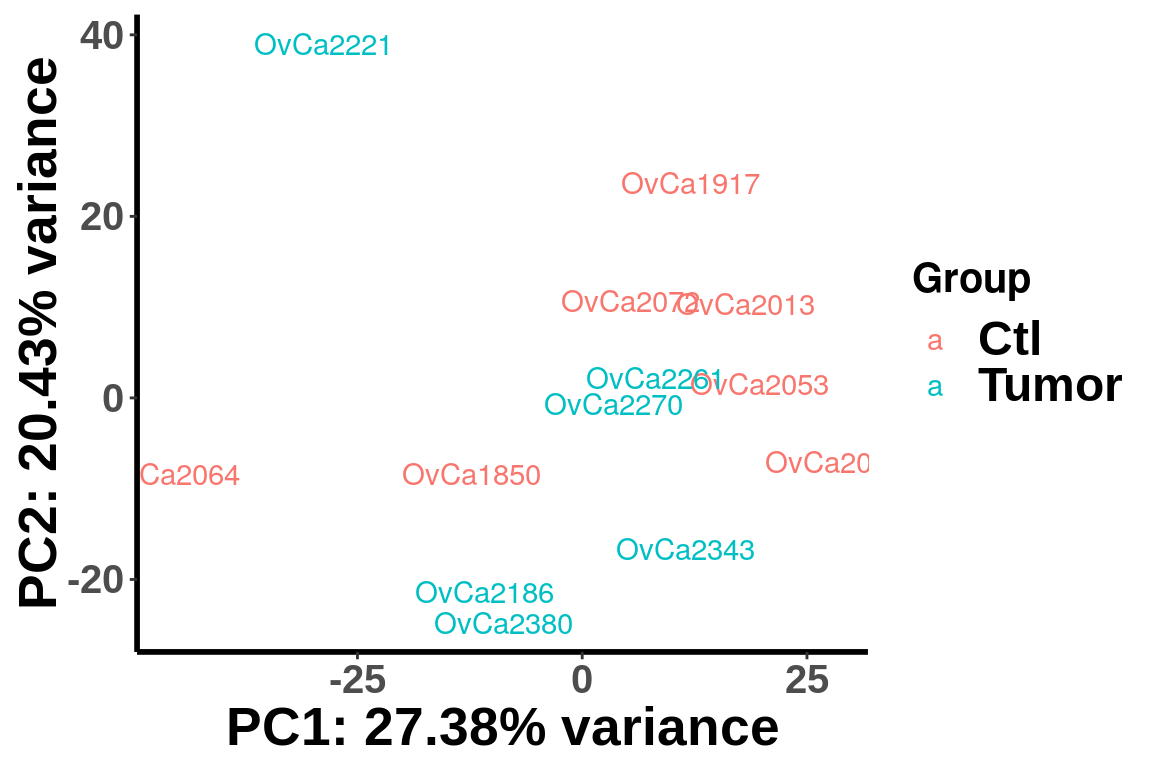
Run PoissonGamma test
PoissonGamma Test
RADAR_cov <- diffIP_parallel(RADAR[1:14],thread = 25, Covariates = age)$all.est
RADAR_pos_cov <- diffIP_parallel(RADAR_pos[1:14],thread = 25, Covariates = age)$all.est
RADAR_cov_bin100 <- diffIP_parallel(RADAR_bin100[1:14],thread = 25, Covariates = age)$all.estOther method
In order to compare performance of other method on this data set, we run other three methods on default mode on this dataset.
cov <- matrix(age,ncol = 1)
colnames(cov) <- "age"
deseq2.res_cov <- DESeq2(countdata = RADAR$ip_adjExpr_filtered,pheno = matrix(X,ncol = 1),covariates = cov )
edgeR.res_cov <- edgeR(countdata = RADAR$ip_adjExpr_filtered,pheno = matrix(X,ncol = 1),covariates = cov )
deseq2.res_pos_cov <- DESeq2(countdata = RADAR_pos$ip_adjExpr_filtered,pheno = matrix(X,ncol = 1),covariates = cov )
edgeR.res_pos_cov <- edgeR(countdata = RADAR_pos$ip_adjExpr_filtered,pheno = matrix(X,ncol = 1),covariates = cov )Compare distribution of p value.
pvalues <- data.frame(pvalue = c(RADAR_cov[,"p_value3"],deseq2.res_cov$pvalue,edgeR.res_cov$pvalue,QNB.res$pvalue),
method = factor(rep(c("RADAR","DESeq2","edgeR","QNB"),c(length(RADAR_cov[,"p_value3"]),
length(deseq2.res_cov$pvalue),
length(edgeR.res_cov$pvalue),
length(QNB.res$pvalue)
)
),levels =c("RADAR","DESeq2","edgeR","QNB")
)
)
ggplot(pvalues, aes(x = pvalue))+geom_histogram(breaks = seq(0,1,0.04),col=I("black"))+facet_grid(.~method)+theme_bw()+xlab("p-value")+theme( axis.title = element_text(size = 22, face = "bold"),strip.text = element_text(size = 25, face = "bold"),axis.text.x = element_text(size = 18, face = "bold",colour = "black"),axis.text.y = element_text(size = 15, face = "bold",colour = "black",angle = 90),panel.grid = element_blank(), axis.line = element_line(size = 0.7 ,colour = "black"),axis.ticks = element_line(colour = "black"), panel.spacing = unit(0.4, "lines") )+ scale_x_continuous(breaks = seq(0,1,0.2),labels=function(x) sprintf("%.1f", x))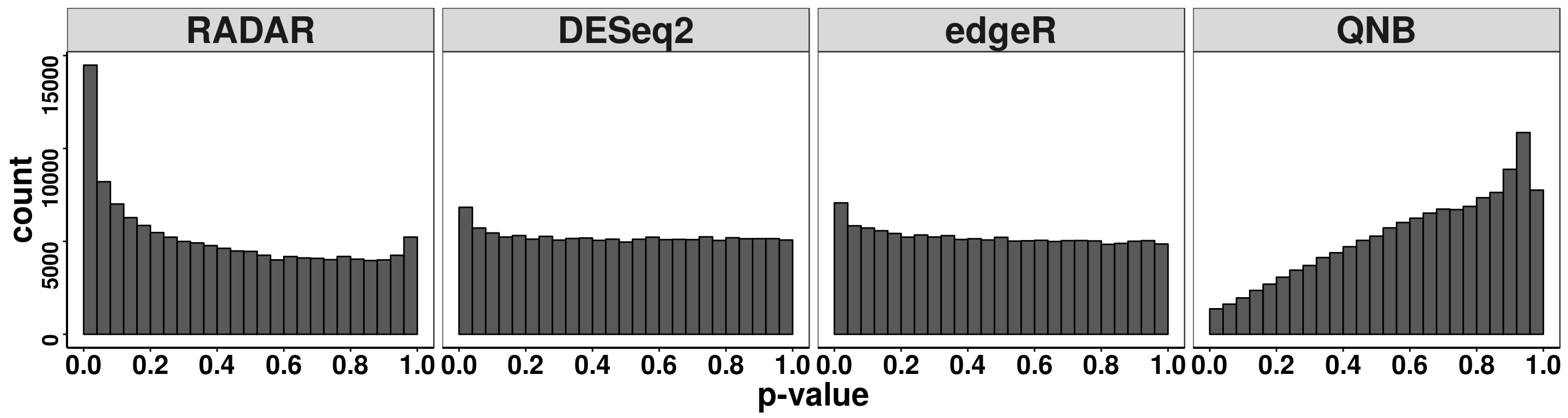
Compare adjust expression level by geneSum vs. bin read counts.
par(mfrow = c(1,3))
tmp <- hist(c(RADAR_pos_cov[,"p_value3"]),plot = F)
tmp$counts <- tmp$counts*(nrow(RADAR$ip_adjExpr_filtered)/nrow(RADAR_pos$ip_adjExpr_filtered))
hist(RADAR_cov[,"p_value3"],main = "RADAR",xlab = "p value",col =rgb(1,0,0,0.4),cex.main = 2.5,cex.axis =2,cex.lab=2, ylim = c(0,max(hist(RADAR_cov[,"p_value3"],plot = F)$counts,tmp$counts)) )
plot(tmp,col=rgb(0,0,1,0.5),add = T)
axis(side = 1, lwd = 1,cex.axis =2)
axis(side = 2, lwd = 1,cex.axis =2)
legend("topright", c("By geneSum", "By position"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 10,bty="n",text.font = 2)
y.scale <- max(hist(RADAR_cov[,"p_value3"],plot = F)$counts)
tmp <- hist(c(deseq2.res_pos_cov$pvalue),plot = F)
tmp$counts <- tmp$counts*(nrow(RADAR$ip_adjExpr_filtered)/nrow(RADAR_pos$ip_adjExpr_filtered))
hist(deseq2.res_cov$pvalue,main = "DESeq2",xlab = "p value",col =rgb(1,0,0,0.4),cex.main = 2.5,cex.axis =2,cex.lab=2, ylim = c(0,max(hist(deseq2.res_cov$pvalue,plot = F)$counts,tmp$counts,y.scale)) )
plot(tmp,col=rgb(0,0,1,0.5),add = T)
axis(side = 1, lwd = 1,cex.axis =2)
axis(side = 2, lwd = 1,cex.axis =2)
legend("topright", c("By geneSum", "By position"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 10,bty="n",text.font = 2)
tmp <- hist(c(edgeR.res_pos_cov$pvalue),plot = F)
tmp$counts <- tmp$counts*(nrow(RADAR$ip_adjExpr_filtered)/nrow(RADAR_pos$ip_adjExpr_filtered))
hist(edgeR.res_cov$pvalue,main = "edgeR",xlab = "p value",col =rgb(1,0,0,0.4),cex.main = 2.5,cex.axis =2,cex.lab=2, ylim = c(0,max(hist(edgeR.res_cov$pvalue,plot = F)$counts,tmp$counts,y.scale)) )
plot(tmp,col=rgb(0,0,1,0.5),add = T)
axis(side = 1, lwd = 1,cex.axis =2)
axis(side = 2, lwd = 1,cex.axis =2)
legend("topright", c("By geneSum", "By position"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 10,bty="n",text.font = 2)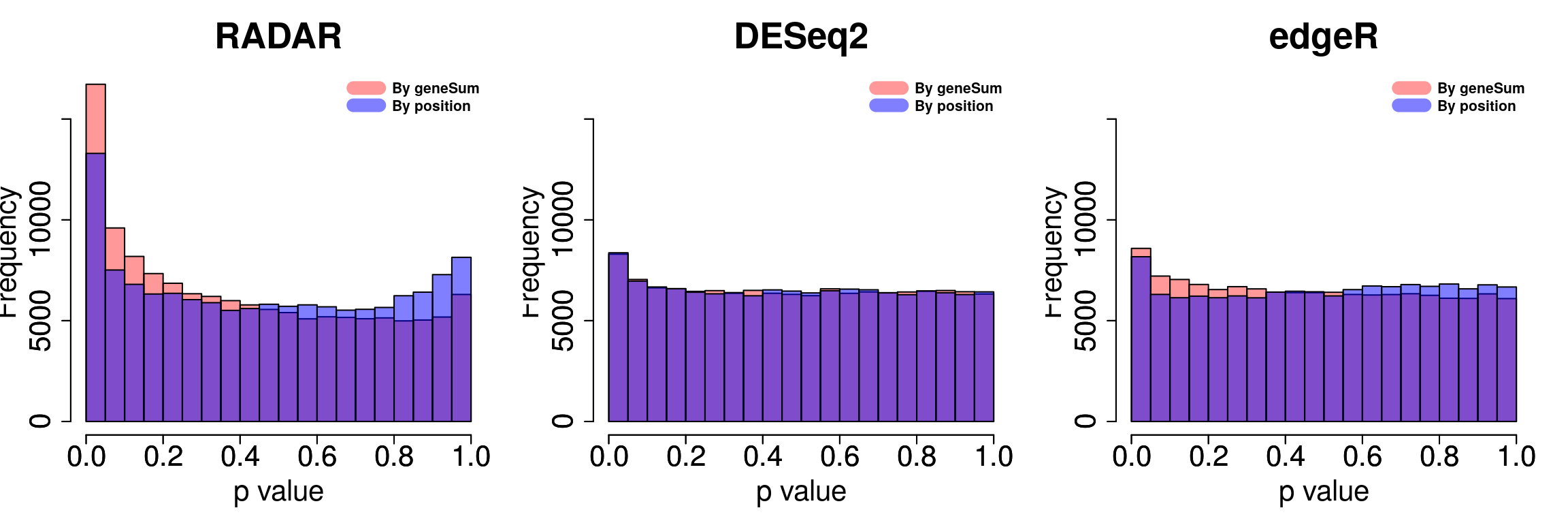
Compare the distibution of beta
par(mfrow = c(1,4))
hist(RADAR_cov[,"beta1"],main = "RADAR",xlab = "beta",breaks = 100,col =rgb(0.2,0.2,0.2,0.5),cex.main = 2.5,cex.axis =2,cex.lab=2)
#hist(radar_byPos$all.est[,"beta1"],main = "Norm by pos",xlab = "beta",breaks = 100)
hist(deseq2.res_cov$log2FC, main = "DESeq2",xlab = "beta",breaks = 100,col =rgb(0.2,0.2,0.2,0.5),cex.main = 2.5,cex.axis =2,cex.lab=2)
hist(edgeR.res_cov$log2FC,main = "edgeR", xlab = "beta",breaks = 100,col =rgb(0.2,0.2,0.2,0.5),cex.main = 2.5,cex.axis =2,cex.lab=2)
hist(QNB.res$log2.OR, main = "QNB",xlab = "beta",breaks = 100,col =rgb(0.2,0.2,0.2,0.5),cex.main = 2.5,cex.axis =2,cex.lab=2)
Number of significant bins detected at qvalue < 0.1.
sigBins <- apply(cbind("RADAR"=RADAR_cov[,"p_value3"],"DESeq2"=deseq2.res_cov$pvalue,"edgeR"=edgeR.res_cov$pvalue,"QNB"=QNB.res$pvalue),2, function(x){
length( which( qvalue::qvalue(x)$qvalue < 0.1 ) )
})
print(sigBins)## RADAR DESeq2 edgeR QNB
## 2736 16 169 13Plot overlap of significant hits
plot.new()
MyTools::venn_diagram4( names(which( qvalue::qvalue(RADAR_cov[,"p_value3"])$qvalue < 0.1 ) ),
rownames(RADAR_cov)[which( qvalue::qvalue(deseq2.res_cov$pvalue)$qvalue < 0.1 ) ],
rownames(RADAR_cov)[which( qvalue::qvalue(edgeR.res_cov$pvalue)$qvalue < 0.1 ) ],
rownames(RADAR_cov)[which( qvalue::qvalue(QNB.res$pvalue)$qvalue < 0.1 ) ],
"RADAR","DESeq2","edgeR","QNB")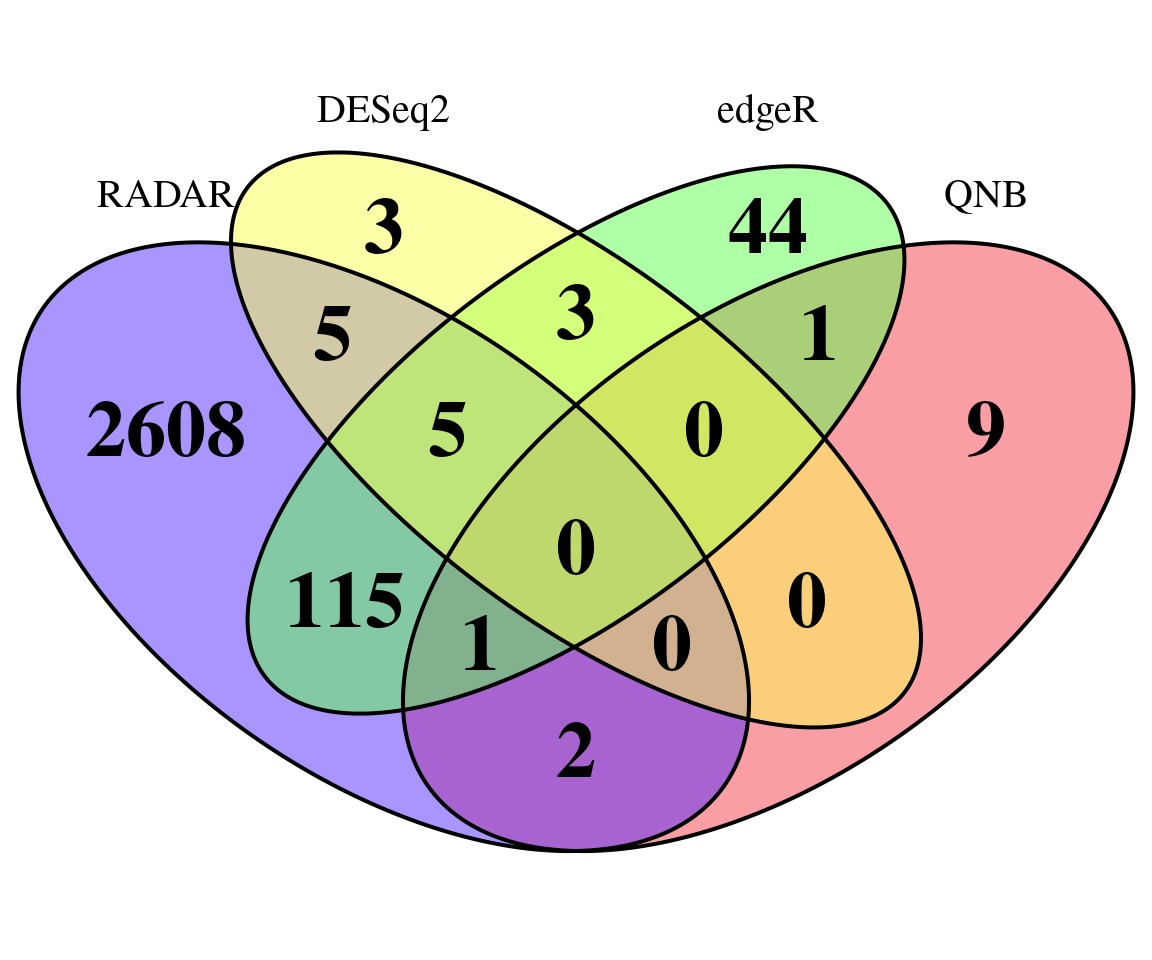
(Adjust expression level by local read count)
plot.new()
MyTools::venn_diagram4( names(which( qvalue::qvalue(RADAR_pos_cov[,"p_value3"])$qvalue < 0.1 ) ),
rownames(RADAR_pos_cov)[which( qvalue::qvalue(deseq2.res_pos_cov$pvalue)$qvalue < 0.1 ) ],
rownames(RADAR_pos_cov)[which( qvalue::qvalue(edgeR.res_pos_cov$pvalue)$qvalue < 0.1 ) ],
rownames(RADAR_pos_cov)[which( qvalue::qvalue(QNB.res$pvalue)$qvalue < 0.1 ) ],
"RADAR","DESeq2","edgeR","QNB")
RADAR_res <- reportPoissonGammaMerge(RADAR,cutoff = 0.1,Beta_cutoff = 0)Self-FDR test
Subset samples and run the test.
selfFDR_cov <- NULL
samplenames <- RADAR$samplenames
set.seed(10)
for(i in 1:15){
## Sample subset
sample_id <- c(sample(x = 1:7, size = 5,replace = F),7+sample(x = 1:6, size = 4,replace = F) )
Truth_id <- setdiff(1:13,sample_id)
## run radar
radar_sample <- diffIP_parallel(RADAR[1:14],Covariates = cov ,exclude = samplenames[setdiff(1:13,sample_id)],thread = 25,plotPvalue = F)$all.est
radar_sample <- cbind(radar_sample, "qvalue"= qvalue::qvalue(radar_sample[,"p_value3"])$qvalue )
radar_truth <- RADAR_cov
radar_truth <- cbind(radar_truth, "qvalue"= qvalue::qvalue(radar_truth[,"p_value3"])$qvalue )
radar_selfFDR <- sapply(c(0.1,0.05,0.01),function(x){length( which(! which(radar_sample[,"qvalue"]<x) %in% which(radar_truth[,"qvalue"]<0.1) ) )/length(which(radar_sample[,"qvalue"]<x))})
## run DESeq2
deseq_sample <- DESeq2(countdata = RADAR$ip_adjExpr_filtered[,sample_id],pheno = matrix(c(rep(0,7),rep(1,6))[sample_id],ncol = 1),covariates = as.matrix(data.frame(age =cov[sample_id])) )
deseq_sample$qvalue <- qvalue::qvalue( deseq_sample$pvalue )$qvalue
deseq_truth <- deseq2.res_cov
deseq_truth$qvalue <- qvalue::qvalue( deseq_truth$pvalue )$qvalue
deseq_selfFDR <- sapply(c(0.1,0.05,0.01),function(x){length( which(! which(deseq_sample[,"qvalue"]<x) %in% which(deseq_truth[,"qvalue"]<0.1) ) )/length(which(deseq_sample[,"qvalue"]<x))})
## run edgeR
edgeR_sample <- edgeR(countdata = RADAR$ip_adjExpr_filtered[,sample_id],pheno = matrix(c(rep(0,7),rep(1,6))[sample_id],ncol = 1),covariates = as.matrix(data.frame(batch =cov[sample_id])) )
edgeR_sample$qvalue <- qvalue::qvalue( edgeR_sample$pvalue )$qvalue
edgeR_truth <- edgeR.res_cov
edgeR_truth$qvalue <- qvalue::qvalue( edgeR_truth$pvalue )$qvalue
edgeR_selfFDR <- sapply(c(0.1,0.05,0.01),function(x){length( which(! which(edgeR_sample[,"qvalue"]<x) %in% which(edgeR_truth[,"qvalue"]<0.1) ) )/length(which(edgeR_sample[,"qvalue"]<x))})
fdr <- rbind( data.frame(self_FDR = radar_selfFDR, qvalue_cutoff=c("0.1","0.05","0.01"), method = rep("RADAR",3) ),
data.frame(self_FDR = deseq_selfFDR, qvalue_cutoff=c("0.1","0.05","0.01"), method = rep("DESeq2",3) ),
data.frame(self_FDR = edgeR_selfFDR, qvalue_cutoff=c("0.1","0.05","0.01"), method = rep("edgeR",3) )
)
selfFDR_cov <- rbind( selfFDR_cov, fdr )
}
selfFDR_cov$qvalue_cutoff <- factor(selfFDR_cov$qvalue_cutoff,levels = c("0.1","0.05","0.01"))
save(selfFDR_cov, file = "~/PoissonGamma_Method/Ovary_Cancer/selfFDR_cov.RData")load("~/PoissonGamma_Method/Ovary_Cancer/selfFDR_cov.RData")
ggplot(data = selfFDR_cov, aes(x=method, y= self_FDR, colour = qvalue_cutoff))+geom_boxplot()+theme_bw() +theme_bw() +xlab("Method")+ylab("Self-FDR")+ theme(panel.border = element_blank(), panel.grid.major = element_blank(),
panel.grid.minor = element_blank(), axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=20, face="bold", hjust=0.5),
axis.title.y=element_text(size=20, face="bold", vjust=0.4, angle=90),
legend.title=element_text(size = 15,face = "bold"),legend.text = element_text(size = 18, face = "bold",family = "arial"),
axis.text = element_text(size = 15,face = "bold",colour = "black") , axis.ticks = element_line(colour = "black",size = 1) )+scale_color_discrete(name = "qvalue\ncutoff")
Compare normalization
selfFDR_cov_pos <- NULL
samplenames <- RADAR$samplenames
set.seed(10)
for(i in 1:15){
## Sample subset
sample_id <- c(sample(x = 1:7, size = 5,replace = F),7+sample(x = 1:6, size = 4,replace = F) )
Truth_id <- setdiff(1:13,sample_id)
## run radar
radar_sample <- diffIP_parallel(RADAR_pos[1:14],Covariates = cov ,exclude = samplenames[setdiff(1:13,sample_id)],thread = 25,plotPvalue = F)$all.est
radar_sample <- cbind(radar_sample, "qvalue"= qvalue::qvalue(radar_sample[,"p_value3"])$qvalue )
radar_truth <- RADAR_pos_cov
radar_truth <- cbind(radar_truth, "qvalue"= qvalue::qvalue(radar_truth[,"p_value3"])$qvalue )
radar_selfFDR <- sapply(c(0.1,0.05,0.01),function(x){length( which(! which(radar_sample[,"qvalue"]<x) %in% which(radar_truth[,"qvalue"]<0.1) ) )/length(which(radar_sample[,"qvalue"]<x))})
## run DESeq2
deseq_sample <- DESeq2(countdata = RADAR_pos$ip_adjExpr_filtered[,sample_id],pheno = matrix(c(rep(0,7),rep(1,6))[sample_id],ncol = 1),covariates = as.matrix(data.frame(age =cov[sample_id])) )
deseq_sample$qvalue <- qvalue::qvalue( deseq_sample$pvalue )$qvalue
deseq_truth <- deseq2.res_pos_cov
deseq_truth$qvalue <- qvalue::qvalue( deseq_truth$pvalue )$qvalue
deseq_selfFDR <- sapply(c(0.1,0.05,0.01),function(x){length( which(! which(deseq_sample[,"qvalue"]<x) %in% which(deseq_truth[,"qvalue"]<0.1) ) )/length(which(deseq_sample[,"qvalue"]<x))})
## run edgeR
edgeR_sample <- edgeR(countdata = RADAR_pos$ip_adjExpr_filtered[,sample_id],pheno = matrix(c(rep(0,7),rep(1,6))[sample_id],ncol = 1),covariates = as.matrix(data.frame(batch =cov[sample_id])) )
edgeR_sample$qvalue <- qvalue::qvalue( edgeR_sample$pvalue )$qvalue
edgeR_truth <- edgeR.res_pos_cov
edgeR_truth$qvalue <- qvalue::qvalue( edgeR_truth$pvalue )$qvalue
edgeR_selfFDR <- sapply(c(0.1,0.05,0.01),function(x){length( which(! which(edgeR_sample[,"qvalue"]<x) %in% which(edgeR_truth[,"qvalue"]<0.1) ) )/length(which(edgeR_sample[,"qvalue"]<x))})
fdr <- rbind( data.frame(self_FDR = radar_selfFDR, qvalue_cutoff=c("0.1","0.05","0.01"), method = rep("RADAR",3) ),
data.frame(self_FDR = deseq_selfFDR, qvalue_cutoff=c("0.1","0.05","0.01"), method = rep("DESeq2",3) ),
data.frame(self_FDR = edgeR_selfFDR, qvalue_cutoff=c("0.1","0.05","0.01"), method = rep("edgeR",3) )
)
selfFDR_cov_pos <- rbind( selfFDR_cov_pos, fdr )
}
selfFDR_cov$qvalue_cutoff <- factor(selfFDR_cov$qvalue_cutoff,levels = c("0.1","0.05","0.01"))
save(selfFDR_cov_pos, file = "~/PoissonGamma_Method/Ovary_Cancer/selfFDR_cov_pos.RData")load("~/PoissonGamma_Method/Ovary_Cancer/selfFDR_cov_pos.RData")
selfFDR_all <- cbind(rbind(selfFDR_cov, selfFDR_cov_pos), "NormBy"= c(rep("By gene-level count",nrow(selfFDR_cov)),rep("By bin-level count",nrow(selfFDR_cov_pos))) )
selfFDR_all$NormBy <- factor(selfFDR_all$NormBy,levels = c("By gene-level count","By bin-level count"))
ggplot(data = selfFDR_all, aes(x=method, y= self_FDR, colour = qvalue_cutoff))+geom_boxplot()+facet_grid(~NormBy)+theme_bw() +xlab("Method")+ylab("Self-FDR")+ theme( panel.grid.major = element_blank(),
axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=20, face="bold", hjust=0.5,family = "arial"),
axis.title.y=element_text(size=20, face="bold", vjust=0.4, angle=90,family = "arial"),
legend.title=element_text(size = 15,face = "bold"),legend.text = element_text(size = 18, face = "bold",family = "arial"),
axis.text = element_text(size = 18,face = "bold",family = "arial",colour = "black") ,strip.text.x = element_text(size = 20,face = "bold") )+scale_color_discrete(name = "qvalue\ncutoff")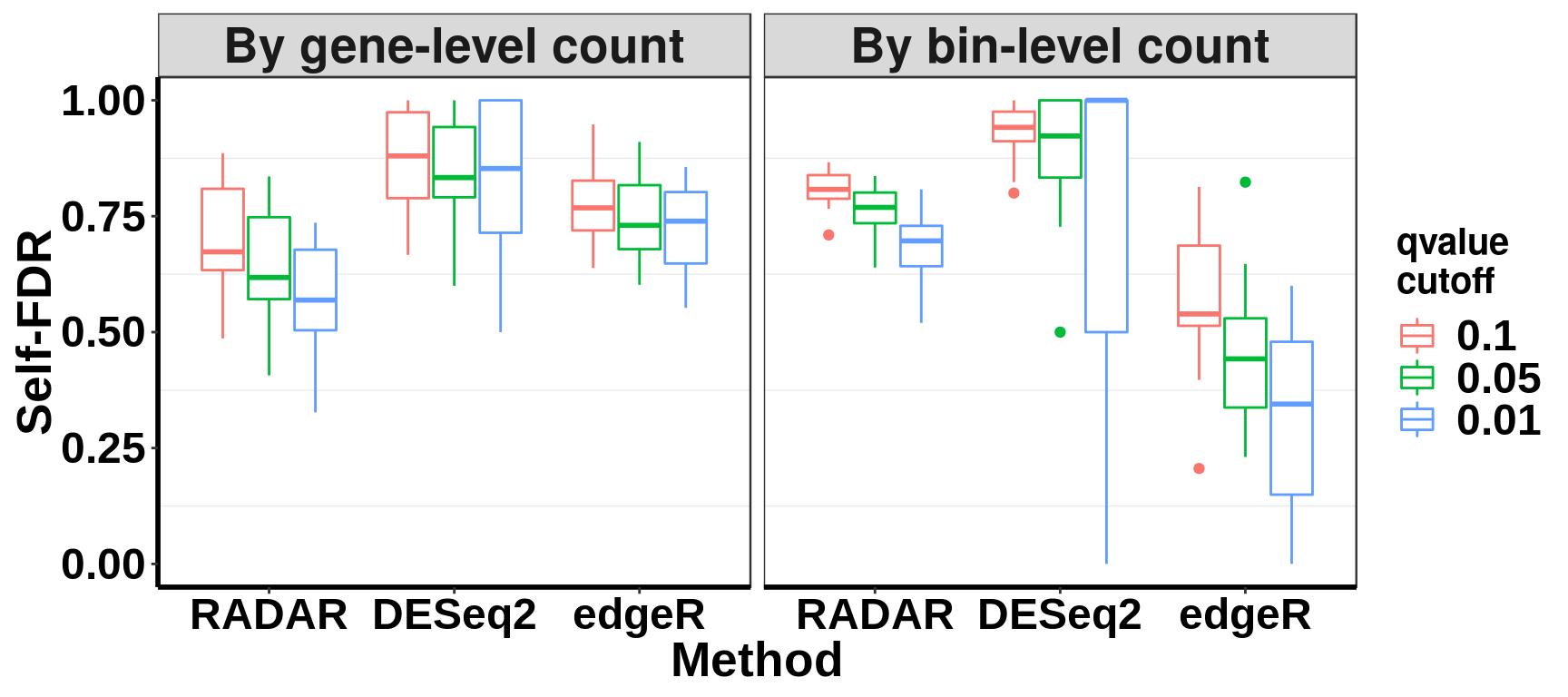
Permutation Test
Because covariate “age” is continuous variable, which cannot be balanced when permuting diseases status. Therefore, permutation test is performed without covariate.
radar_permuted_P <-NULL
deseq_permuted_P <- NULL
edgeR_permuted_P <- NULL
qnb_permuted_P <- NULL
set.seed(2)
for(i in 1:15){
permute_X <- RADAR$X
permute_X[sample(1:7,4)] <- unique(RADAR$X)[2]
permute_X[sample(8:13,3)] <- unique(RADAR$X)[1]
radar_permute <- diffIP_parallel(c(RADAR[1:11],list("X"=permute_X),RADAR[13:14]) ,thread = 15,plotPvalue = F)$all.est[,"p_value"]
deseq_permute <- DESeq2(countdata = RADAR$ip_adjExpr_filtered,pheno = matrix(permute_X,ncol = 1) )$pvalue
edgeR_permute <- edgeR(countdata = RADAR$ip_adjExpr_filtered,pheno = matrix(permute_X,ncol = 1) )$pvalue
qnb_permute <- QNB::qnbtest(control_ip = RADAR$reads[filteredBins,(which(permute_X == "Ctl" )+13)],
treated_ip = RADAR$reads[filteredBins,(which(permute_X == "Tumor" )+13)],
control_input = RADAR$reads[filteredBins,which(permute_X == "Ctl" )],
treated_input = RADAR$reads[filteredBins,which(permute_X == "Tumor" )],plot.dispersion = FALSE)$pvalue
radar_permuted_P <- cbind(radar_permuted_P, radar_permute)
deseq_permuted_P <- cbind(deseq_permuted_P,deseq_permute)
edgeR_permuted_P <- cbind(edgeR_permuted_P,edgeR_permute)
qnb_permuted_P <- cbind(qnb_permuted_P, qnb_permute)
}
for( i in 1:ncol(radar_permuted_P)){
par(mfrow=c(1,3))
hist(RADAR$all.est[,"p_value"], col =rgb(1,0,0,0.4),main = "RADAR", xlab = "p value",cex.main = 2.5,cex.axis =2,cex.lab=2)
hist(radar_permuted_P[,i],col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 10,bty="n")
hist(deseq2.res$pvalue, col =rgb(1,0,0,0.4),main = "DESeq2",xlab = "p value",cex.main = 2.5,cex.axis =2,cex.lab=2)
hist(deseq_permuted_P[,i],col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 10,bty="n")
hist(edgeR.res$pvalue, col =rgb(1,0,0,0.4),main = "edgeR",xlab = "p value",cex.main = 2.5,cex.axis =2,cex.lab=2)
hist(edgeR_permuted_P[,i],col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 10,bty="n")
}par(mfrow=c(1,3))
hist(RADAR$all.est[,"p_value"], col =rgb(1,0,0,0.4),main = "RADAR", xlab = "p value",font=2, cex.lab=2.5, cex.axis = 1.5 , font.lab=2 ,cex.main = 3, lwd = 3)
tmp <- hist(c(radar_permuted_P),plot = F)
tmp$counts <- tmp$counts/15
plot(tmp,col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 12,bty="n",text.font = 2, cex = 1.5)
y.scale <- max(hist(RADAR$all.est[,"p_value"],plot = F)$counts)
tmp <- hist(c(deseq_permuted_P),plot = F)
tmp$counts <- tmp$counts/15
hist(deseq2.res$pvalue, col =rgb(1,0,0,0.4),main = "DESeq2",xlab = "p value" ,font=2, cex.lab=2.5, cex.axis = 1.5 , font.lab=2 ,cex.main = 3, lwd = 3, ylim = c(0,y.scale) )
plot(tmp,col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 12,bty="n",text.font = 2, cex = 1.5)
hist(edgeR.res$pvalue, col =rgb(1,0,0,0.4),main = "edgeR",xlab = "p value",font=2, cex.lab=2.5, cex.axis = 1.5 , font.lab=2 ,cex.main = 3,lwd = 3,ylim = c(0,y.scale))
tmp <- hist(c(edgeR_permuted_P),plot = F)
tmp$counts <- tmp$counts/15
plot(tmp,col=rgb(0,0,1,0.5),add = T)
legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 12,bty="n",text.font = 2, cex = 1.5)
#tmp <- hist(c(qnb_permuted_P),plot = F)
#tmp$counts <- tmp$counts/15
#hist(QNB.res$pvalue, col =rgb(1,0,0,0.4),main = "QNB",xlab = "p value",font=2, cex.lab=1.5, font.lab=2 ,cex.main = 2, ylim = c(0,max(tmp$counts) ) )
#plot(tmp,col=rgb(0,0,1,0.5),add = T)
#legend("topright", c("Original", "Permuted"), col=c(rgb(1,0,0,0.4), rgb(0,0,1,0.5)), lwd = 10,bty="n",text.font = 2)Power analysis with different sample size
radar_truth <- RADAR_cov
radar_truth <- cbind(radar_truth, "qvalue"= qvalue::qvalue(radar_truth[,"p_value3"])$qvalue )
deseq_truth <- deseq2.res_cov
deseq_truth$qvalue <- qvalue::qvalue( deseq_truth$pvalue )$qvalue
edgeR_truth <- edgeR.res_cov
edgeR_truth$qvalue <- qvalue::qvalue( edgeR_truth$pvalue )$qvalue
## test N=6
selfConsist_cov_6s <- NULL
num_sigBins_6s <- NULL
selfFDR_cov_6s <- NULL
selfPower_cov_6s <- NULL
set.seed(11)
for(i in 1:10){
## Sample subset
sample_id1 <- c(sample(x = 1:7, size = 6,replace = F),7+sample(x = 1:6, size = 5,replace = F) )
sample_id2 <- c(sample(x = 1:7, size = 6,replace = F),7+sample(x = 1:6, size = 5,replace = F) )
if(sample_id1 == sample_id2) next
## run radar
radar_sample1 <- diffIP_parallel(RADAR[1:14],Covariates = cov ,exclude = samplenames[setdiff(1:13,sample_id1)],thread = 25,plotPvalue = F)$all.est
radar_sample1 <- cbind(radar_sample1, "qvalue"= qvalue::qvalue(radar_sample1[,"p_value3"])$qvalue )
radar_sample2 <- diffIP_parallel(RADAR[1:14],Covariates = cov ,exclude = samplenames[setdiff(1:13,sample_id2)],thread = 25,plotPvalue = F)$all.est
radar_sample2 <- cbind(radar_sample2, "qvalue"= qvalue::qvalue(radar_sample2[,"p_value3"])$qvalue )
radar_selfCon <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ 2 * length( intersect( which(radar_sample1[,"qvalue"]<x), which(radar_sample2[,"qvalue"]<x) ) )/(length(which(radar_sample1[,"qvalue"]<x))+length(which(radar_sample2[,"qvalue"]<x)) ) } )
radar_num_sigBins <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ round( (length(which(radar_sample1[,"qvalue"]<x))+length(which(radar_sample2[,"qvalue"]<x)) )/2 ) } )
radar_selfFDR <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which(! which(radar_sample1[,"qvalue"]<x) %in% which(radar_truth[,"qvalue"]<0.1) ) )/length(which(radar_sample1[,"qvalue"]<x))})
radar_power <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which( which(radar_truth[,"qvalue"]<0.1) %in% which(radar_sample1[,"qvalue"]<x) ) )/length(which(radar_truth[,"qvalue"]<0.1) ) } )
## run DESeq2
deseq_sample1 <- DESeq2(countdata = RADAR$ip_adjExpr_filtered[,sample_id1],pheno = matrix(c(rep(0,7),rep(1,6))[sample_id1],ncol = 1),covariates = as.matrix(data.frame(age =cov[sample_id1])) )
deseq_sample1$qvalue <- qvalue::qvalue( deseq_sample1$pvalue )$qvalue
deseq_sample2 <- DESeq2(countdata = RADAR$ip_adjExpr_filtered[,sample_id2],pheno = matrix(c(rep(0,7),rep(1,6))[sample_id2],ncol = 1),covariates = as.matrix(data.frame(age =cov[sample_id2])) )
deseq_sample2$qvalue <- qvalue::qvalue( deseq_sample2$pvalue )$qvalue
deseq_selfCon <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ 2 * length( intersect( which(deseq_sample1[,"qvalue"]<x), which(deseq_sample2[,"qvalue"]<x) ) )/(length(which(deseq_sample1[,"qvalue"]<x)) + (length(which(deseq_sample2[,"qvalue"]<x)) )) })
deseq_num_sigBins <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ round( (length(which(deseq_sample1[,"qvalue"]<x))+length(which(deseq_sample2[,"qvalue"]<x)) )/2 ) } )
deseq_selfFDR <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which(! which(deseq_sample1[,"qvalue"]<x) %in% which(deseq_truth[,"qvalue"]<0.1) ) )/length(which(deseq_sample1[,"qvalue"]<x))})
deseq_power <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which( which(deseq_truth[,"qvalue"]<0.1) %in% which(deseq_sample1[,"qvalue"]<x) ) )/length(which(deseq_truth[,"qvalue"]<0.1) ) })
## run edgeR
edgeR_sample1 <- edgeR(countdata = RADAR$ip_adjExpr_filtered[,sample_id1],pheno = matrix(c(rep(0,7),rep(1,6))[sample_id1],ncol = 1),covariates = as.matrix(data.frame(age =cov[sample_id1])) )
edgeR_sample1$qvalue <- qvalue::qvalue( edgeR_sample1$pvalue )$qvalue
edgeR_sample2 <- edgeR(countdata = RADAR$ip_adjExpr_filtered[,sample_id2],pheno = matrix(c(rep(0,7),rep(1,6))[sample_id2],ncol = 1),covariates = as.matrix(data.frame(age =cov[sample_id2])) )
edgeR_sample2$qvalue <- qvalue::qvalue( edgeR_sample2$pvalue )$qvalue
edgeR_selfCon <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ 2 * length( intersect( which(edgeR_sample1[,"qvalue"]<x), which(edgeR_sample2[,"qvalue"]<x) ) )/(length(which(edgeR_sample1[,"qvalue"]<x)) + (length(which(edgeR_sample2[,"qvalue"]<x)) )) })
edgeR_num_sigBins <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ round( (length(which(edgeR_sample1[,"qvalue"]<x))+length(which(edgeR_sample2[,"qvalue"]<x)) )/2 ) } )
edgeR_selfFDR <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which(! which(edgeR_sample1[,"qvalue"]<x) %in% which(edgeR_truth[,"qvalue"]<0.1) ) )/length(which(edgeR_sample1[,"qvalue"]<x))})
edgeR_power <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which( which(edgeR_truth[,"qvalue"]<0.1) %in% which(edgeR_sample1[,"qvalue"]<x) ) )/length(which(edgeR_truth[,"qvalue"]<0.1) ) })
consistency <- rbind( data.frame(self_Consistency = radar_selfCon, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "RADAR" ),
data.frame(self_Consistency = deseq_selfCon, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "DESeq2" ),
data.frame(self_Consistency = edgeR_selfCon, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "edgeR" )
)
num_hits <- rbind( data.frame(num_sigBins = radar_num_sigBins, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "RADAR" ),
data.frame(num_sigBins = deseq_num_sigBins, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "DESeq2" ),
data.frame(num_sigBins = edgeR_num_sigBins, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "edgeR" )
)
fdr <- rbind( data.frame(self_FDR = radar_selfFDR, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "RADAR" ),
data.frame(self_FDR = deseq_selfFDR, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "DESeq2" ),
data.frame(self_FDR = edgeR_selfFDR, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "edgeR" )
)
power <- rbind( data.frame(power = radar_power, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "RADAR" ),
data.frame(power = deseq_power, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "DESeq2" ),
data.frame(power = edgeR_power, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "edgeR" )
)
selfConsist_cov_6s <- rbind( selfConsist_cov_6s, consistency )
num_sigBins_6s <- rbind(num_sigBins_6s, num_hits)
selfFDR_cov_6s <- rbind(selfFDR_cov_6s, fdr)
selfPower_cov_6s <- rbind(selfPower_cov_6s, power)
}
## test N=5
selfConsist_cov_5s <- NULL
num_sigBins_5s <- NULL
selfFDR_cov_5s <- NULL
selfPower_cov_5s <- NULL
set.seed(11)
for(i in 1:10){
## Sample subset
sample_id1 <- c(sample(x = 1:7, size = 5,replace = F),7+sample(x = 1:6, size = 5,replace = F) )
sample_id2 <- c(sample(x = 1:7, size = 5,replace = F),7+sample(x = 1:6, size = 5,replace = F) )
if(sample_id1 == sample_id2) next
## run radar
radar_sample1 <- diffIP_parallel(RADAR[1:14],Covariates = cov ,exclude = samplenames[setdiff(1:13,sample_id1)],thread = 25,plotPvalue = F)$all.est
radar_sample1 <- cbind(radar_sample1, "qvalue"= qvalue::qvalue(radar_sample1[,"p_value3"])$qvalue )
radar_sample2 <- diffIP_parallel(RADAR[1:14],Covariates = cov ,exclude = samplenames[setdiff(1:13,sample_id2)],thread = 25,plotPvalue = F)$all.est
radar_sample2 <- cbind(radar_sample2, "qvalue"= qvalue::qvalue(radar_sample2[,"p_value3"])$qvalue )
radar_selfCon <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ 2 * length( intersect( which(radar_sample1[,"qvalue"]<x), which(radar_sample2[,"qvalue"]<x) ) )/(length(which(radar_sample1[,"qvalue"]<x))+length(which(radar_sample2[,"qvalue"]<x)) ) } )
radar_num_sigBins <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ round( (length(which(radar_sample1[,"qvalue"]<x))+length(which(radar_sample2[,"qvalue"]<x)) )/2 ) } )
radar_selfFDR <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which(! which(radar_sample1[,"qvalue"]<x) %in% which(radar_truth[,"qvalue"]<0.1) ) )/length(which(radar_sample1[,"qvalue"]<x))})
radar_power <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which( which(radar_truth[,"qvalue"]<0.1) %in% which(radar_sample1[,"qvalue"]<x) ) )/length(which(radar_truth[,"qvalue"]<0.1) ) } )
## run DESeq2
deseq_sample1 <- DESeq2(countdata = RADAR$ip_adjExpr_filtered[,sample_id1],pheno = matrix(c(rep(0,7),rep(1,6))[sample_id1],ncol = 1),covariates = as.matrix(data.frame(age =cov[sample_id1])) )
deseq_sample1$qvalue <- qvalue::qvalue( deseq_sample1$pvalue )$qvalue
deseq_sample2 <- DESeq2(countdata = RADAR$ip_adjExpr_filtered[,sample_id2],pheno = matrix(c(rep(0,7),rep(1,6))[sample_id2],ncol = 1),covariates = as.matrix(data.frame(age =cov[sample_id2])) )
deseq_sample2$qvalue <- qvalue::qvalue( deseq_sample2$pvalue )$qvalue
deseq_selfCon <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ 2 * length( intersect( which(deseq_sample1[,"qvalue"]<x), which(deseq_sample2[,"qvalue"]<x) ) )/(length(which(deseq_sample1[,"qvalue"]<x)) + (length(which(deseq_sample2[,"qvalue"]<x)) )) })
deseq_num_sigBins <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ round( (length(which(deseq_sample1[,"qvalue"]<x))+length(which(deseq_sample2[,"qvalue"]<x)) )/2 ) } )
deseq_selfFDR <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which(! which(deseq_sample1[,"qvalue"]<x) %in% which(deseq_truth[,"qvalue"]<0.1) ) )/length(which(deseq_sample1[,"qvalue"]<x))})
deseq_power <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which( which(deseq_truth[,"qvalue"]<0.1) %in% which(deseq_sample1[,"qvalue"]<x) ) )/length(which(deseq_truth[,"qvalue"]<0.1) ) })
## run edgeR
edgeR_sample1 <- edgeR(countdata = RADAR$ip_adjExpr_filtered[,sample_id1],pheno = matrix(c(rep(0,7),rep(1,6))[sample_id1],ncol = 1),covariates = as.matrix(data.frame(age =cov[sample_id1])) )
edgeR_sample1$qvalue <- qvalue::qvalue( edgeR_sample1$pvalue )$qvalue
edgeR_sample2 <- edgeR(countdata = RADAR$ip_adjExpr_filtered[,sample_id2],pheno = matrix(c(rep(0,7),rep(1,6))[sample_id2],ncol = 1),covariates = as.matrix(data.frame(age =cov[sample_id2])) )
edgeR_sample2$qvalue <- qvalue::qvalue( edgeR_sample2$pvalue )$qvalue
edgeR_selfCon <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ 2 * length( intersect( which(edgeR_sample1[,"qvalue"]<x), which(edgeR_sample2[,"qvalue"]<x) ) )/(length(which(edgeR_sample1[,"qvalue"]<x)) + (length(which(edgeR_sample2[,"qvalue"]<x)) )) })
edgeR_num_sigBins <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ round( (length(which(edgeR_sample1[,"qvalue"]<x))+length(which(edgeR_sample2[,"qvalue"]<x)) )/2 ) } )
edgeR_selfFDR <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which(! which(edgeR_sample1[,"qvalue"]<x) %in% which(edgeR_truth[,"qvalue"]<0.1) ) )/length(which(edgeR_sample1[,"qvalue"]<x))})
edgeR_power <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which( which(edgeR_truth[,"qvalue"]<0.1) %in% which(edgeR_sample1[,"qvalue"]<x) ) )/length(which(edgeR_truth[,"qvalue"]<0.1) ) })
consistency <- rbind( data.frame(self_Consistency = radar_selfCon, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "RADAR" ),
data.frame(self_Consistency = deseq_selfCon, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "DESeq2" ),
data.frame(self_Consistency = edgeR_selfCon, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "edgeR" )
)
num_hits <- rbind( data.frame(num_sigBins = radar_num_sigBins, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "RADAR" ),
data.frame(num_sigBins = deseq_num_sigBins, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "DESeq2" ),
data.frame(num_sigBins = edgeR_num_sigBins, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "edgeR" )
)
fdr <- rbind( data.frame(self_FDR = radar_selfFDR, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "RADAR" ),
data.frame(self_FDR = deseq_selfFDR, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "DESeq2" ),
data.frame(self_FDR = edgeR_selfFDR, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "edgeR" )
)
power <- rbind( data.frame(power = radar_power, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "RADAR" ),
data.frame(power = deseq_power, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "DESeq2" ),
data.frame(power = edgeR_power, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "edgeR" )
)
selfConsist_cov_5s <- rbind( selfConsist_cov_5s, consistency )
num_sigBins_5s <- rbind(num_sigBins_5s, num_hits)
selfFDR_cov_5s <- rbind(selfFDR_cov_5s, fdr)
selfPower_cov_5s <- rbind(selfPower_cov_5s, power)
}
## test N=4
selfConsist_cov_4s <- NULL
num_sigBins_4s <- NULL
selfFDR_cov_4s <- NULL
selfPower_cov_4s <- NULL
set.seed(11)
for(i in 1:10){
## Sample subset
tmp_sample <- sample(x = 1:6, size = 5,replace = F)
sample_id1 <- c(sample(x = 1:7, size = 4,replace = F),7+sample(x = 1:6, size = 4,replace = F) )
sample_id2 <- c(sample(x = 1:7, size = 4,replace = F),7+sample(x = 1:6, size = 4,replace = F) )
if(sample_id1 == sample_id2) next
## run radar
radar_sample1 <- diffIP_parallel(RADAR[1:14],Covariates = cov ,exclude = samplenames[setdiff(1:13,sample_id1)],thread = 25,plotPvalue = F)$all.est
radar_sample1 <- cbind(radar_sample1, "qvalue"= qvalue::qvalue(radar_sample1[,"p_value3"])$qvalue )
radar_sample2 <- diffIP_parallel(RADAR[1:14],Covariates = cov ,exclude = samplenames[setdiff(1:13,sample_id2)],thread = 25,plotPvalue = F)$all.est
radar_sample2 <- cbind(radar_sample2, "qvalue"= qvalue::qvalue(radar_sample2[,"p_value3"])$qvalue )
radar_selfCon <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ 2 * length( intersect( which(radar_sample1[,"qvalue"]<x), which(radar_sample2[,"qvalue"]<x) ) )/(length(which(radar_sample1[,"qvalue"]<x))+length(which(radar_sample2[,"qvalue"]<x)) ) } )
radar_num_sigBins <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ round( (length(which(radar_sample1[,"qvalue"]<x))+length(which(radar_sample2[,"qvalue"]<x)) )/2 ) } )
radar_selfFDR <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which(! which(radar_sample1[,"qvalue"]<x) %in% which(radar_truth[,"qvalue"]<0.1) ) )/length(which(radar_sample1[,"qvalue"]<x))})
radar_power <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which( which(radar_truth[,"qvalue"]<0.1) %in% which(radar_sample1[,"qvalue"]<x) ) )/length(which(radar_truth[,"qvalue"]<0.1) ) } )
## run DESeq2
deseq_sample1 <- DESeq2(countdata = RADAR$ip_adjExpr_filtered[,sample_id1],pheno = matrix(c(rep(0,7),rep(1,6))[sample_id1],ncol = 1),covariates = as.matrix(data.frame(age =cov[sample_id1])) )
deseq_sample1$qvalue <- qvalue::qvalue( deseq_sample1$pvalue )$qvalue
deseq_sample2 <- DESeq2(countdata = RADAR$ip_adjExpr_filtered[,sample_id2],pheno = matrix(c(rep(0,7),rep(1,6))[sample_id2],ncol = 1),covariates = as.matrix(data.frame(age =cov[sample_id2])) )
deseq_sample2$qvalue <- qvalue::qvalue( deseq_sample2$pvalue )$qvalue
deseq_selfCon <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ 2 * length( intersect( which(deseq_sample1[,"qvalue"]<x), which(deseq_sample2[,"qvalue"]<x) ) )/(length(which(deseq_sample1[,"qvalue"]<x)) + (length(which(deseq_sample2[,"qvalue"]<x)) )) })
deseq_num_sigBins <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ round( (length(which(deseq_sample1[,"qvalue"]<x))+length(which(deseq_sample2[,"qvalue"]<x)) )/2 ) } )
deseq_selfFDR <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which(! which(deseq_sample1[,"qvalue"]<x) %in% which(deseq_truth[,"qvalue"]<0.1) ) )/length(which(deseq_sample1[,"qvalue"]<x))})
deseq_power <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which( which(deseq_truth[,"qvalue"]<0.1) %in% which(deseq_sample1[,"qvalue"]<x) ) )/length(which(deseq_truth[,"qvalue"]<0.1) ) })
## run edgeR
edgeR_sample1 <- edgeR(countdata = RADAR$ip_adjExpr_filtered[,sample_id1],pheno = matrix(c(rep(0,7),rep(1,6))[sample_id1],ncol = 1),covariates = as.matrix(data.frame(age =cov[sample_id1])) )
edgeR_sample1$qvalue <- qvalue::qvalue( edgeR_sample1$pvalue )$qvalue
edgeR_sample2 <- edgeR(countdata = RADAR$ip_adjExpr_filtered[,sample_id2],pheno = matrix(c(rep(0,7),rep(1,6))[sample_id2],ncol = 1),covariates = as.matrix(data.frame(age =cov[sample_id2])) )
edgeR_sample2$qvalue <- qvalue::qvalue( edgeR_sample2$pvalue )$qvalue
edgeR_selfCon <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ 2 * length( intersect( which(edgeR_sample1[,"qvalue"]<x), which(edgeR_sample2[,"qvalue"]<x) ) )/(length(which(edgeR_sample1[,"qvalue"]<x)) + (length(which(edgeR_sample2[,"qvalue"]<x)) )) })
edgeR_num_sigBins <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ round( (length(which(edgeR_sample1[,"qvalue"]<x))+length(which(edgeR_sample2[,"qvalue"]<x)) )/2 ) } )
edgeR_selfFDR <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which(! which(edgeR_sample1[,"qvalue"]<x) %in% which(edgeR_truth[,"qvalue"]<0.1) ) )/length(which(edgeR_sample1[,"qvalue"]<x))})
edgeR_power <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which( which(edgeR_truth[,"qvalue"]<0.1) %in% which(edgeR_sample1[,"qvalue"]<x) ) )/length(which(edgeR_truth[,"qvalue"]<0.1) ) })
consistency <- rbind( data.frame(self_Consistency = radar_selfCon, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "RADAR" ),
data.frame(self_Consistency = deseq_selfCon, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "DESeq2" ),
data.frame(self_Consistency = edgeR_selfCon, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "edgeR" )
)
num_hits <- rbind( data.frame(num_sigBins = radar_num_sigBins, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "RADAR" ),
data.frame(num_sigBins = deseq_num_sigBins, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "DESeq2" ),
data.frame(num_sigBins = edgeR_num_sigBins, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "edgeR" )
)
fdr <- rbind( data.frame(self_FDR = radar_selfFDR, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "RADAR" ),
data.frame(self_FDR = deseq_selfFDR, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "DESeq2" ),
data.frame(self_FDR = edgeR_selfFDR, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "edgeR" )
)
power <- rbind( data.frame(power = radar_power, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "RADAR" ),
data.frame(power = deseq_power, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "DESeq2" ),
data.frame(power = edgeR_power, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "edgeR" )
)
selfConsist_cov_4s <- rbind( selfConsist_cov_4s, consistency )
num_sigBins_4s <- rbind(num_sigBins_4s, num_hits)
selfFDR_cov_4s <- rbind(selfFDR_cov_4s, fdr)
selfPower_cov_4s <- rbind(selfPower_cov_4s, power)
}
## test N=3
selfConsist_cov_3s <- NULL
num_sigBins_3s <- NULL
selfFDR_cov_3s <- NULL
selfPower_cov_3s <- NULL
set.seed(11)
for(i in 1:10){
## Sample subset
tmp_sample <- sample(x = 1:6, size = 4,replace = F)
sample_id1 <- c(sample(x = 1:7, size = 3,replace = F),7+sample(x = 1:6, size = 3,replace = F) )
sample_id2 <- c(sample(x = 1:7, size = 3,replace = F),7+sample(x = 1:6, size = 3,replace = F) )
if(sample_id1 == sample_id2) next
## run radar
radar_sample1 <- diffIP_parallel(RADAR[1:14],Covariates = cov ,exclude = samplenames[setdiff(1:13,sample_id1)],thread = 25,plotPvalue = F)$all.est
radar_sample1 <- cbind(radar_sample1, "qvalue"= qvalue::qvalue(radar_sample1[,"p_value3"])$qvalue )
radar_sample2 <- diffIP_parallel(RADAR[1:14],Covariates = cov ,exclude = samplenames[setdiff(1:13,sample_id2)],thread = 25,plotPvalue = F)$all.est
radar_sample2 <- cbind(radar_sample2, "qvalue"= qvalue::qvalue(radar_sample2[,"p_value3"])$qvalue )
radar_selfCon <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ 2 * length( intersect( which(radar_sample1[,"qvalue"]<x), which(radar_sample2[,"qvalue"]<x) ) )/(length(which(radar_sample1[,"qvalue"]<x))+length(which(radar_sample2[,"qvalue"]<x)) ) } )
radar_num_sigBins <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ round( (length(which(radar_sample1[,"qvalue"]<x))+length(which(radar_sample2[,"qvalue"]<x)) )/2 ) } )
radar_selfFDR <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which(! which(radar_sample1[,"qvalue"]<x) %in% which(radar_truth[,"qvalue"]<0.1) ) )/length(which(radar_sample1[,"qvalue"]<x))})
radar_power <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which( which(radar_truth[,"qvalue"]<0.1) %in% which(radar_sample1[,"qvalue"]<x) ) )/length(which(radar_truth[,"qvalue"]<0.1) ) } )
## run DESeq2
deseq_sample1 <- DESeq2(countdata = RADAR$ip_adjExpr_filtered[,sample_id1],pheno = matrix(c(rep(0,7),rep(1,6))[sample_id1],ncol = 1),covariates = as.matrix(data.frame(age =cov[sample_id1])) )
deseq_sample1$qvalue <- qvalue::qvalue( deseq_sample1$pvalue )$qvalue
deseq_sample2 <- DESeq2(countdata = RADAR$ip_adjExpr_filtered[,sample_id2],pheno = matrix(c(rep(0,7),rep(1,6))[sample_id2],ncol = 1),covariates = as.matrix(data.frame(age =cov[sample_id2])) )
deseq_sample2$qvalue <- qvalue::qvalue( deseq_sample2$pvalue )$qvalue
deseq_selfCon <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ 2 * length( intersect( which(deseq_sample1[,"qvalue"]<x), which(deseq_sample2[,"qvalue"]<x) ) )/(length(which(deseq_sample1[,"qvalue"]<x)) + (length(which(deseq_sample2[,"qvalue"]<x)) )) })
deseq_num_sigBins <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ round( (length(which(deseq_sample1[,"qvalue"]<x))+length(which(deseq_sample2[,"qvalue"]<x)) )/2 ) } )
deseq_selfFDR <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which(! which(deseq_sample1[,"qvalue"]<x) %in% which(deseq_truth[,"qvalue"]<0.1) ) )/length(which(deseq_sample1[,"qvalue"]<x))})
deseq_power <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which( which(deseq_truth[,"qvalue"]<0.1) %in% which(deseq_sample1[,"qvalue"]<x) ) )/length(which(deseq_truth[,"qvalue"]<0.1) ) })
## run edgeR
edgeR_sample1 <- edgeR(countdata = RADAR$ip_adjExpr_filtered[,sample_id1],pheno = matrix(c(rep(0,7),rep(1,6))[sample_id1],ncol = 1),covariates = as.matrix(data.frame(age =cov[sample_id1])) )
edgeR_sample1$qvalue <- qvalue::qvalue( edgeR_sample1$pvalue )$qvalue
edgeR_sample2 <- edgeR(countdata = RADAR$ip_adjExpr_filtered[,sample_id2],pheno = matrix(c(rep(0,7),rep(1,6))[sample_id2],ncol = 1),covariates = as.matrix(data.frame(age =cov[sample_id2])) )
edgeR_sample2$qvalue <- qvalue::qvalue( edgeR_sample2$pvalue )$qvalue
edgeR_selfCon <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ 2 * length( intersect( which(edgeR_sample1[,"qvalue"]<x), which(edgeR_sample2[,"qvalue"]<x) ) )/(length(which(edgeR_sample1[,"qvalue"]<x)) + (length(which(edgeR_sample2[,"qvalue"]<x)) )) })
edgeR_num_sigBins <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){ round( (length(which(edgeR_sample1[,"qvalue"]<x))+length(which(edgeR_sample2[,"qvalue"]<x)) )/2 ) } )
edgeR_selfFDR <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which(! which(edgeR_sample1[,"qvalue"]<x) %in% which(edgeR_truth[,"qvalue"]<0.1) ) )/length(which(edgeR_sample1[,"qvalue"]<x))})
edgeR_power <- sapply(c(0.5,0.2,0.1,0.05,0.01),function(x){length( which( which(edgeR_truth[,"qvalue"]<0.1) %in% which(edgeR_sample1[,"qvalue"]<x) ) )/length(which(edgeR_truth[,"qvalue"]<0.1) ) })
consistency <- rbind( data.frame(self_Consistency = radar_selfCon, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "RADAR" ),
data.frame(self_Consistency = deseq_selfCon, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "DESeq2" ),
data.frame(self_Consistency = edgeR_selfCon, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "edgeR" )
)
num_hits <- rbind( data.frame(num_sigBins = radar_num_sigBins, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "RADAR" ),
data.frame(num_sigBins = deseq_num_sigBins, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "DESeq2" ),
data.frame(num_sigBins = edgeR_num_sigBins, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "edgeR" )
)
fdr <- rbind( data.frame(self_FDR = radar_selfFDR, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "RADAR" ),
data.frame(self_FDR = deseq_selfFDR, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "DESeq2" ),
data.frame(self_FDR = edgeR_selfFDR, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "edgeR" )
)
power <- rbind( data.frame(power = radar_power, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "RADAR" ),
data.frame(power = deseq_power, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "DESeq2" ),
data.frame(power = edgeR_power, qvalue_cutoff=c("0.5","0.2","0.1","0.05","0.01"), method = "edgeR" )
)
selfConsist_cov_3s <- rbind( selfConsist_cov_3s, consistency )
num_sigBins_3s <- rbind(num_sigBins_3s, num_hits)
selfFDR_cov_3s <- rbind(selfFDR_cov_3s, fdr)
selfPower_cov_3s <- rbind(selfPower_cov_3s, power)
}
all_num_sigBins <- cbind(rbind(num_sigBins_3s,num_sigBins_4s,num_sigBins_5s,num_sigBins_6s),
"Replicate_Number"=rep(c("N=3","N=4","N=5","N=6"),c(nrow(num_sigBins_3s),nrow(num_sigBins_4s),nrow(num_sigBins_5s),nrow(num_sigBins_6s)))
)
all_selfConsist_cov <- cbind(rbind(selfConsist_cov_3s,selfConsist_cov_4s,selfConsist_cov_5s,selfConsist_cov_6s),
"Replicate_Number"=rep(c("N=3","N=4","N=5","N=6"),c(nrow(selfConsist_cov_3s),nrow(selfConsist_cov_4s),nrow(selfConsist_cov_5s),nrow(selfConsist_cov_6s)))
)
all_selfFDR_cov <- cbind(rbind(selfFDR_cov_3s,selfFDR_cov_4s,selfFDR_cov_5s,selfFDR_cov_6s),
"Replicate_Number"=rep(c("N=3","N=4","N=5","N=6"),c(nrow(selfFDR_cov_3s),nrow(selfFDR_cov_4s),nrow(selfFDR_cov_5s),nrow(selfFDR_cov_6s)))
)
all_selfPower_cov <- cbind(rbind(selfPower_cov_3s,selfPower_cov_4s,selfPower_cov_5s,selfPower_cov_6s),
"Replicate_Number"=rep(c("N=3","N=4","N=5","N=6"),c(nrow(selfPower_cov_3s),nrow(selfPower_cov_4s),nrow(selfPower_cov_5s),nrow(selfPower_cov_6s)))
)
save(all_num_sigBins,all_selfConsist_cov,all_selfFDR_cov,all_selfPower_cov,file = "~/PoissonGamma_Method/Ovary_Cancer/powerAnalysis.RData")Sample size and number of detected bins
load("~/PoissonGamma_Method/Ovary_Cancer/powerAnalysis.RData")
ggplot( all_num_sigBins,aes(x = Replicate_Number, y = log10(num_sigBins+1) ,colour = qvalue_cutoff ) ) + ylab("Log10(number of significant bins) ") + xlab("Number of replicates") + geom_boxplot()+ facet_grid(. ~ method) +theme_bw() + theme( panel.grid.major = element_blank(),
panel.grid.minor =element_line(colour="grey", size=0.5),
# axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=20, face="bold", hjust=0.5,family = "arial"),
axis.title.y=element_text(size=20, face="bold", vjust=0.4, angle=90,family = "arial"),
legend.title=element_text(size = 15,face = "bold"),legend.text = element_text(size = 18, face = "bold",family = "arial"),
axis.text = element_text(size = 15,face = "bold",family = "arial",colour = "black") , strip.text.x = element_text(size = 15,face = "bold") ) + scale_color_discrete(name = "qvalue\ncutoff")
Number of replicates and consistency
ggplot( all_selfConsist_cov,aes(x = Replicate_Number, y = self_Consistency ,colour = qvalue_cutoff ) )+geom_boxplot()+ylab("Self-consistency ") + xlab("Number of replicates") + facet_grid(. ~ method) +theme_bw() + theme( panel.grid.major = element_blank(),
panel.grid.minor =element_line(colour="grey", size=0.5),
# axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=20, face="bold", hjust=0.5,family = "arial"),
axis.title.y=element_text(size=20, face="bold", vjust=0.4, angle=90,family = "arial"),
legend.title=element_text(size = 15,face = "bold"),legend.text = element_text(size = 18, face = "bold",family = "arial"),
axis.text.x = element_text(size = 15,face = "bold",family = "arial",colour = "black") ,axis.text.y = element_text(size = 15,face = "bold",family = "arial",colour = "black") ,strip.text.x = element_text(size = 15,face = "bold") ) + scale_color_discrete(name = "qvalue\ncutoff")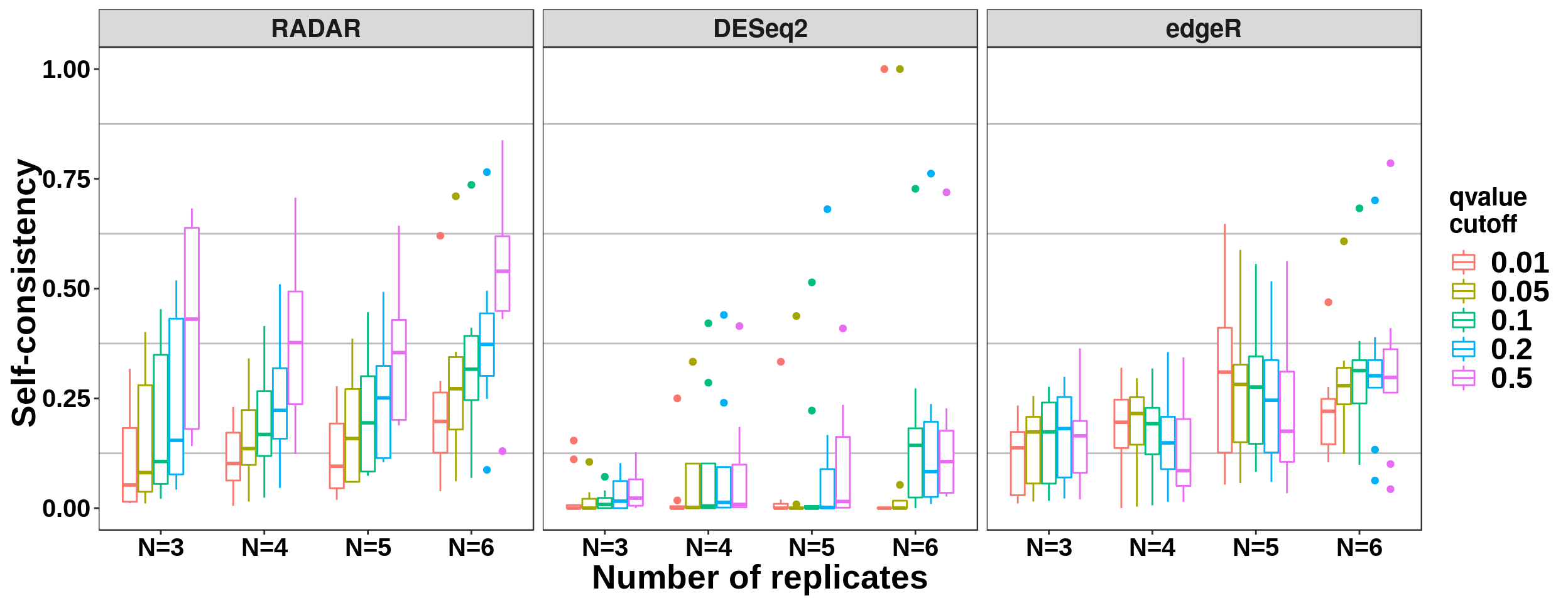
Number of replicates and self-FDR
all_selfFDR_cov$qvalue_cutoff <- factor(all_selfFDR_cov$qvalue_cutoff, levels = c(0.5,0.2,0.1,0.05,0.01))
ggplot( all_selfFDR_cov,aes(x = Replicate_Number, y = self_FDR ,colour = qvalue_cutoff ) )+geom_boxplot()+ylab("Self-FDR") + xlab("Number of replicates") + facet_grid(. ~ method) +theme_bw() + theme( panel.grid.major = element_blank(),
panel.grid.minor =element_line(colour="grey", size=0.5),
# axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=20, face="bold", hjust=0.5,family = "arial"),
axis.title.y=element_text(size=20, face="bold", vjust=0.4, angle=90,family = "arial"),
legend.title=element_text(size = 15,face = "bold"),legend.text = element_text(size = 18, face = "bold",family = "arial"),
axis.text.x = element_text(size = 15,face = "bold",family = "arial",colour = "black") ,axis.text.y = element_text(size = 15,face = "bold",family = "arial",colour = "black") ,strip.text.x = element_text(size = 15,face = "bold") ) + scale_color_discrete(name = "qvalue\ncutoff")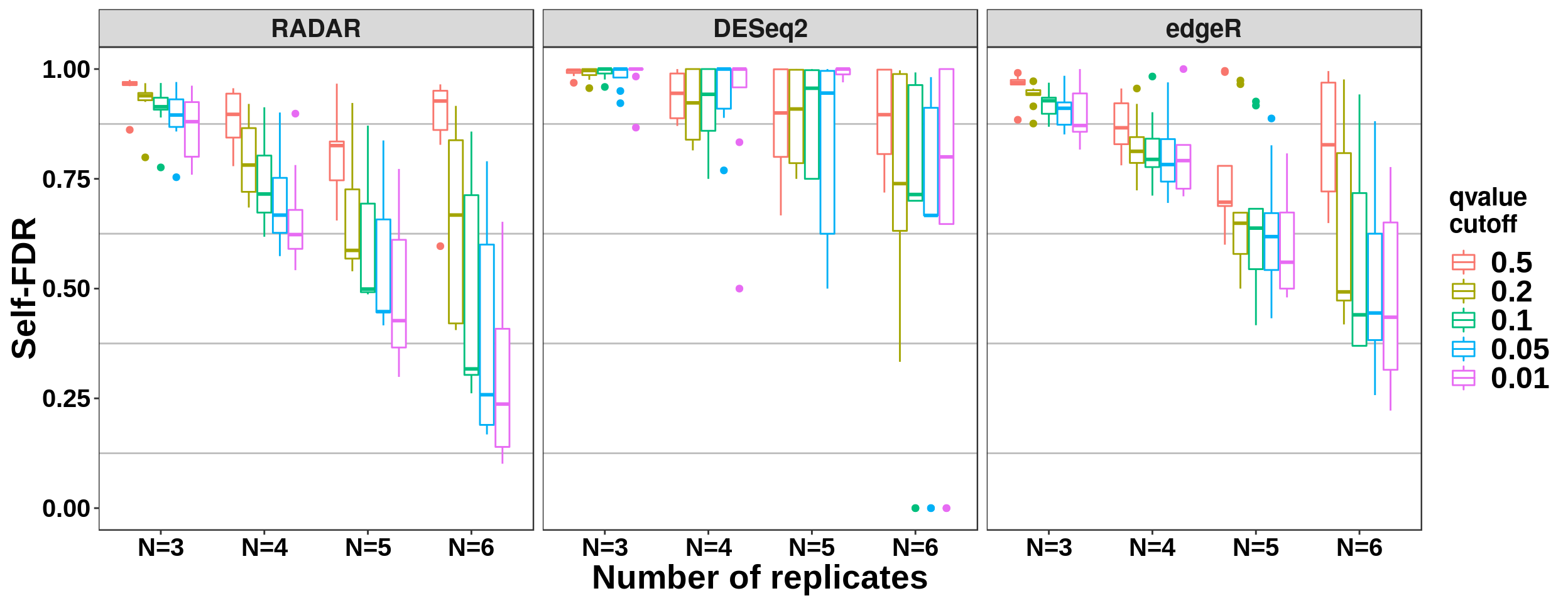
Number of replicates and self-power
ggplot( all_selfPower_cov,aes(x = Replicate_Number, y = power ,colour = qvalue_cutoff ) )+geom_boxplot()+ylab("Self-power") + xlab("Number of replicates") + facet_grid(. ~ method) +theme_bw() + theme( panel.grid.major = element_blank(),
panel.grid.minor =element_line(colour="grey", size=0.5),
# axis.line = element_line(colour = "black",size = 1),
axis.title.x=element_text(size=20, face="bold", hjust=0.5,family = "arial"),
axis.title.y=element_text(size=20, face="bold", vjust=0.4, angle=90,family = "arial"),
legend.title=element_text(size = 15,face = "bold"),legend.text = element_text(size = 18, face = "bold",family = "arial"),
axis.text.x = element_text(size = 15,face = "bold",family = "arial",colour = "black") ,axis.text.y = element_text(size = 15,face = "bold",family = "arial",colour = "black") ,strip.text.x = element_text(size = 15,face = "bold") ) + scale_color_discrete(name = "qvalue\ncutoff")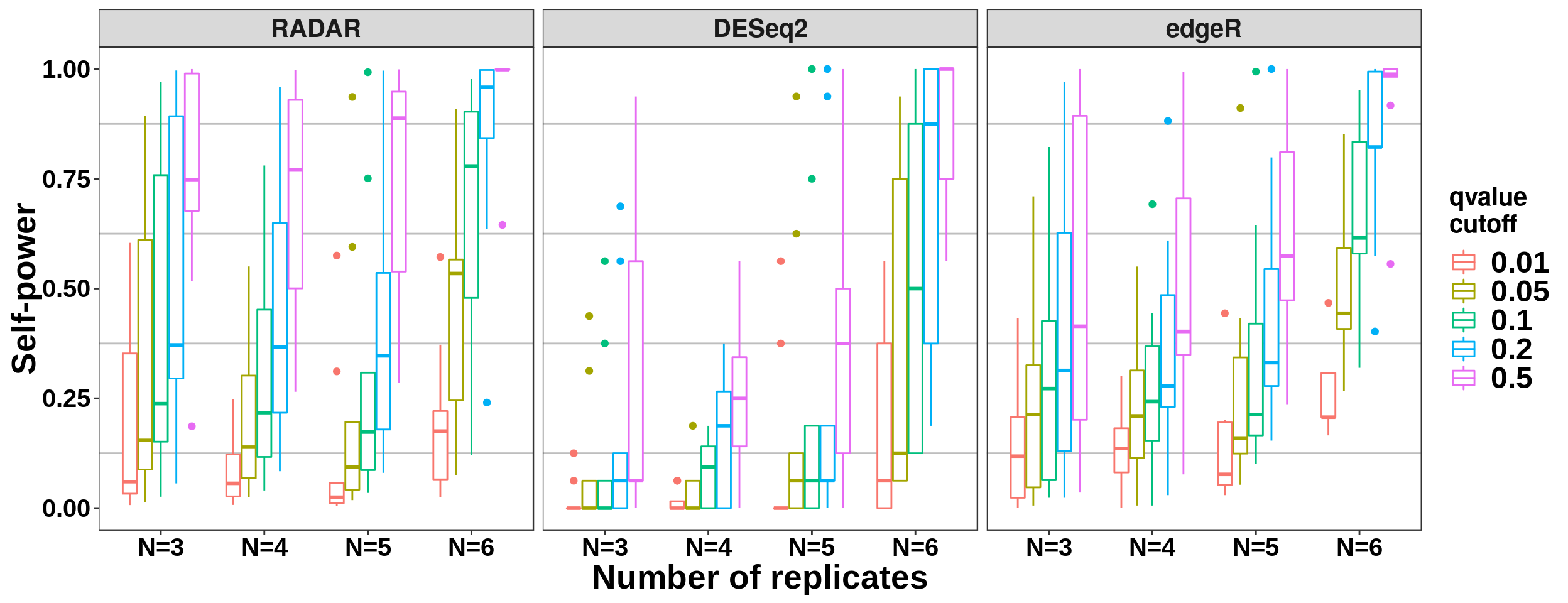
load("~/PoissonGamma_Method/Ovary_Cancer/powerAnalysis.RData")
combined_power <- NULL
for(x in unique( paste0(all_selfFDR_cov$qvalue_cutoff,all_selfFDR_cov$method) ) ){
tmp_fdr <- all_selfFDR_cov[paste0(all_selfFDR_cov$qvalue_cutoff,all_selfFDR_cov$method) == x, ]
tmp_power <- all_selfPower_cov[paste0(all_selfPower_cov$qvalue_cutoff,all_selfPower_cov$method) == x,]
tmp_all <- data.frame( self_FDR = tapply(tmp_fdr$self_FDR, tmp_fdr$Replicate_Number, median), power = tapply(tmp_power$power, tmp_power$Replicate_Number, median) , Replicate_Number = names( tapply(tmp_fdr$self_FDR, tmp_fdr$Replicate_Number, median)), qvalue_cutoff = unique(tmp_fdr$qvalue_cutoff) , method = unique(tmp_fdr$method) )
combined_power <- rbind(combined_power,tmp_all)
}
ggplot( combined_power,aes(x = self_FDR, y = power, colour = Replicate_Number, shape = Replicate_Number ) )+geom_point(size = 3)+geom_line()+facet_grid(~ method )+ylab("Self-Power") + xlab("Self-FDR") + facet_grid(. ~ method) +theme_bw() + theme( panel.grid.major = element_blank(),
panel.grid.minor =element_line(colour="grey", size=0.1),
axis.line = element_line(colour = "black",size = 0.75),
axis.ticks = element_line(colour = "black",size = 0.75),
axis.title.x=element_text(size=20, face="bold", hjust=0.5,family = "arial"),
axis.title.y=element_text(size=20, face="bold", vjust=0.4, angle=90,family = "arial"),
legend.title=element_text(size = 18,face = "bold"),legend.text = element_text(size = 20, face = "bold",family = "arial"),
axis.text = element_text(size = 16,face = "bold",family = "arial",colour = "black") ,strip.text.x = element_text(size = 20,face = "bold"), panel.spacing = unit(1.5, "lines"))+scale_color_discrete(name = "Number of\nreplicates")+scale_shape_discrete(name = "Number of\nreplicates")+scale_y_continuous(breaks = seq(0,1,0.2))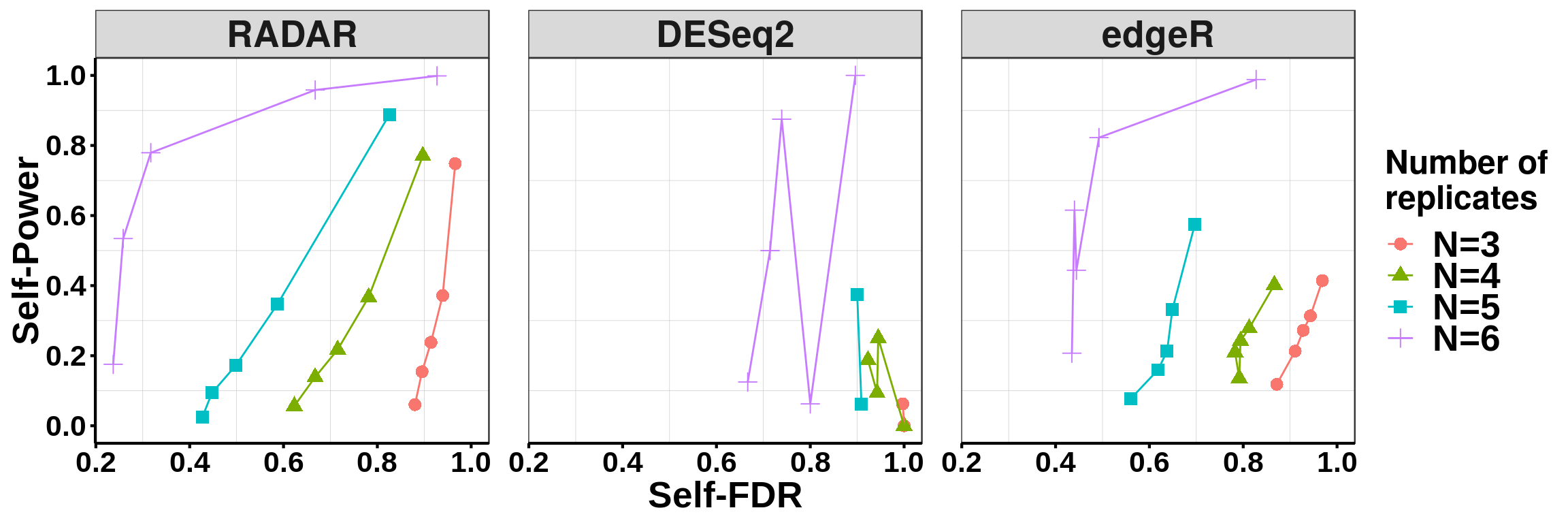
Downstream analysis of the differential peaks
Report differential peaks
Diff_peaks_cov <- reportPoissonGammaMerge(RADAR, cutoff = 0.1, est = RADAR_cov)
Diff_peaks_bin100 <- reportPoissonGammaMerge(RADAR_bin100,cutoff = 0.1, est = RADAR_cov_bin100)
write.table(Diff_peaks_cov,"~/PoissonGamma_Method/Ovary_Cancer/Diff_peaks_ageCov.xls",sep = "\t", col.names = T,row.names = F,quote = F)library(Vennerable)
Diff_peaks_cov.gr <- makeGRangesFromDataFrame(Diff_peaks_cov)
Diff_peaks_bin100.gr <- makeGRangesFromDataFrame(Diff_peaks_bin100)
findOverlaps(Diff_peaks_cov.gr,Diff_peaks_bin100.gr)## Hits object with 793 hits and 0 metadata columns:
## queryHits subjectHits
## <integer> <integer>
## [1] 3 12
## [2] 6 3
## [3] 9 7
## [4] 16 13
## [5] 17 1
## ... ... ...
## [789] 2204 1348
## [790] 2206 1332
## [791] 2207 1246
## [792] 2229 1137
## [793] 2241 1340
## -------
## queryLength: 2245 / subjectLength: 1357plot(Venn(list("bin50"=Diff_peaks_cov$name, "bin100"= Diff_peaks_bin100$name)))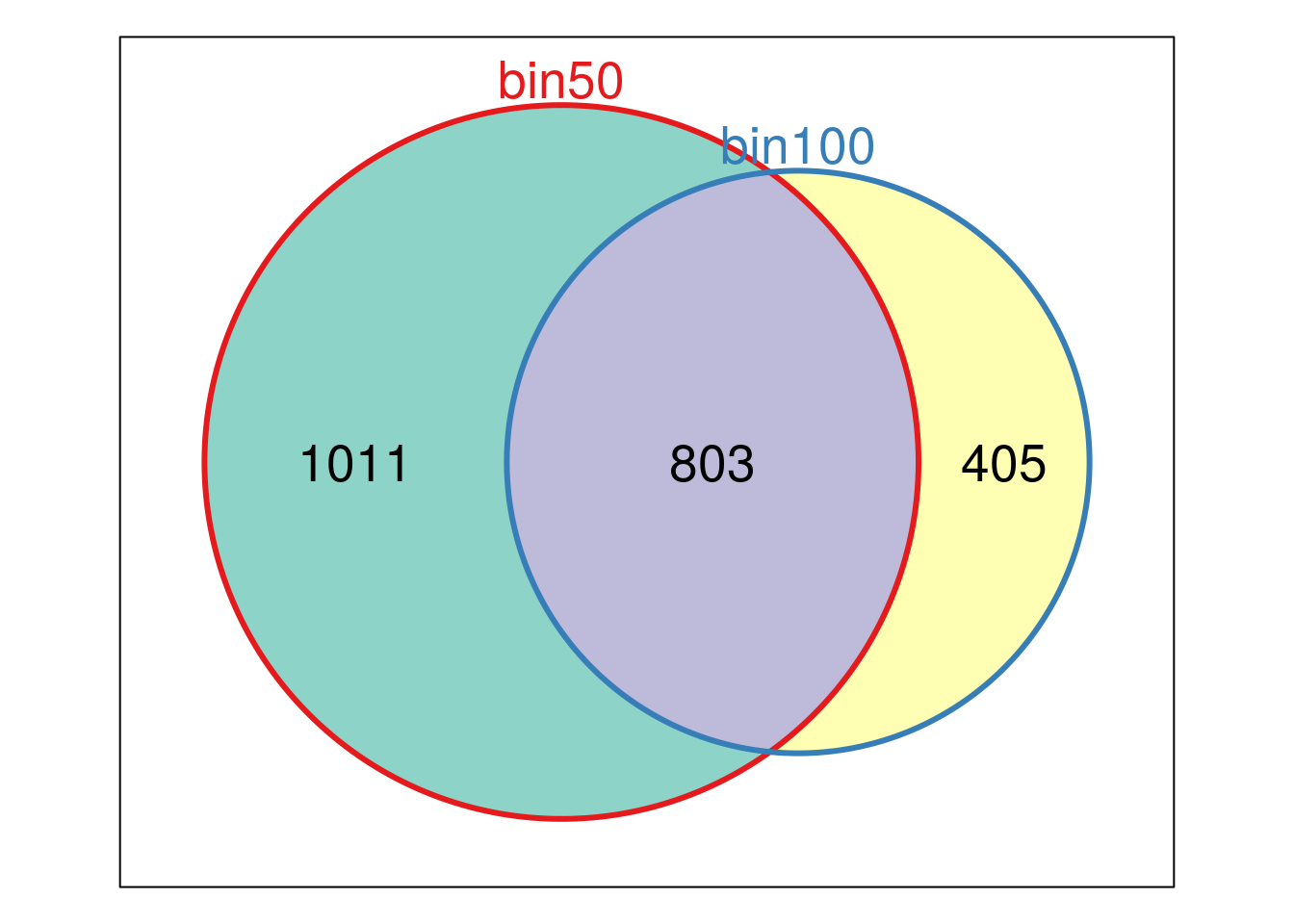
GO and KEGG enrichment analysis
library(clusterProfiler)
library(org.Hs.eg.db)eg_diffPeak_cov <- bitr(unique(Diff_peaks_cov$name), fromType="SYMBOL", toType="ENTREZID", OrgDb="org.Hs.eg.db")
KEGG_diffPeak_cov <- enrichKEGG(eg_diffPeak_cov$ENTREZID,organism = "hsa",pAdjustMethod = "none", pvalueCutoff = 0.05,minGSSize = 3)
ego_diffPeak_cov <- enrichGO(gene = eg_diffPeak_cov$ENTREZID,
universe = bitr(rownames(RADAR$geneSum), fromType="SYMBOL", toType="ENTREZID", OrgDb="org.Hs.eg.db")$ENTREZID,
OrgDb = org.Hs.eg.db,
ont = "BP",
pAdjustMethod = "BH",
pvalueCutoff = 0.01,
qvalueCutoff = 0.05,
readable = TRUE)Plot enriched pathways
enrichMap(KEGG_diffPeak_cov,n = 20,vertex.label.font = 2, layout=igraph::layout.fruchterman.reingold)
KEGG_diffPeak_cov_df <- as.data.frame(KEGG_diffPeak_cov)
KEGG_diffPeak_cov_df$geneID <- unlist( lapply(strsplit(KEGG_diffPeak_cov_df$geneID,"/"),function(x){ paste( bitr(x,fromType="ENTREZID", toType="SYMBOL",OrgDb="org.Hs.eg.db")$SYMBOL, collapse = "/") }) )
write.table(KEGG_diffPeak_cov_df, "~/PoissonGamma_Method/Ovary_Cancer/kegg_diffPeak_covariates.xls",sep = "\t", col.names = T, row.names = F,quote = F)
knitr::kable( KEGG_diffPeak_cov_df[KEGG_diffPeak_cov_df$Description %in% c("PI3K-Akt signaling pathway","MAPK signaling pathway","
Prostate cancer","Small cell lung cancer","Transcriptional misregulation in cancer","TNF signaling pathway"),c("ID","Description","pvalue","geneID")],format = "pandoc",align = 'c' , row.names = F)Plot enriched biological process
enrichMap(ego_diffPeak_cov,n = 25,vertex.label.font = 2, layout=igraph::layout.circle)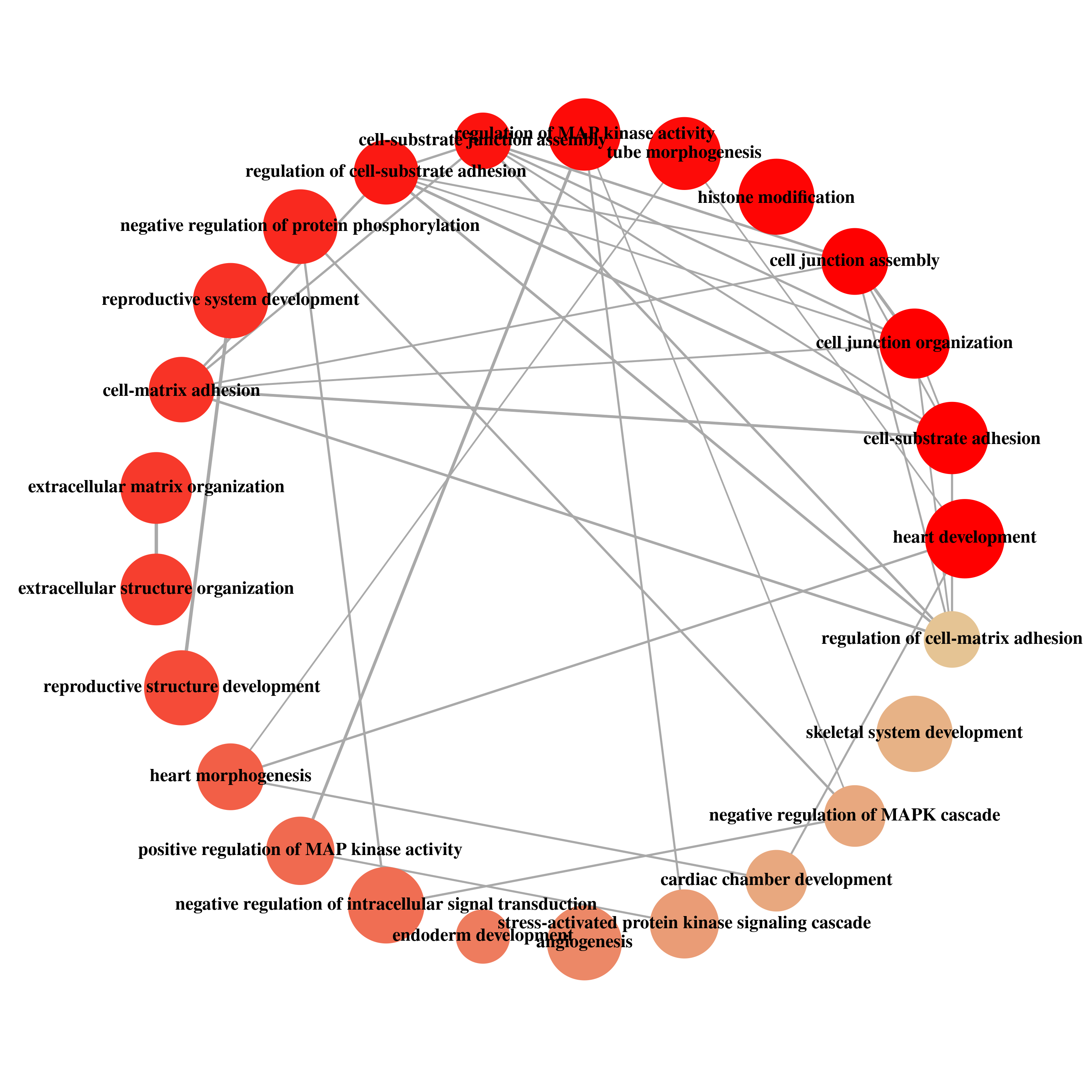
edgeR result GO and KEGG enrichment analysis
diff_peak_edgeR <- reportPoissonGammaMerge(RADAR,cutoff = 0.1, est = data.frame(row.names = edgeR.res_cov$GeneID, beta = edgeR.res_cov$log2FC, p_value = edgeR.res_cov$pvalue, padj = edgeR.res_cov$p.adjust) )## Getting significant bins....
## Reporting 156 bins in 135 genes...
## Reporting 153 peaks...
## Hyper-thread registered: TRUE
## Using 6 thread(s) to report merged report...
## Time used to report peaks: 0.0787798364957174 mins...eg_diffPeak_edgeR <- bitr(unique(diff_peak_edgeR$name), fromType="SYMBOL", toType="ENTREZID", OrgDb="org.Hs.eg.db")
KEGG_diffPeak_edgeR <- enrichKEGG(eg_diffPeak_edgeR$ENTREZID,organism = "hsa",pAdjustMethod = "none", pvalueCutoff = 0.05,minGSSize = 3)
ego_diffPeak_edgeR <- enrichGO(gene = eg_diffPeak_edgeR$ENTREZID,
universe = bitr(rownames(RADAR$geneSum), fromType="SYMBOL", toType="ENTREZID", OrgDb="org.Hs.eg.db")$ENTREZID,
OrgDb = org.Hs.eg.db,
ont = "BP",
pAdjustMethod = "BH",
pvalueCutoff = 0.01,
qvalueCutoff = 0.05,
readable = TRUE)
ego_diffPeak_edgeR## #
## # over-representation test
## #
## #...@organism Homo sapiens
## #...@ontology BP
## #...@keytype ENTREZID
## #...@gene chr [1:130] "10216" "90381" "55567" "104797538" "364" "10686" "3240" ...
## #...pvalues adjusted by 'BH' with cutoff <0.01
## #...0 enriched terms found
## 'data.frame': 0 obs. of 8 variables:
## $ ID : chr
## $ Description: chr
## $ GeneRatio : chr
## $ BgRatio : chr
## $ pvalue : num
## $ p.adjust : num
## $ qvalue : num
## $ Count : int
## #...Citation
## Guangchuang Yu, Li-Gen Wang, Yanyan Han and Qing-Yu He.
## clusterProfiler: an R package for comparing biological themes among
## gene clusters. OMICS: A Journal of Integrative Biology
## 2012, 16(5):284-287Plot differential peaks detected.
PTEN
plotGeneCov(RADAR,"PTEN",GTF = GTF ,adjustExprLevel = T)+scale_x_continuous(expand = c(0.02,0))+theme(legend.position = "none")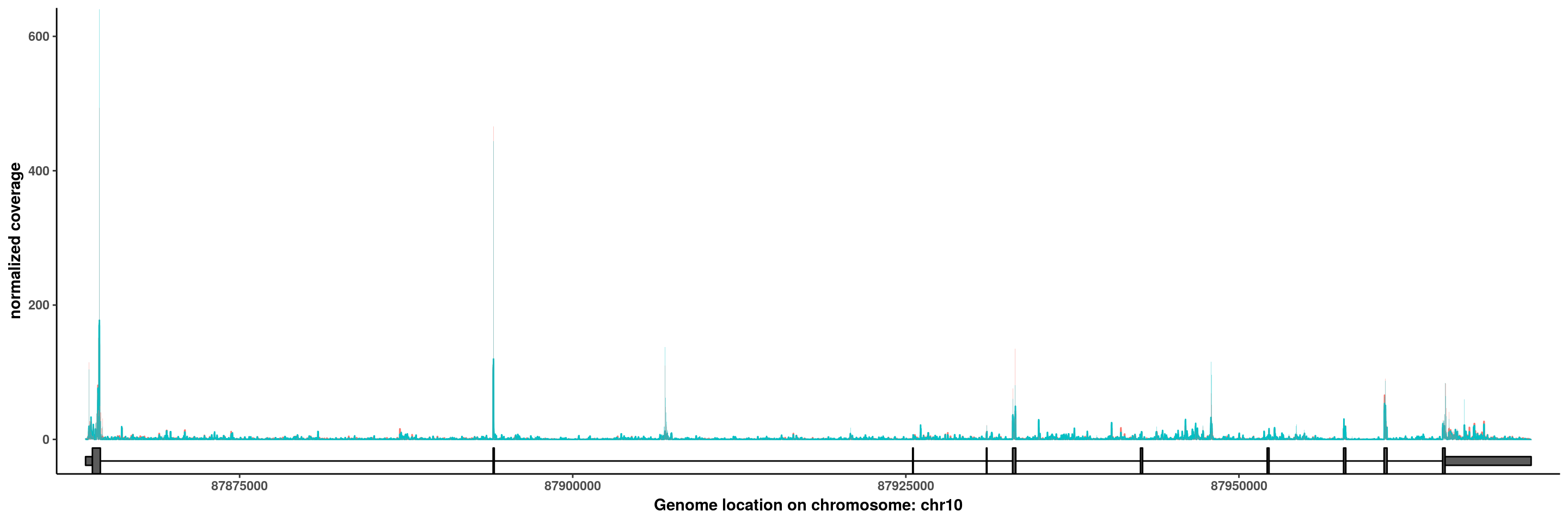
plotGeneCov(RADAR,"PTEN",GTF = GTF ,ZoomIn = c(87965095,87966657),adjustExprLevel = T)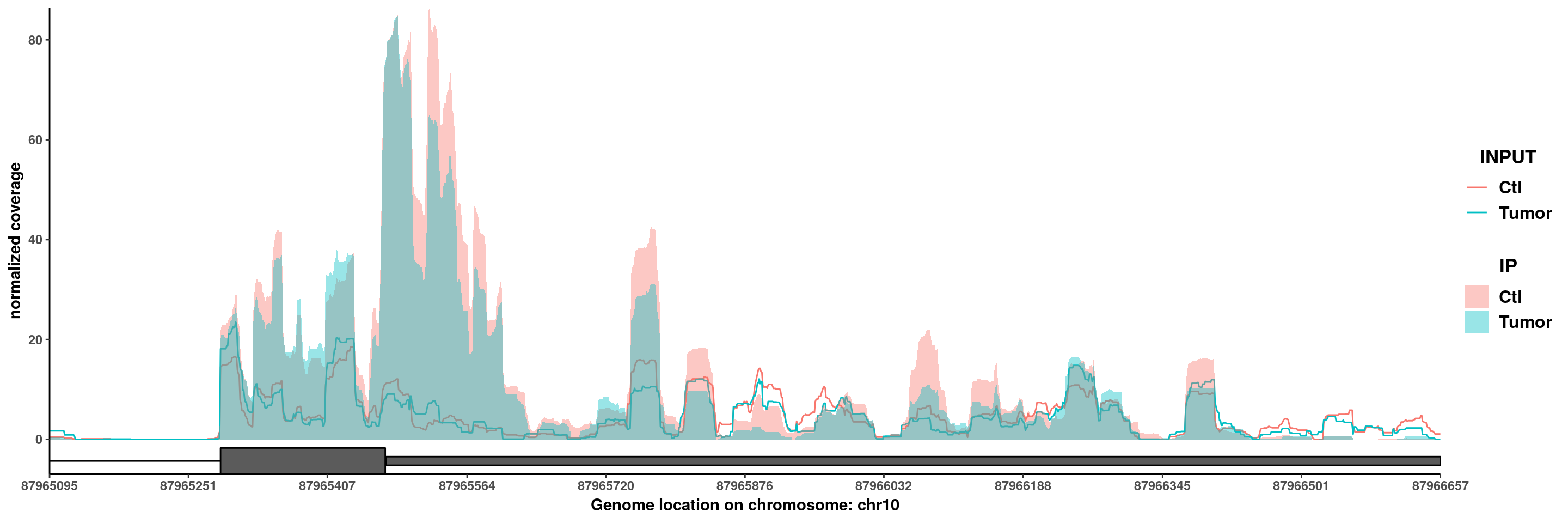
plotGeneCov(RADAR,"PTEN",GTF = GTF , ZoomIn = c(87929633,87936182),adjustExprLevel = T)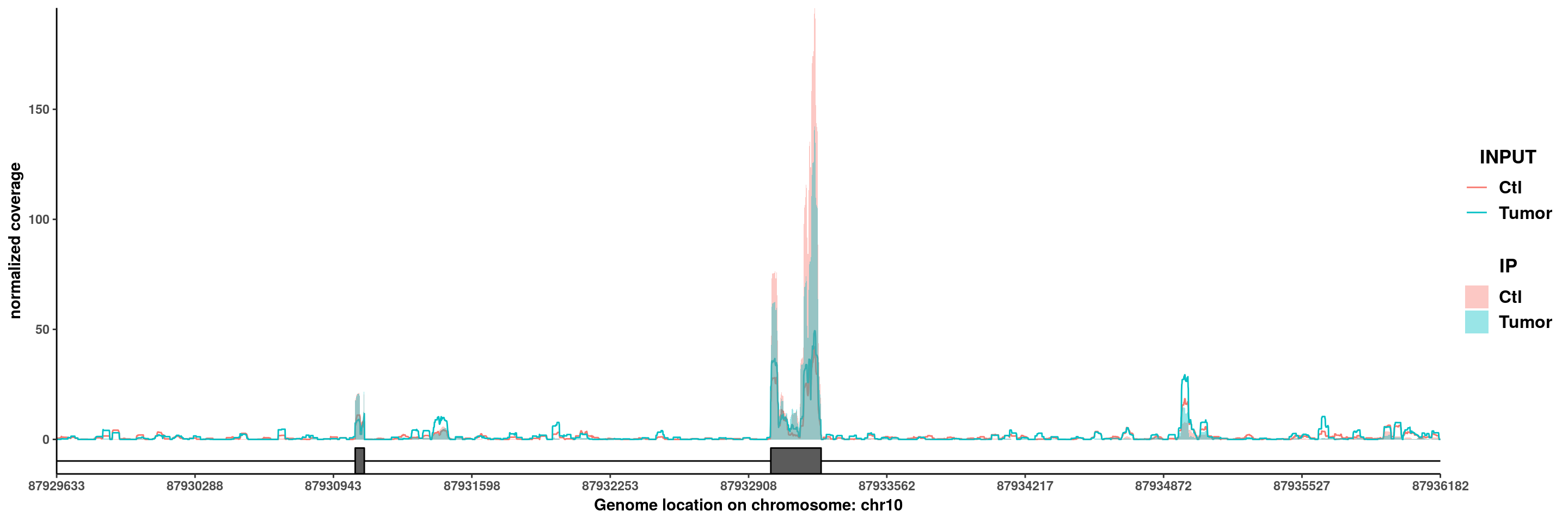
BCL2
plotGeneCov(RADAR,"BCL2",GTF = GTF ,adjustExprLevel =T )+scale_x_continuous(expand = c(0.012,0))+theme(legend.position = "none")
plotGeneCov(RADAR,"BCL2",GTF = GTF ,ZoomIn = c(63317900,63318363),adjustExprLevel = T)
PIK3AP1
plotGeneCov(RADAR,"PIK3AP1",GTF = GTF ,adjustExprLevel =T )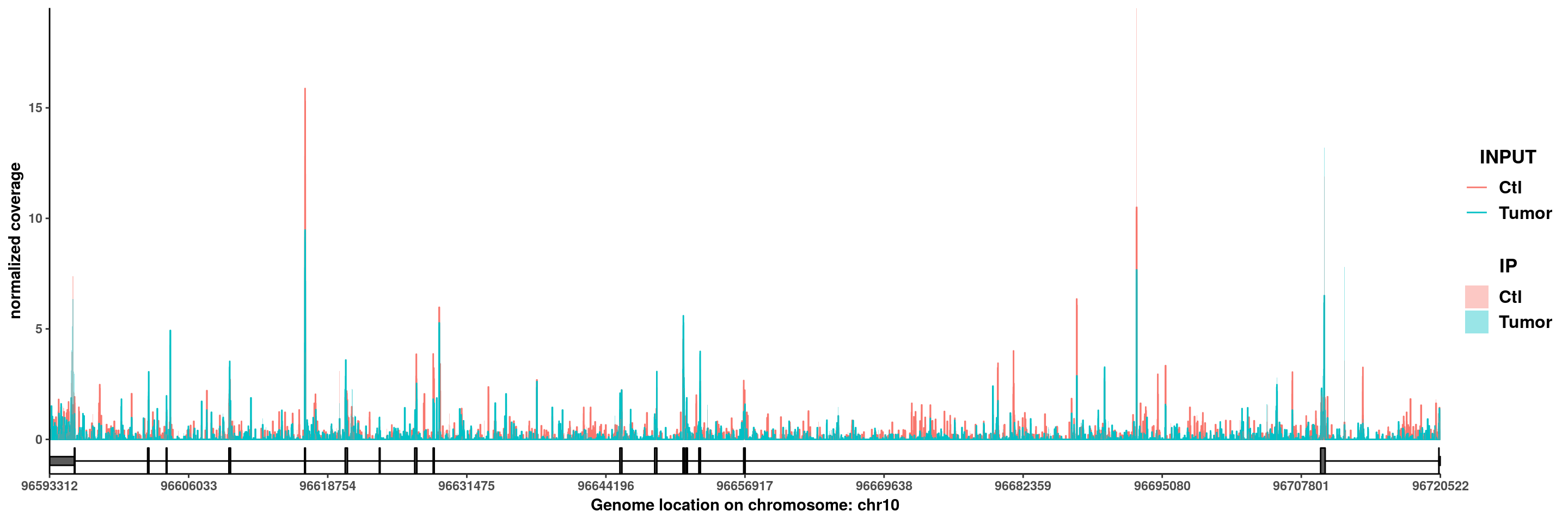
plotGeneCov(RADAR,"PIK3AP1",GTF = GTF ,ZoomIn = c(96708976, 96710100),adjustExprLevel = T)
Heatmap for differential loci
topBins <- RADAR$ip_adjExpr_filtered[rownames(RADAR_cov[RADAR_cov[,"padj"]<0.1,]),]
log_topBins <- log(topBins+1)
log_topBins_center <- t(apply(log_topBins,1,function(x){x-mean(x)}) )
colnames(log_topBins_center) <- RADAR$X
dist.pear <- function(x) as.dist(1-cor(t(x)))
hclust.ave <- function(x) hclust(x, method="average")
gplots::heatmap.2(log_topBins_center,scale="row",trace="none",labRow=NA,main = "Methylation level of significant bins",
distfun=dist.pear, hclustfun=hclust.ave,col=rev(RColorBrewer::brewer.pal(9,"RdBu")))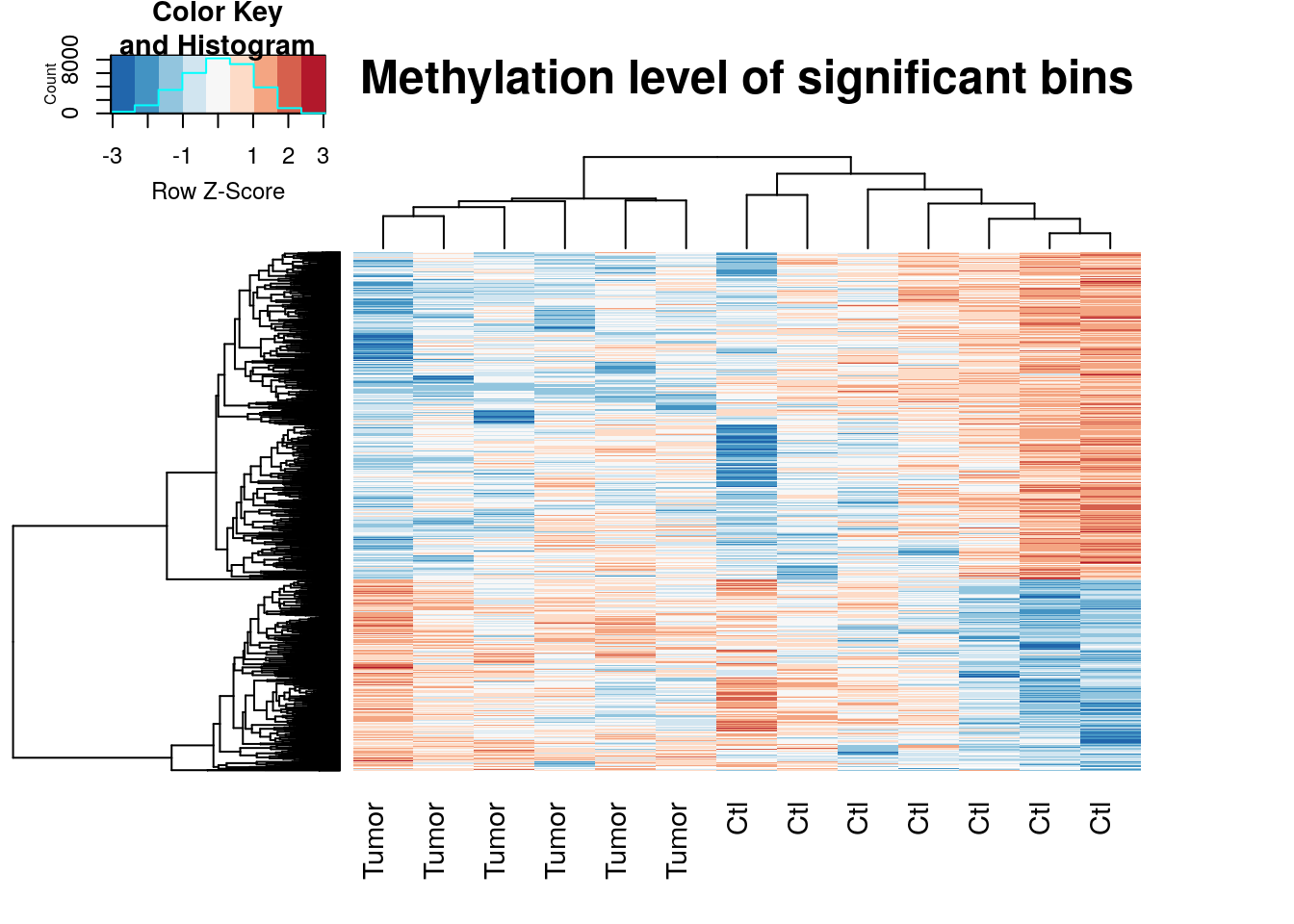
Session information
sessionInfo()## R version 3.4.4 (2018-03-15)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 17.10
##
## Matrix products: default
## BLAS: /usr/local/lib/R/lib/libRblas.so
## LAPACK: /usr/local/lib/R/lib/libRlapack.so
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] grid stats4 parallel stats graphics grDevices utils
## [8] datasets methods base
##
## other attached packages:
## [1] org.Hs.eg.db_3.5.0 clusterProfiler_3.6.0
## [3] DOSE_3.4.0 Vennerable_3.1.0.9000
## [5] VennDiagram_1.6.20 futile.logger_1.4.3
## [7] RADAR_0.1.5 RcppArmadillo_0.9.100.5.0
## [9] Rcpp_0.12.19 RColorBrewer_1.1-2
## [11] gplots_3.0.1 doParallel_1.0.14
## [13] iterators_1.0.10 foreach_1.4.4
## [15] ggplot2_3.0.0 Rsamtools_1.30.0
## [17] Biostrings_2.46.0 XVector_0.18.0
## [19] GenomicFeatures_1.30.3 AnnotationDbi_1.40.0
## [21] Biobase_2.38.0 GenomicRanges_1.30.3
## [23] GenomeInfoDb_1.14.0 IRanges_2.12.0
## [25] S4Vectors_0.16.0 BiocGenerics_0.24.0
##
## loaded via a namespace (and not attached):
## [1] nlme_3.1-131.1 bitops_1.0-6
## [3] matrixStats_0.54.0 bit64_0.9-7
## [5] progress_1.2.0 httr_1.3.1
## [7] rprojroot_1.3-2 tools_3.4.4
## [9] backports_1.1.2 R6_2.2.2
## [11] KernSmooth_2.23-15 mgcv_1.8-23
## [13] DBI_1.0.0 lazyeval_0.2.1
## [15] colorspace_1.3-2 withr_2.1.2
## [17] tidyselect_0.2.4 gridExtra_2.3
## [19] prettyunits_1.0.2 RMySQL_0.10.15
## [21] bit_1.1-14 compiler_3.4.4
## [23] graph_1.56.0 formatR_1.5
## [25] DelayedArray_0.4.1 labeling_0.3
## [27] rtracklayer_1.38.3 caTools_1.17.1
## [29] scales_0.5.0 RBGL_1.54.0
## [31] stringr_1.3.1 digest_0.6.15
## [33] rmarkdown_1.10 pkgconfig_2.0.1
## [35] htmltools_0.3.6 highr_0.7
## [37] rlang_0.2.1 RSQLite_2.1.1
## [39] bindr_0.1.1 BiocParallel_1.12.0
## [41] gtools_3.8.1 GOSemSim_2.4.1
## [43] dplyr_0.7.6 RCurl_1.95-4.10
## [45] magrittr_1.5 GO.db_3.5.0
## [47] GenomeInfoDbData_1.0.0 Matrix_1.2-12
## [49] munsell_0.5.0 stringi_1.2.3
## [51] yaml_2.1.19 SummarizedExperiment_1.8.1
## [53] zlibbioc_1.24.0 plyr_1.8.4
## [55] qvalue_2.10.0 blob_1.1.1
## [57] gdata_2.18.0 DO.db_2.9
## [59] crayon_1.3.4 lattice_0.20-35
## [61] splines_3.4.4 hms_0.4.2
## [63] knitr_1.20 pillar_1.2.3
## [65] fgsea_1.4.1 igraph_1.2.1
## [67] reshape2_1.4.3 codetools_0.2-15
## [69] biomaRt_2.34.2 futile.options_1.0.1
## [71] fastmatch_1.1-0 XML_3.98-1.11
## [73] glue_1.2.0 evaluate_0.10.1
## [75] lambda.r_1.2.3 data.table_1.11.4
## [77] tidyr_0.8.1 gtable_0.2.0
## [79] purrr_0.2.5 assertthat_0.2.0
## [81] tibble_1.4.2 rvcheck_0.1.0
## [83] GenomicAlignments_1.14.2 memoise_1.1.0
## [85] MyTools_0.0.0 bindrcpp_0.2.2This R Markdown site was created with workflowr